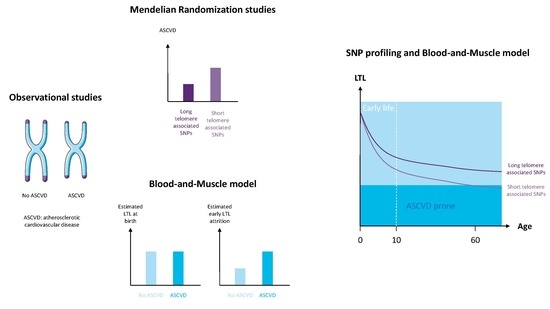
TERC Variants Associated with Short Leukocyte Telomeres: Implication of Higher Early Life Leukocyte Telomere Attrition as Assessed by the Blood-and-Muscle Model
Abstract
1. Introduction
2. Materials and Methods
2.1. The Cohort
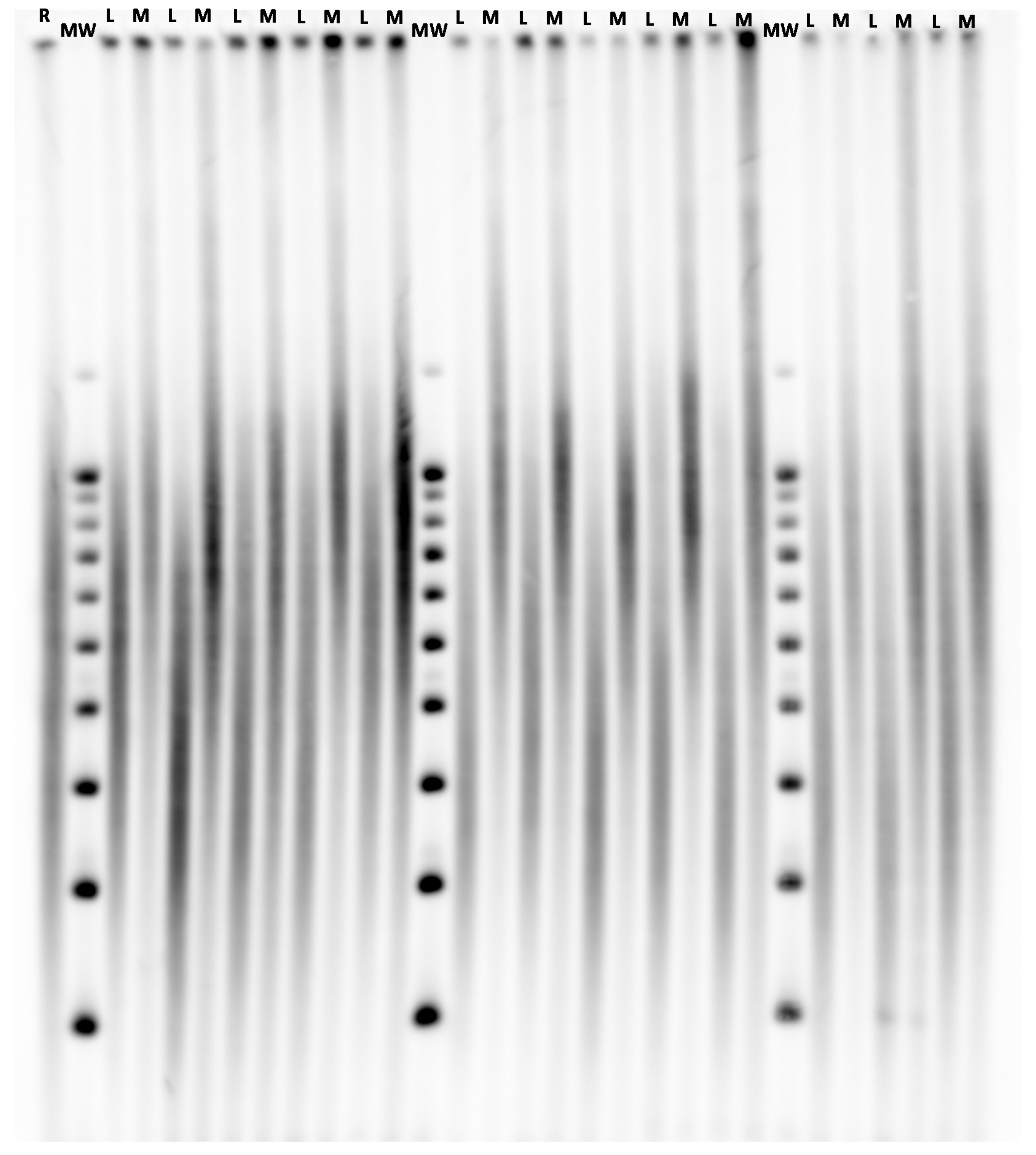
2.2. Telomere Length Measurements
2.3. Single Nucleotide Polymorphism Genotyping
2.4. Statistical Analyses
3. Results
3.1. Population Characteristics
3.2. Telomere Length Dynamics
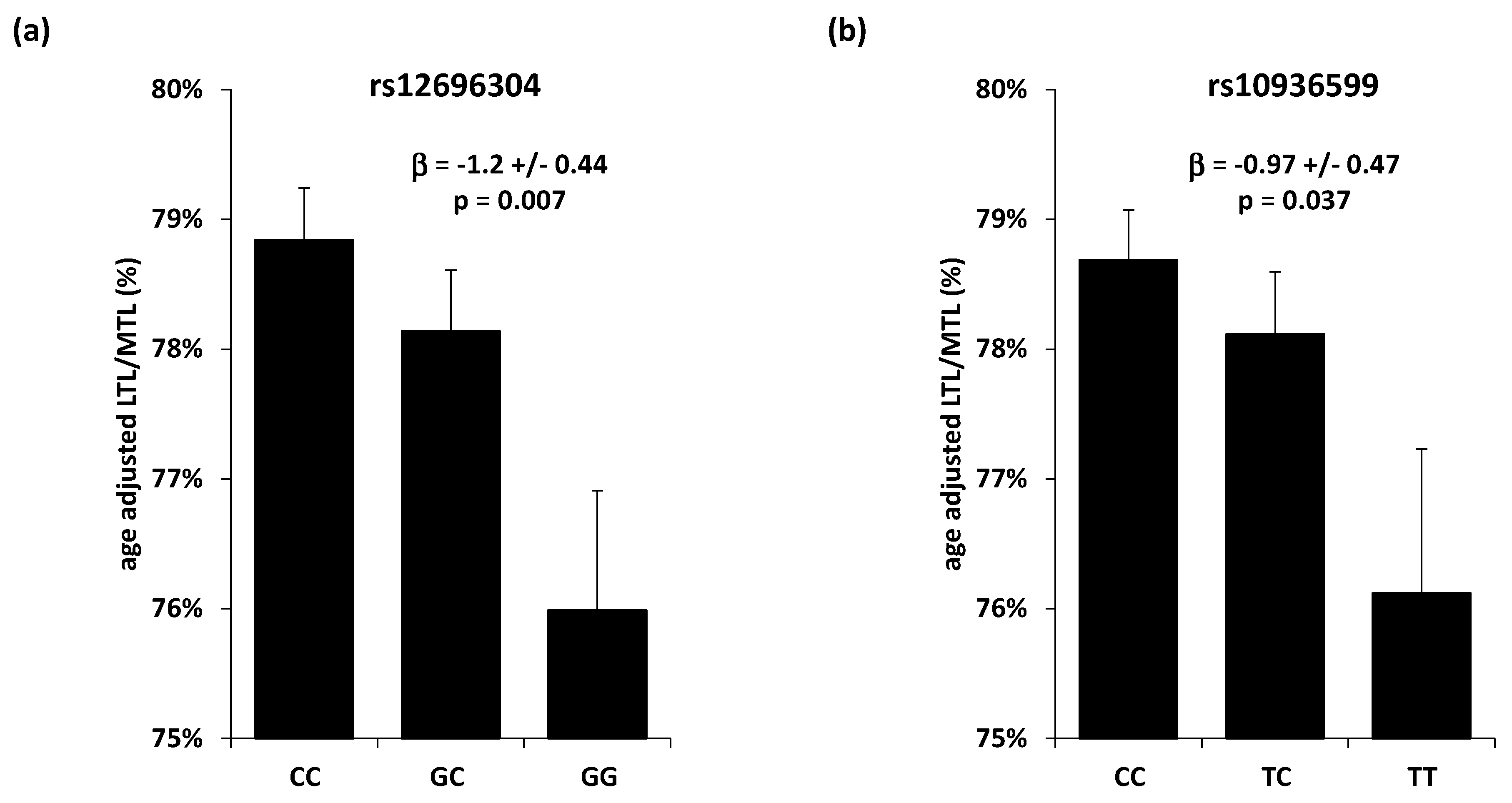
3.3. Single Nucleotide Polymorphisms Association with Telomere Length
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- D’Mello, M.J.J.; Ross, S.A.; Briel, M.; Anand, S.S.; Gerstein, H.; Paré, G. Association between shortened leukocyte telomere length and cardiometabolic outcomes: Systematic review and meta-analysis. Circ. Cardiovasc. Genet. 2015, 8, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Haycock, P.C.; Heydon, E.E.; Kaptoge, S.; Butterworth, A.S.; Thompson, A.; Willeit, P. Leucocyte telomere length and risk of cardiovascular disease: Systematic review and meta-analysis. BMJ 2014, 349, g4227. [Google Scholar] [CrossRef] [PubMed]
- Codd, V.; Nelson, C.P.; Albrecht, E.; Mangino, M.; Deelen, J.; Buxton, J.L.; Hottenga, J.J.; Fischer, K.; Esko, T.; Surakka, I.; et al. Identification of seven loci affecting mean telomere length and their association with disease. Nat. Genet. 2013, 45, 422–427. [Google Scholar] [CrossRef] [PubMed]
- Mangino, M.; Hwang, S.-J.; Spector, T.D.; Hunt, S.C.; Kimura, M.; Fitzpatrick, A.L.; Christiansen, L.; Petersen, I.; Elbers, C.C.; Harris, T.; et al. Genome-wide meta-analysis points to CTC1 and ZNF676 as genes regulating telomere homeostasis in humans. Hum. Mol. Genet. 2012, 21, 5385–5394. [Google Scholar] [CrossRef] [PubMed]
- Haycock, P.C.; Burgess, S.; Nounu, A.; Zheng, J.; Okoli, G.N.; Bowden, J.; Wade, K.H.; Timpson, N.J.; Evans, D.M.; Willeit, P.; et al. Association Between Telomere Length and Risk of Cancer and Non-Neoplastic Diseases: A Mendelian Randomization Study. JAMA Oncol. 2017, 3, 636–651. [Google Scholar] [CrossRef] [PubMed]
