Integration of ATAC-Seq and RNA-Seq Identifies Key Genes in Light-Induced Primordia Formation of Sparassis latifolia
Abstract
1. Introduction
2. Results
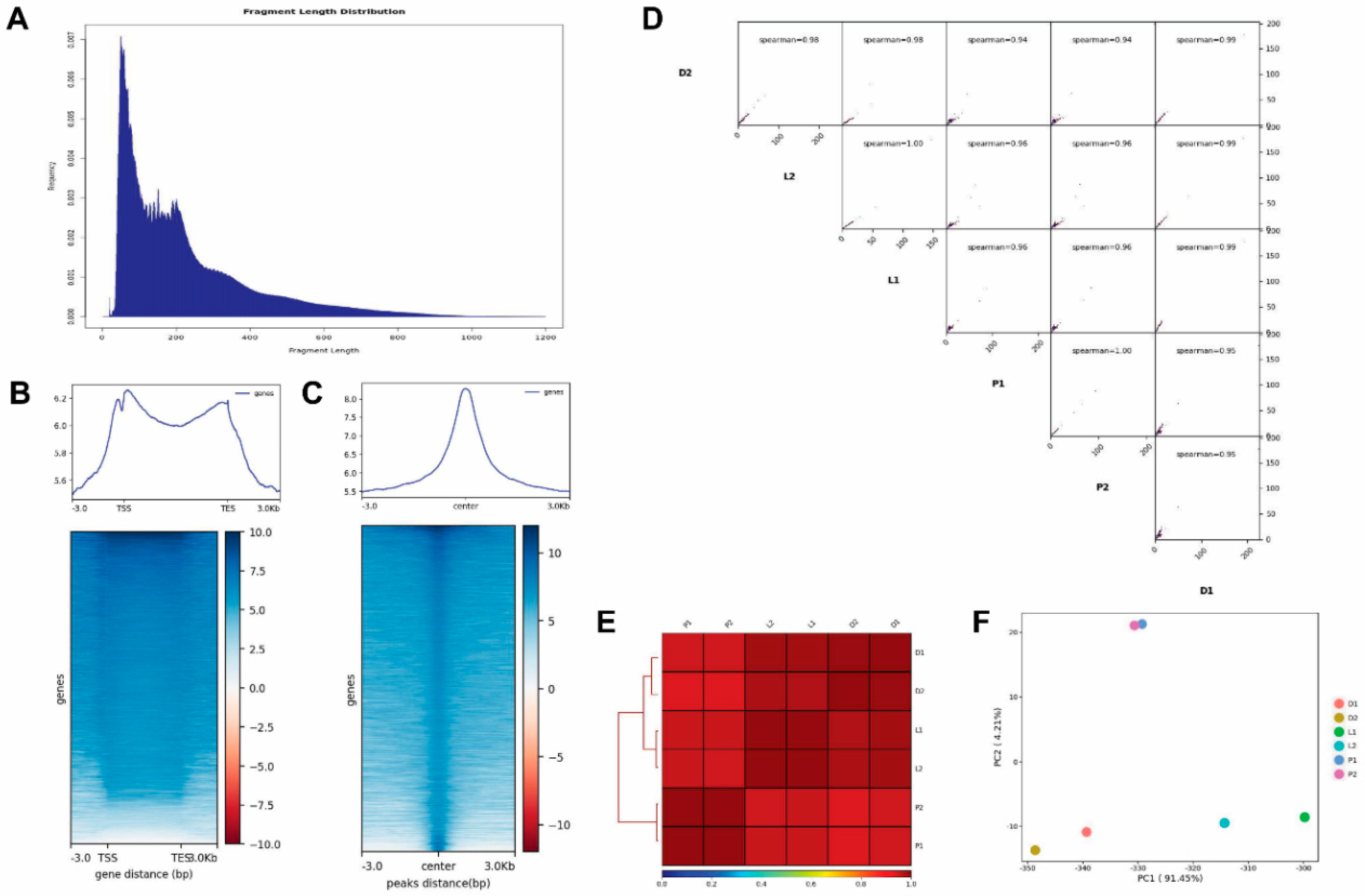
2.1. Results of ATAC-Seq
2.2. Differential Chromatin Accessibility in LIPF
2.3. Integration of ATAC-Seq Results with RNA-Seq
2.4. Validation of the Results by qRT-PCR
3. Discussion
4. Materials and Methods
4.1. Strains, Culture Conditions, and Isolation of Nucleic Acids
4.2. ATAC-Seq
4.3. Integration Analysis of ATAC-Seq and RNA-Seq
4.4. Function Annotation
4.5. Gene Expression Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability
Conflicts of Interest
Abbreviations
| LIPF | light induced primordia formation |
| S. latifolia | Sparassis latifolia |
| ATAC-seq | assay for transposase accessible chromatin by sequencing |
| DEPs | differentially expressed peaks |
| DEGs | differentially expressed genes |
| PDA | potato dextrose agar |
| PCA | Principal Components Analysis |
| GEO | Gene Expression Omnibus |
| GAPDH | glyceraldehyde-3-phosphate dehydrogenase |
| qRT-PCR | real-time quantitative PCR |
| FDR | false discovery rate |
References
- Busch, S.; Braus, G. How to build a fungal fruit body: From uniform cells to specialized tissue. Mol. Microbiol. 2010, 64, 873–876. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.K.; Au, C.H.; Wilke, S.K.; Stajich, J.E.; Zolan, M.E.; Pukkila, P.J.; Kwan, H.S. 5’-Serial Analysis of Gene Expression studies reveal a transcriptomic switch during fruiting body development in Coprinopsis cinerea. BMC Genom. 2013, 14, 195. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Dai, Y.; Yang, C.; Wei, P.; Song, B.; Yang, Y.; Sun, L.; Zhang, Z.W.; Li, Y. Comparative Transcriptome Analysis Identified Candidate Genes Related to Bailinggu Mushroom Formation and Genetic Markers for Genetic Analyses and Breeding. Sci. Rep. 2017, 7, 9266. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Gong, W.; Zhu, Z.; Yan, L.; Hu, Z.; Peng, Y. Comparative transcriptomics of Pleurotus eryngii reveals blue-light regulation of carbohydrate-active enzymes (CAZymes) expression at primordium differentiated into fruiting body stage. Genomics 2017, 110, 201–209. [Google Scholar] [CrossRef]
- Wang, Y.; Zeng, X.; Liu, W. De novo transcriptomic analysis during Lentinula edodes fruiting body growth. Gene 2018, 641, 326–334. [Google Scholar] [CrossRef]
- Ohm, R.A.; de Jong, J.F.; Lugones, L.G.; Aerts, A.; Kothe, E.; Stajich, J.E.; de Vries, R.P.; Record, E.; Levasseur, A.; Baker, S.E.; et al. Genome sequence of the model mushroom Schizophyllum commune. Nat. Biotechnol. 2010, 28, 957–963. [Google Scholar] [CrossRef]
