Rapid Characterization of Chemical Components in Edible Mushroom Sparassis crispa by UPLC-Orbitrap MS Analysis and Potential Inhibitory Effects on Allergic Rhinitis
Abstract
1. Introduction
2. Results and Discussion
2.1. Optimization of UPLC and MS Conditions
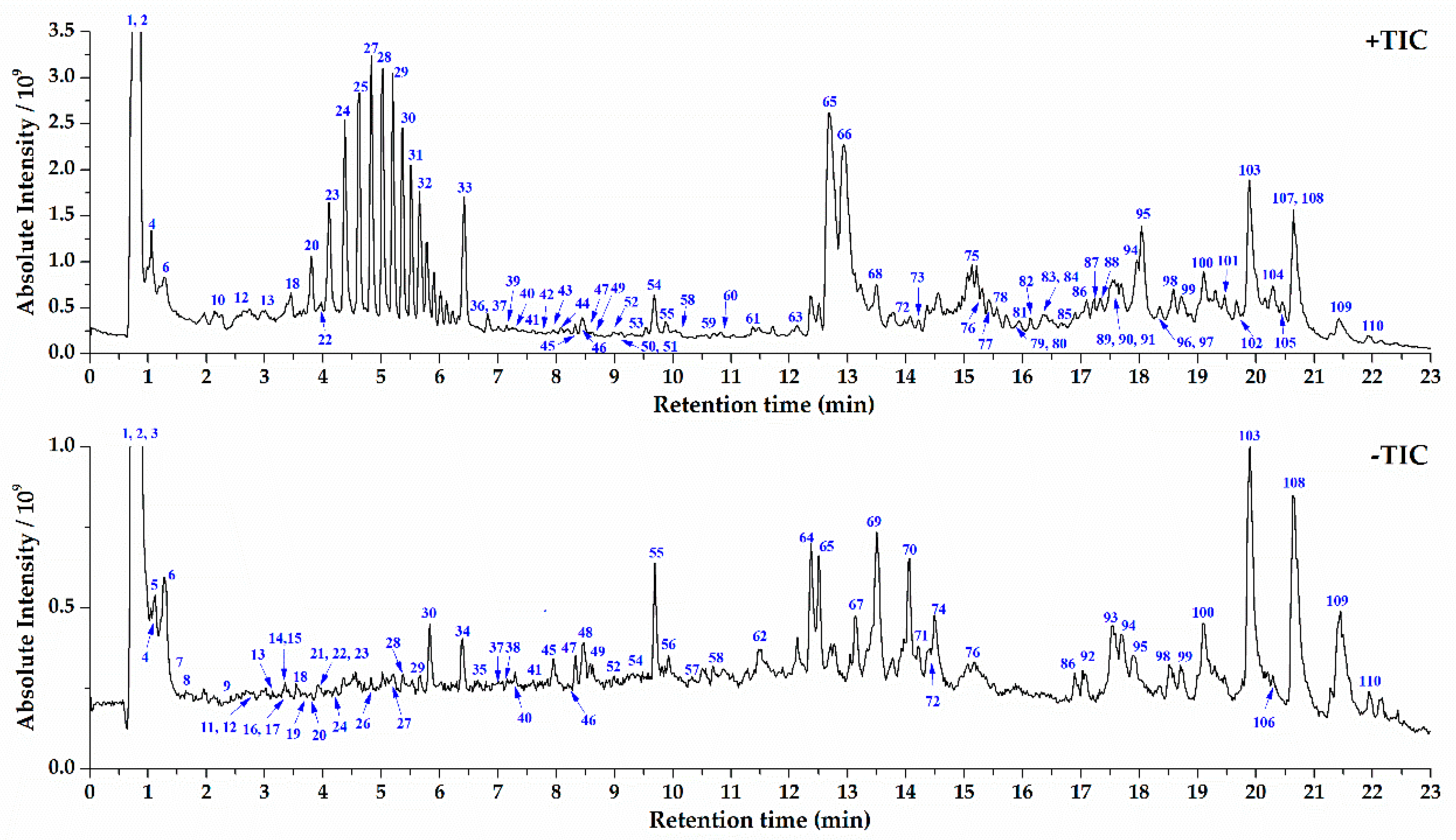
2.2. UPLC-Orbitrap MS Analysis of S. crispa and Component Identification
2.3. Structural Characterization and Identification of Various Type of Components in S. crispa
2.3.1. Structural Characterization and Identification of Alkaloids
2.3.2. Structural Characterization and Identification of Organic Acids
2.3.3. Structural Characterization and Identification of Sesquiterpenes
2.3.4. Structural Characterization and Identification of Sterols
2.3.5. Structural Characterization and Identification of Phthalides and Others Types of Compounds
2.4. In Silico Prediction of Potential SFKs Inhibitors
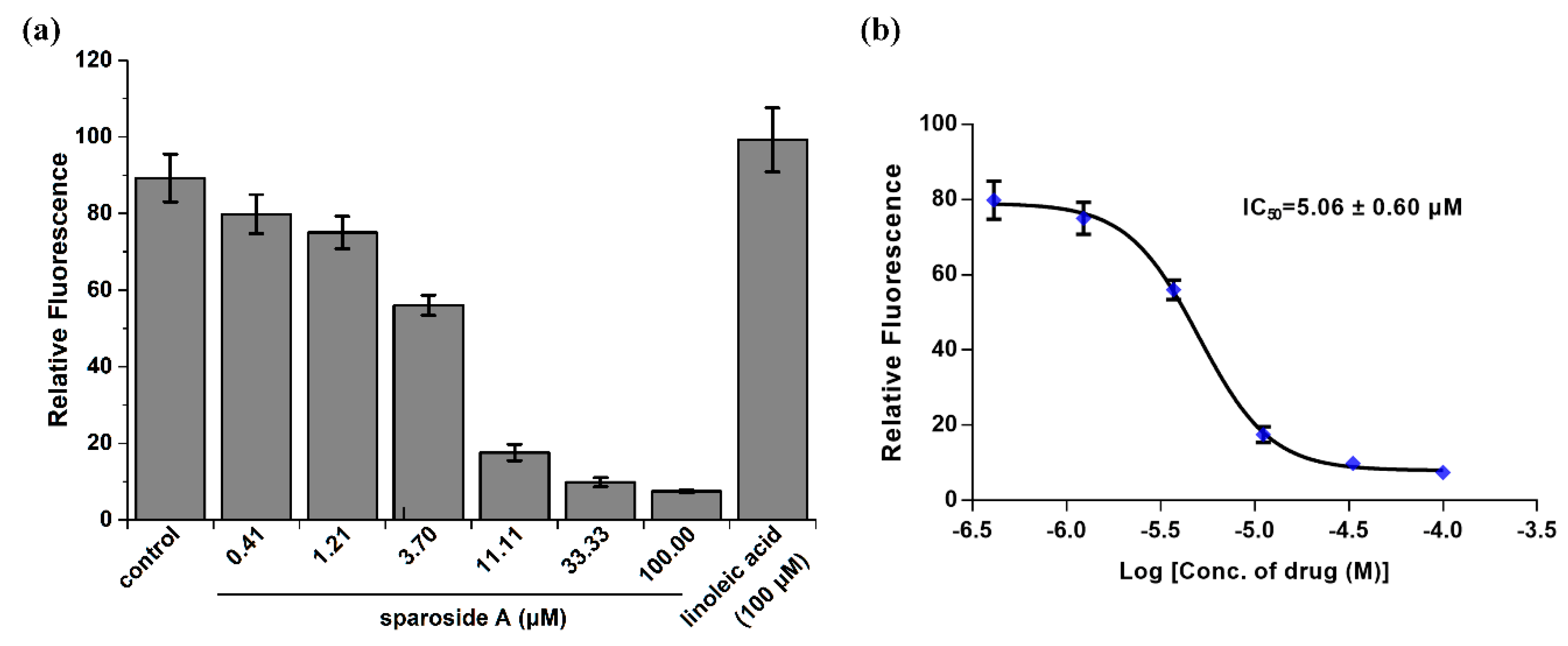
2.5. Anti-inflammatory Activity Confirmation of Two Predicted Components
3. Materials and Methods
3.1. Reagents and Materials
3.2. Sample Preparation
3.3. UPLC Separation
3.4. Orbitrap MS Analysis and Data Processing
3.5. Virtual Screening for Potential SFKs Inhibitors
3.6. Anti-inflammatory Activity Evaluation by Intracellular Calcium Mobilization Assay
3.7. Data Process and Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| UPLC | ultra-high performance liquid chromatography |
| GC | gas chromatography |
| MS | mass spectrometer |
| CID | collision-induced dissociation |
| NCE | normalized collision energy |
| AR | allergic rhinitis |
| SFKs | Src family kinases |
| DMEM | Dulbecco’s modified Eagle’s medium |
| FBS | fetal bovine serums |
| PBS | phosphate buffer solution |
| DMSO | dimethylsulfoxide |
References
- Ohno, N.; Miura, N.N.; Nakajima, M.; Yadomae, T. Antitumor 1,3-beta-glucan from cultured fruit body of Sparassis crispa. Biol. Pharm. Bull. 2000, 23, 866–872. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Lee, S.; Singh, T.S.K.; Choi, J.K.; Shin, T.; Kim, S. Sparassis crispa suppresses mast cell-mediated allergic inflammation: Role of calcium, mitogen-activated protein kinase and nuclear factor-kappa B. Int. J. Mol. Med. 2012, 30, 344–350. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Kim, J.Y.; Ryu, H.S.; Park, H.; Kim, Y.O.; Kang, J.S.; Kim, H.M.; Hong, J.T.; Kim, Y.; Han, S. Induction of dendritic cell maturation by β-glucan isolated from Sparassis crispa. Int. Immunopharmacol. 2010, 10, 1284–1294. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Kimura, T. Dietary Sparassis crispa (Hanabiratake) ameliorates plasma levels of adiponectin and glucose in type 2 diabetic mice. J. Health Sci. 2010, 56, 541–546. [Google Scholar] [CrossRef]
- Kimura, T.; Hashimoto, M.; Yamada, M.; Nishikawa, Y. Sparassis crispa (Hanabiratake) ameliorates skin conditions in rats and humans. Biosci. Biotech. Bioch. 2013, 77, 1961–1963. [Google Scholar] [CrossRef] [PubMed]
- Bang, S.; Chae, H.S.; Lee, C.; Choi, H.G.; Ryu, J.; Li, W.; Lee, H.; Jeong, G.; Chin, Y.; Shim, S.H. New aromatic compounds from the fruiting body of Sparassis crispa (Wulf.) and their inhibitory activities on proprotein convertase subtilisin/kexin type 9 mRNA expression. J. Agr. Food Chem. 2017, 65, 6152–6157. [Google Scholar] [CrossRef]
- Yoshikawa, K.; Kokudo, N.; Hashimoto, T.; Yamamoto, K.; Inose, T.; Kimura, T. Novel phthalide compounds from Sparassis crispa (Hanabiratake), hanabiratakelide A–C, exhibiting anti-cancer related activity. Biol. Pharm. Bull. 2010, 33, 1355–1359. [Google Scholar] [CrossRef]
- Kodani, S.; Hayashi, K.; Hashimoto, M.; Kimura, T.; Dombo, M.; Kawagishi, H. New sesquiterpenoid from the mushroom Sparassis crispa. Biosci. Biotech. Bioch. 2009, 73, 228–229. [Google Scholar] [CrossRef][Green Version]
- Lee, Y.; Thi, N.N.; Kim, H.G.; Lee, D.Y. Ergosterol peroxides from the fruit body of Sparassis crispa. J. Biol. Appl. Chem. 2016, 59, 313–316. [Google Scholar] [CrossRef]
- Woodward, S.; Sultan, H.Y.; Barrett, D.K.; Pearce, R.B. Two new antifungal metabolites produced by Sparassis crispa in culture and in decayed trees. J. Gen. Microbiol. 1993, 139, 153–159. [Google Scholar] [CrossRef]