- Scheller Madrid, A.; Rode, L.; Nordestgaard, B.G.; Bojesen, S.E. Short Telomere Length and Ischemic Heart Disease: Observational and Genetic Studies in 290 022 Individuals. Clin. Chem. 2016, 62, 1140–1149. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Y.; Karlsson, I.K.; Karlsson, R.; Tillander, A.; Reynolds, C.A.; Pedersen, N.L.; Hägg, S. Exploring the Causal Pathway From Telomere Length to Coronary Heart Disease: A Network Mendelian Randomization Study. Circ. Res. 2017, 121, 214–219. [Google Scholar] [CrossRef]
- Benetos, A.; Toupance, S.; Gautier, S.; Labat, C.; Kimura, M.; Rossi, P.M.; Settembre, N.; Hubert, J.; Frimat, L.; Bertrand, B.; et al. Short Leukocyte Telomere Length Precedes Clinical Expression of Atherosclerosis: The Blood-and-Muscle Model. Circ. Res. 2018, 122, 616–623. [Google Scholar] [CrossRef]
- Kimura, M.; Stone, R.C.; Hunt, S.C.; Skurnick, J.; Lu, X.; Cao, X.; Harley, C.B.; Aviv, A. Measurement of telomere length by the Southern blot analysis of terminal restriction fragment lengths. Nat. Protoc. 2010, 5, 1596–1607. [Google Scholar] [CrossRef]
- He, C.; Holme, J.; Anthony, J. SNP genotyping: The KASP assay. Methods Mol. Biol. 2014, 1145, 75–86. [Google Scholar] [CrossRef]
- Sabharwal, S.; Verhulst, S.; Guirguis, G.; Kark, J.D.; Labat, C.; Roche, N.E.; Martimucci, K.; Patel, K.; Heller, D.S.; Kimura, M.; et al. Telomere length dynamics in early life: The blood-and-muscle model. FASEB J. 2018, 32, 529–534. [Google Scholar] [CrossRef] [PubMed]
- Barraclough, J.Y.; Skilton, M.R.; Garden, F.L.; Toelle, B.G.; Marks, G.B.; Celermajer, D.S. Early and late childhood telomere length predict subclinical atherosclerosis at age 14 yrs.-The CardioCAPS study. Int. J. Cardiol. 2019, 278, 250–253. [Google Scholar] [CrossRef] [PubMed]
- Skilton, M.R.; Nakhla, S.; Ayer, J.G.; Harmer, J.A.; Toelle, B.G.; Leeder, S.R.; Jones, G.; Marks, G.B.; Celermajer, D.S.; Childhood Asthma Prevention Study Group. Telomere length in early childhood: Early life risk factors and association with carotid intima-media thickness in later childhood. Eur. J. Prev. Cardiol. 2016, 23, 1086–1092. [Google Scholar] [CrossRef] [PubMed]