- Liu, J.Y.; Chang, M.C.; Meng, J.L.; Feng, C.P.; Zhao, H.; Zhang, M.L. Comparative Proteome Reveals Metabolic Changes during the Fruiting Process in Flammulina velutipes. J. Agric. Food Chem. 2017, 65, 5091–5100. [Google Scholar] [CrossRef]
- Zhang, G.; Sun, Z.; Ren, A.; Shi, L.; Shi, D.; Li, X.; Zhao, M. The mitogen-activated protein kinase GlSlt2 regulates fungal growth, fruiting body development, cell wall integrity, oxidative stress and ganoderic acid biosynthesis in Ganoderma lucidum. Fungal Genet. Biol. 2017, 104, 6–15. [Google Scholar] [CrossRef]
- Wang, F.; Song, X.H.; Dong, X.M.; Zhang, J.J.; Dong, C.H. DASH-type cryptochromes regulate fruiting body development and secondary metabolism differently than CmWC-1 in the fungus Cordyceps militaris. Appl. Microbiol. Biot. 2017, 101, 4645–4657. [Google Scholar] [CrossRef]
- Zhang, J.; Ren, A.; Chen, H.; Zhao, M.; Shi, L.; Chen, M.; Wang, H.; Feng, Z. Transcriptome analysis and its application in identifying genes associated with fruiting body development in basidiomycete Hypsizygus marmoreus. PLoS ONE 2015, 10, e0123025. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, H.; Chen, M.; Ren, A.; Huang, J.; Wang, H.; Zhao, M.; Feng, Z. Cloning and functional analysis of a laccase gene during fruiting body formation in Hypsizygus marmoreus. Microbiol. Res. 2015, 179, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Rahmad, N.; Al-Obaidi, J.R.; Nor Rashid, N.M.; Zean, N.B.; Mohd Yusoff, M.H.; Shaharuddin, N.S.; Mohd Jamil, N.A.; Mohd Saleh, N. Comparative proteomic analysis of different developmental stages of the edible mushroom Termitomyces heimii. Biol. Res. 2014, 47, 30. [Google Scholar] [CrossRef] [PubMed]
- Plaza, D.F.; Lin, C.W.; van der Velden, N.S.; Aebi, M.; Kunzler, M. Comparative transcriptomics of the model mushroom Coprinopsis cinerea reveals tissue-specific armories and a conserved circuitry for sexual development. BMC Genom. 2014, 15, 492. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wu, X.; Gao, W.; Zhao, M.; Zhang, J.; Huang, C. Differential Expression Patterns of Pleurotus ostreatus Catalase Genes during Developmental Stages and under Heat Stress. Genes (Basel) 2017, 8, 335. [Google Scholar] [CrossRef]
- Dai, Y.C.; Wang, Z.; Binder, M.; Hibbett, D.S. Phylogeny and a new species of Sparassis (Polyporales, Basidiomycota): Evidence from mitochondrial atp6, nuclear rDNA and rpb2 genes. Mycologia 2006, 98, 584–592. [Google Scholar] [CrossRef]
- Thi Nhu Ngoc, L.; Oh, Y.K.; Lee, Y.J.; Lee, Y.C. Effects of Sparassis crispa in Medical Therapeutics: A Systematic Review and Meta-Analysis of Randomized Controlled Trials. Int. J. Mol. Sci. 2018, 19, 1487. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, J.; Zhong, X.; Li, J.; Wang, X.; Ji, L.; Shang, X. Rapid Characterization of Chemical Components in Edible Mushroom Sparassis crispa by UPLC-Orbitrap MS Analysis and Potential Inhibitory Effects on Allergic Rhinitis. Molecules 2019, 24, 2014. [Google Scholar] [CrossRef]