- Kiyama, R.; Furutani, Y.; Kawaguchi, K.; Nakanishi, T. Genome sequence of the cauliflower mushroom Sparassis crispa (Hanabiratake) and its association with beneficial usage. Sci. Rep.-UK 2018, 8, 16053. [Google Scholar] [CrossRef] [PubMed]
- Bang, S.; Lee, C.; Ryu, J.; Li, W.; Koh, Y.; Jeon, J.; Lee, J.; Shim, S.H. Simultaneous determination of the bioactive compounds from Sparassis crispa (Wulf.) by HPLC-DAD and their inhibitory effects on LPS-stimulated cytokine production in bone marrow-derived dendritic cell. Arch. Pharm. Res. 2018, 41, 823–829. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.; Park, S.; Kim, E.; Son, H. GC-MS based metabolomics study of fermented stipe of Sparassis crispa. Food Sci. Biotechnol. 2018, 27, 1111–1118. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Qu, Y.; Wang, L.; Zhang, X.; Xiao, H. Ultra-high performance liquid chromatography with linear ion trap-Orbitrap hybrid mass spectrometry combined with a systematic strategy based on fragment ions for the rapid separation and characterization of components in Stellera chamaejasme extracts. J. Sep. Sci. 2016, 39, 1379–1388. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Lu, Y.; Chen, Z.; Tan, J.; Bai, J.; Li, P.; Wang, Z.; Du, S. Rapid characterization of components in Bolbostemma paniculatum by UPLC/LTQ-Orbitrap MSn analysis and multivariate statistical analysis for herb discrimination. Molecules 2018, 23, 1155. [Google Scholar] [CrossRef] [PubMed]
- Wheatley, L.M.; Togias, A. Allergic rhinitis. New Engl. J. Med. 2015, 372, 456–463. [Google Scholar] [CrossRef] [PubMed]
- Kimura, T. Studies on pharmacological activities of the cauliflower mushroom Sparassis crispa. S. Afr. Med. J. 2013, 92, 97–108. [Google Scholar]
- Han, J.M.; Lee, E.K.; Gong, S.Y.; Sohng, J.K.; Kang, Y.J.; Jung, H.J. Sparassis crispa exerts anti-inflammatory activity via suppression of TLR-mediated NF-κB and MAPK signaling pathways in LPS-induced RAW264.7 macrophage cells. J. Ethnopharmacol. 2019, 231, 10–18. [Google Scholar] [CrossRef]
- Kawa, A. The role of mast cells in allergic inflammation. Resp. Med. 2012, 106, 9–14. [Google Scholar]