- Toupance, S.; Labat, C.; Temmar, M.; Rossignol, P.; Kimura, M.; Aviv, A.; Benetos, A. Short Telomeres, but Not Telomere Attrition Rates, Are Associated With Carotid Atherosclerosis. Hypertension 2017, 70, 420–425. [Google Scholar] [CrossRef] [PubMed]
- Chiriacò, M.; Georgiopoulos, G.; Duranti, E.; Antonioli, L.; Puxeddu, I.; Nannipieri, M.; Rosada, J.; Blandizzi, C.; Taddei, S.; Virdis, A.; et al. Inflammation and Vascular Ageing: From Telomeres to Novel Emerging Mechanisms. High Blood Press. Cardiovasc. Prev. Off. J. Ital. Soc. Hypertens. 2019, 26, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Zhang, H.; Zhao, Y.; Wang, Y.; Wang, W.; He, Y.; Zhang, W.; Zhang, W.; Zhu, X.; Zhou, Y.; et al. Telomere Dysfunction Disturbs Macrophage Mitochondrial Metabolism and the NLRP3 Inflammasome through the PGC-1α/TNFAIP3 Axis. Cell Rep. 2018, 22, 3493–3506. [Google Scholar] [CrossRef]
- Grebe, A.; Hoss, F.; Latz, E. NLRP3 Inflammasome and the IL-1 Pathway in Atherosclerosis. Circ. Res. 2018, 122, 1722–1740. [Google Scholar] [CrossRef]
- Amorós-Pérez, M.; Fuster, J.J. Clonal hematopoiesis driven by somatic mutations: A new player in atherosclerotic cardiovascular disease. Atherosclerosis 2020, 297, 120–126. [Google Scholar] [CrossRef]
- Jaiswal, S.; Natarajan, P.; Silver, A.J.; Gibson, C.J.; Bick, A.G.; Shvartz, E.; McConkey, M.; Gupta, N.; Gabriel, S.; Ardissino, D.; et al. Clonal Hematopoiesis and Risk of Atherosclerotic Cardiovascular Disease. N. Engl. J. Med. 2017, 377, 111–121. [Google Scholar] [CrossRef]
- Zink, F.; Stacey, S.N.; Norddahl, G.L.; Frigge, M.L.; Magnusson, O.T.; Jonsdottir, I.; Thorgeirsson, T.E.; Sigurdsson, A.; Gudjonsson, S.A.; Gudmundsson, J.; et al. Clonal hematopoiesis, with and without candidate driver mutations, is common in the elderly. Blood 2017, 130, 742–752. [Google Scholar] [CrossRef]