- Uchida, M.; Horii, N.; Hasegawa, N.; Oyanagi, E.; Yano, H.; Iemitsu, M. Sparassis crispa Intake Improves the Reduced Lipopolysaccharide-Induced TNF-alpha Production That Occurs upon Exhaustive Exercise in Mice. Nutrients 2019, 11, 2049. [Google Scholar] [CrossRef]
- Xiao, D.L.; Ma, L.; Yang, C.; Ying, Z.H.; Jiang, X.L.; Lin, Y.Q. De Novo Sequencing of a Sparassis latifolia Genome and Its Associated Comparative Analyses. Can. J. Infect. Dis. Med. 2018, 2018, 12. [Google Scholar] [CrossRef]
- Xiao, D.; Ma, L.; Ying, Z.; Jiang, X.; Lin, Y. Analysis of Sparassis latifolia proteomes at the early fruiting and fruiting stages of mushroom development using iTRAQ-coupled 2D LC-MS/MS. Acta Edulis Fungi 2016, 23, 1–9. (In Chinese) [Google Scholar] [CrossRef]
- Carmady, B.; Smith, C.A. Use of Chinese medicine by cancer patients: A review of surveys. Chin. Med. 2011, 6, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Xiao, D.; Zhang, D.; Ma, L.; Wang, H.; Lin, Y. Preliminary study on differentially expressed genes of Sparassis latifolia under light inducing. Edible Fungi China 2017, 36, 60–63. (In Chinese) [Google Scholar] [CrossRef]
- Yang, C.; Ma, L.; Xiao, D.L.; Ying, Z.H.; Jiang, X.L.; Lin, Y.Q. Identification and Evaluation of Reference Genes for qRT-PCR Normalization in Sparassis latifolia (Agaricomycetes). Int. J. Med. Mushrooms 2019, 21, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Ma, L.; Ying, Z.H.; Jiang, X.L.; Lin, Y.Q. Sequence Analysis and Expression of a Blue-light Photoreceptor Gene, Slwc-1 from the Cauliflower Mushroom Sparassis latifolia. Curr. Microbiol. 2017, 74, 469–475. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Ma, L.; Xiao, D.; Ying, Z.; Jiang, X.; Lin, Y. Sequence and expression analysis of DASH-type cryptochrome gene, Slcry1 from the Sparassis latifolia. Acta Edulis Fungi 2018, 25, 9–16. (In Chinese) [Google Scholar] [CrossRef]
- Buenrostro, J.D.; Wu, B.; Chang, H.Y.; Greenleaf, W.J. ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr. Protoc. Mol. Biol. 2015, 109, 21–29. [Google Scholar] [CrossRef]
- Buenrostro, J.D.; Giresi, P.G.; Zaba, L.C.; Chang, H.Y.; Greenleaf, W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 2013, 10, 1213–1218. [Google Scholar] [CrossRef]
- Idnurm, A.; Heitman, J. Light controls growth and development via a conserved pathway in the fungal kingdom. PLoS Biol. 2005, 3, e95. [Google Scholar] [CrossRef]
- Kiyama, R.; Furutani, Y.; Kawaguchi, K.; Nakanishi, T. Genome sequence of the cauliflower mushroom Sparassis crispa (Hanabiratake) and its association with beneficial usage. Sci. Rep. 2018, 8, 16053. [Google Scholar] [CrossRef]