- Engen, J.R.; Wales, T.E.; Hochrein, J.M.; Meyn, M.A.; Banu Ozkan, S.; Bahar, I.; Smithgall, T.E. Structure and dynamic regulation of Src-family kinases. Cell. Mol. Life Sci. 2008, 65, 3058–3073. [Google Scholar] [CrossRef]
- Rossi, A.; Herlaar, E.; Braselmann, S.; Huynh, S.; Taylor, V.; Frances, R.; Issakani, S.; Argade, A.; Singh, R.; Payan, D. Identification of the Syk kinase inhibitor R112 by a human mast cell screen. J. Allergy Clin. Immun. 2006, 118, 749–755. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Cai, S.F.; Nie, J.; Li, Y.Y.; Shi, G.; Hao, J.; Fu, W.; Tan, H.; Chen, S.; Li, B.; et al. The natural compound nujiangexanthone A suppresses mast cell activation and allergic asthma. Biochem. Pharmacol. 2016, 100, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Masuda, E.S.; Payan, D.G. Discovery and development of spleen tyrosine kinase (Syk) inhibitors. J. Med. Chem. 2012, 55, 3614–3643. [Google Scholar] [CrossRef] [PubMed]
- Jelić, D.; Mildner, B.; Koštrun, S.; Nujić, K.; Verbanac, D.; Culić, O.; Antolović, R.; Brandt, W. Homology modeling of human Fyn kinase structure: Discovery of rosmarinic acid as a new Fyn kinase inhibitor and in silico study of its possible binding modes. J. Med. Chem. 2007, 50, 1090–1100. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Yang, J.H.; Li, X.; Hwangbo, K.; Hwang, S.; Taketomi, Y.; Murakami, M.; Chang, Y.; Kim, C.; Son, J.; et al. Emodin, a naturally occurring anthraquinone derivative, suppresses IgE-mediated anaphylactic reaction and mast cell activation. Biochem. Pharmacol. 2011, 82, 1700–1708. [Google Scholar] [CrossRef] [PubMed]
- Han, S.Y.; Bae, J.Y.; Park, S.H.; Kim, Y.H.; Park, J.H.Y.; Kang, Y.H. Resveratrol inhibits IgE-mediated basophilic mast cell degranulation and passive cutaneous anaphylaxis in mice. J. Nutr. 2013, 143, 632–639. [Google Scholar] [CrossRef] [PubMed]
- Dunn, W.B.; Erban, A.; Weber, R.J.M.; Creek, D.J.; Brown, M.; Breitling, R.; Hankemeier, T.; Goodacre, R.; Neumann, S.; Kopka, J.; et al. Mass appeal: Metabolite identification in mass spectrometry-focused untargeted metabolomics. Metabolomics 2013, 9, 44–66. [Google Scholar] [CrossRef]