- Schratz, K.E.; Haley, L.; Danoff, S.K.; Blackford, A.; DeZern, A.; Gocke, C.D.; Duffield, A.S.; Armanios, M. Cancer spectrum and outcomes in the Mendelian short telomere syndromes. Blood 2020. [Google Scholar] [CrossRef] [PubMed]
- Aviv, A.; Levy, D. Hemothelium, Clonal Hematopoiesis of Indeterminate Potential, and Atherosclerosis. Circulation 2019, 139, 7–9. [Google Scholar] [CrossRef] [PubMed]
- Steensma, D.P. Clinical Implications of Clonal Hematopoiesis. Mayo Clin. Proc. 2018, 93, 1122–1130. [Google Scholar] [CrossRef] [PubMed]
- Lacaud, G.; Kouskoff, V. Hemangioblast, hemogenic endothelium, and primitive versus definitive hematopoiesis. Exp. Hematol. 2017, 49, 19–24. [Google Scholar] [CrossRef]
- Aviv, A.; Levy, D. Telomeres, atherosclerosis, and the hemothelium: The longer view. Annu. Rev. Med. 2012, 63, 293–301. [Google Scholar] [CrossRef]
- Aviv, A.; Hunt, S.C.; Lin, J.; Cao, X.; Kimura, M.; Blackburn, E. Impartial comparative analysis of measurement of leukocyte telomere length/DNA content by Southern blots and qPCR. Nucleic Acids Res. 2011, 39, e134. [Google Scholar] [CrossRef]
- Nettle, D.; Seeker, L.; Nussey, D.; Froy, H.; Bateson, M. Consequences of measurement error in qPCR telomere data: A simulation study. PLoS ONE 2019, 14, e0216118. [Google Scholar] [CrossRef]
| Log LTL | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| SNP | CHR | Position | Gene | Effect Allele | Other Allele | MAF | BETA | SE | P |
| rs11125529 | 2 | 54248729 | ACYP2 | A | C | 0.12 | 0.000024 | 0.0050 | 0.99 |
| rs6772228 | 3 | 58390292 | PXK | A | T | 0.045 | −0.0094 | 0.0073 | 0.20 |
| rs12696304 | 3 | 169763483 | TERC | G | C | 0.29 | −0.0074 | 0.0033 | 0.027 |
| rs10936599 | 3 | 169774313 | TERC | T | C | 0.25 | −0.0080 | 0.0036 | 0.025 |
| rs7675998 | 4 | 163086668 | NAF1 | A | G | 0.24 | −0.0012 | 0.0037 | 0.74 |
| rs2736100 | 5 | 1286401 | TERT | A | C | 0.47 | −0.0038 | 0.0032 | 0.24 |
| rs9419958 | 10 | 103916188 | OBFC1 | T | C | 0.16 | −0.00093 | 0.0044 | 0.84 |
| rs9420907 | 10 | 103916707 | OBFC1 | C | A | 0.17 | 0.00032 | 0.0042 | 0.94 |
| rs4387287 | 10 | 103918139 | OBFC1 | A | C | 0.21 | 0.0038 | 0.0039 | 0.32 |