- Ying, Z.; Lin, Y.; Ma, L.; Jiang, X. Effects of different light quality and quantity on the mycelial growth and primordium formation of Sparassis crispa. Fujian J. Agric. Sci. 2013, 28, 538–540. (In Chinese) [Google Scholar]
- Kouzarides, T. Chromatin Modifications and Their Function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Lowe, E.K.; Cuomo, C.; Voronov, D.; Arnone, M.I. Using ATAC-seq and RNA-seq to increase resolution in GRN connectivity. Methods Cell Biol. 2019, 151, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Hendrickson, D.G.; Soifer, I.; Wranik, B.J.; Kim, G.; Robles, M.; Gibney, P.A.; McIsaac, R.S. A new experimental platform facilitates assessment of the transcriptional and chromatin landscapes of aging yeast. Elife 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Chereji, R.V.; Ocampo, J.; Clark, D.J. MNase-Sensitive Complexes in Yeast: Nucleosomes and Non-histone Barriers. Mol. Cell 2017, 65, 565–577. [Google Scholar] [CrossRef]
- Yang, T.; Xiong, W.; Dong, C. Cloning and analysis of the Oswc-1 gene encoding a putative blue light photoreceptor from Ophiocordyceps sinensis. Mycoscience 2013, 55, 241–245. [Google Scholar] [CrossRef]
- Yang, T.; Dong, C.H. Photo morphogenesis and photo response of the blue-light receptor gene Cmwc-1 in different strains of Cordyceps militaris. FEMS Microbiol. Lett. 2014, 352, 190–197. [Google Scholar] [CrossRef][Green Version]
- Song, H.Y.; Kim, D.H.; Kim, J.M. Comparative transcriptome analysis of dikaryotic mycelia and mature fruiting bodies in the edible mushroom Lentinula edodes. Sci. Rep. 2018, 8, 8983. [Google Scholar] [CrossRef]
- Tang, L.H.; Jian, H.H.; Song, C.Y.; Bao, D.P.; Shang, X.D.; Wu, D.Q.; Tan, Q.; Zhang, X.H. Transcriptome analysis of candidate genes and signaling pathways associated with light-induced brown film formation in Lentinula edodes. Appl. Microbiol. Biotechnol. 2013, 97, 4977–4989. [Google Scholar] [CrossRef]
- Terashima, K.; Yuki, K.; Muraguchi, H.; Akiyama, M.; Kamada, T. The dst1 gene involved in mushroom photomorphogenesis of Coprinus cinereus encodes a putative photoreceptor for blue light. Genetics 2005, 171, 101–108. [Google Scholar] [CrossRef]
- Yang, T.; Guo, M.M.; Yang, H.J.; Guo, S.; Dong, C.H. The blue-light receptor CmWC-1 mediates fruit body development and secondary metabolism in Cordyceps militaris. Appl. Microbiol. Biot. 2015, 100, 743–755. [Google Scholar] [CrossRef]
- Banerjee, G.; Robertson, D.L.; Leonard, T.J. Hydrophobins Sc3 and Sc4 gene expression in mounds, fruiting bodies and vegetative hyphae of Schizophyllum commune. Fungal Genet. Biol. 2008, 45, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.I.; Lee, C.S.; Park, Y.J. Further characterization of hydrophobin genes in genome of Flammulina velutipes. Mycoscience 2016, 57, 320–325. [Google Scholar] [CrossRef]