- Lu, M.; Lin, Y.Q.; Jiang, X.L.; Ying, Z. Effect of inorganic salts, vitamins and phytohormones on mycelial growth of Sparassis crispa. J. Fungal Res. 2011, 9, 172–175. [Google Scholar]
- Lee, M.R.; Hou, J.G.; Begum, S.; Xue, J.J.; Wang, Y.B.; Sung, C.K. Comparison of constituents, antioxidant potency, and acetylcholinesterase inhibition in Lentinus edodes, Sparassis crispa, and Mycoleptodonoides aitchisonii. Food Sci. Biotechnol. 2013, 22, 1747–1751. [Google Scholar] [CrossRef]
- Moreira Lacerda, R.B.; Freitas, T.R.; Martins, M.M.; Teixeira, T.L.; Da Silva, C.V.; Candido, P.A.; de Oliveira, R.J.; Viegas Junior, C.; Bolzani, V.D.S.; Danuello, A.; et al. Isolation, leishmanicidal evaluation and molecular docking simulations of piperidine alkaloids from Senna spectabilis. Bioorgan. Med. Chem. 2018, 26, 5816–5823. [Google Scholar] [CrossRef]
- Chen, L.; Liu, Y.; Guo, Q.; Zheng, Q.; Zhang, W. Metabolomic comparison between wild Ophiocordyceps sinensis and artificial cultured Cordyceps militaris. Biomed. Chromatogr. 2018, 32, e4279. [Google Scholar] [CrossRef] [PubMed]
- Carlyle, R.F. The occurrence in and actions of amino acids on isolated supra oral sphincter preparations of the sea anemone Actinia equina. J. Physiol. 1974, 236, 635–652. [Google Scholar] [CrossRef] [PubMed]
- Aiello, A.; Fattorusso, E.; Giordano, A.; Menna, M.; Navarrete, C.; Muñoz, E. Clavaminols G–N, six new marine sphingoids from the Mediterranean ascidian Clavelina phlegraea. Tetrahedron 2009, 65, 4384–4388. [Google Scholar] [CrossRef]
- Itokawa, H.; Morita, H.; Kayashita, T.; Shimomura, M.; Takeya, K. Cyclic peptides from higher plants. Part 30. Three novel cyclic peptides, yunnanins D, E and F from Stellaria yunnanensis. Heterocycles 1996, 43, 1279–1286. [Google Scholar] [CrossRef]
- Kariotoglou, D.M.A.M. Sphingophosphonolipids, phospholipids, and fatty acids from Aegean jellyfish Aurelia aurita. Lipids 2001, 36, 1255. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Bao, H.; Wang, A.; Tang, C.; Dien, P.; Ye, Y. Two new N-oxide alkaloids from Stemona cochinchinensis. Molecules 2014, 19, 20257–20265. [Google Scholar] [CrossRef]