| rs3027234 | 17 | 8232774 | CTC1 | T | C | 0.21 | 0.0017 | 0.0038 | 0.66 |
| rs8105767 | 19 | 22032639 | ZNF208 | G | A | 0.30 | −0.0027 | 0.0034 | 0.42 |
| rs412658 | 19 | 22176638 | ZNF676 | T | C | 0.39 | −0.0016 | 0.0032 | 0.63 |
| rs6028466 | 20 | 39500359 | DHX35 | A | G | 0.070 | −0.0022 | 0.0062 | 0.72 |
| MTL | ||||||||
|---|---|---|---|---|---|---|---|---|
| SNP | CHR | Position | Gene | Effect Allele | Other Allele | BETA | SE | P |
| rs12696304 | 3 | 169763483 | TERC | G | C | −0.014 | 0.051 | 0.78 |
| rs10936599 | 3 | 169774313 | TERC | T | C | −0.054 | 0.054 | 0.32 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toupance, S.; Stathopoulou, M.G.; Petrelis, A.M.; Gorenjak, V.; Labat, C.; Lai, T.-P.; Visvikis-Siest, S.; Benetos, A. TERC Variants Associated with Short Leukocyte Telomeres: Implication of Higher Early Life Leukocyte Telomere Attrition as Assessed by the Blood-and-Muscle Model. Cells 2020, 9, 1360. https://doi.org/10.3390/cells9061360
Toupance S, Stathopoulou MG, Petrelis AM, Gorenjak V, Labat C, Lai T-P, Visvikis-Siest S, Benetos A. TERC Variants Associated with Short Leukocyte Telomeres: Implication of Higher Early Life Leukocyte Telomere Attrition as Assessed by the Blood-and-Muscle Model. Cells. 2020; 9(6):1360. https://doi.org/10.3390/cells9061360
Chicago/Turabian StyleToupance, Simon, Maria G. Stathopoulou, Alexandros M. Petrelis, Vesna Gorenjak, Carlos Labat, Tsung-Po Lai, Sophie Visvikis-Siest, and Athanase Benetos. 2020. "TERC Variants Associated with Short Leukocyte Telomeres: Implication of Higher Early Life Leukocyte Telomere Attrition as Assessed by the Blood-and-Muscle Model" Cells 9, no. 6: 1360. https://doi.org/10.3390/cells9061360
APA StyleToupance, S., Stathopoulou, M. G., Petrelis, A. M., Gorenjak, V., Labat, C., Lai, T.-P., Visvikis-Siest, S., & Benetos, A. (2020). TERC Variants Associated with Short Leukocyte Telomeres: Implication of Higher Early Life Leukocyte Telomere Attrition as Assessed by the Blood-and-Muscle Model. Cells, 9(6), 1360. https://doi.org/10.3390/cells9061360