- Tao, Y.; Chen, R.; Yan, J.; Long, Y.; Tong, Z.; Song, H.; Xie, B. A hydrophobin gene, Hyd9, plays an important role in the formation of aerial hyphae and primordia in Flammulina filiformis. Gene 2019, 706, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Lin, Y.Q.; Yang, C.; Ying, Z.H.; Jiang, X.L. Production of liquid spawn of an edible mushroom, Sparassis latifolia by submerged fermentation and mycelial growth on pine wood sawdust. Sci. Hortic. 2016, 209, 22–30. [Google Scholar] [CrossRef]
- Corces, M.R.; Trevino, A.E.; Hamilton, E.G.; Greenside, P.G.; Sinnott-Armstrong, N.A.; Vesuna, S.; Satpathy, A.T.; Rubin, A.J.; Montine, K.S.; Wu, B.; et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 2017, 14, 959–962. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBNET J. 2011, 17. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nusbaum, C.; Myers, R.M.; Brown, M.; Li, W.; et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef]
- Heinz, S.; Benner, C.; Spann, N.; Bertolino, E.; Lin, Y.C.; Laslo, P.; Cheng, J.X.; Murre, C.; Singh, H.; Glass, C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 2010, 38, 576–589. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Draghici, S.; Khatri, P.; Tarca, A.L.; Amin, K.; Done, A.; Voichita, C.; Georgescu, C.; Romero, R. A systems biology approach for pathway level analysis. Genome Res. 2007, 17, 1537–1545. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]





© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, C.; Ma, L.; Xiao, D.; Ying, Z.; Jiang, X.; Lin, Y. Integration of ATAC-Seq and RNA-Seq Identifies Key Genes in Light-Induced Primordia Formation of Sparassis latifolia. Int. J. Mol. Sci. 2020, 21, 185. https://doi.org/10.3390/ijms21010185
Yang C, Ma L, Xiao D, Ying Z, Jiang X, Lin Y. Integration of ATAC-Seq and RNA-Seq Identifies Key Genes in Light-Induced Primordia Formation of Sparassis latifolia. International Journal of Molecular Sciences. 2020; 21(1):185. https://doi.org/10.3390/ijms21010185
Chicago/Turabian StyleYang, Chi, Lu Ma, Donglai Xiao, Zhenghe Ying, Xiaoling Jiang, and Yanquan Lin. 2020. "Integration of ATAC-Seq and RNA-Seq Identifies Key Genes in Light-Induced Primordia Formation of Sparassis latifolia" International Journal of Molecular Sciences 21, no. 1: 185. https://doi.org/10.3390/ijms21010185
APA StyleYang, C., Ma, L., Xiao, D., Ying, Z., Jiang, X., & Lin, Y. (2020). Integration of ATAC-Seq and RNA-Seq Identifies Key Genes in Light-Induced Primordia Formation of Sparassis latifolia. International Journal of Molecular Sciences, 21(1), 185. https://doi.org/10.3390/ijms21010185