- Kim, S.; Shin, Y.; Lee, S.; Oh, K.; Lee, S.K.; Shin, J.; Oh, D. Salternamides A–D from a halophilic Streptomyces sp. Actinobacterium. J. Nat. Prod. 2015, 78, 836–843. [Google Scholar] [CrossRef]
- Bodor, A.; Barabás, A. Geometrical isomerism of the O-substituted oximes of some keto-steroids. Tetrahedron 1979, 35, 233–240. [Google Scholar] [CrossRef]
- Kawagishi, H.; Hayashi, K.; Tokuyama, S.; Hashimoto, N.; Kimura, T.; Dombo, M. Novel bioactive compound from the Sparassis crispa mushroom. Biosci. Biotechnol. Biochem. 2007, 71, 1804–1806. [Google Scholar] [CrossRef]
- Kasal, A.; Cerny, V.; Sorm, F. Preparation of some 18-substituted steroids of androstane type from Conessine. Collect. Czechoslov. Chem. Commun. 1963, 28, 411. [Google Scholar] [CrossRef]
- Pentegova, V.A.A.K. Composition of the neutral fraction of the oleoresin of Pinus sibirica. Chem. Nat. Compd. 1966, 2, 193–196. [Google Scholar] [CrossRef]
- Alarcon, J.; Villalobos, N.; Lamilla, C.; Cespedes, C.L. Ceramides and terpenoids from Russula austrodelica Singer. B. Latinoam. Caribe Pl. 2013, 12, 493–498. [Google Scholar]
- Neumann, A.; Patzelt, D.; Wagner-Doebler, I.; Schulz, S. Identification of new N-acylhomoserine lactone signalling compounds of dinoroseobacter shibae DFL-12(T) by overexpression of luxi genes. Chembiochem 2013, 14, 2355–2361. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Wang, H.; Li, F.; Zhu, T.; Gu, Q.; Li, D. Stachybotrin G, a sulfate meroterpenoid from a sponge derived fungus Stachybotrys chartarum MXH-X73. Tetrahedron Lett. 2015, 56, 7053–7055. [Google Scholar] [CrossRef]
- Chapman, K.D.A.V. Identification and quantification of neuroactive N-acylethanolamines in cottonseed processing fractions. J. Am. Oil Chem. Soc. 2003, 80, 223–229. [Google Scholar] [CrossRef]
- Guo, Y.; Gavagnin, M.; Mollo, E.; Cimino, G.; Hamdy, N.A.; Fakhr, I.; Pansini, M. Hurghamides A–D, new N-acyl-2-methylene-β-alanine methyl esters from Red Sea Hippospongia sp. Nat. Prod. Lett. 1997, 9, 281–288. [Google Scholar] [CrossRef]
- Ohnishi, M.; Kawase, S.; Kondo, Y.; Fujino, Y.; Ito, S. Identification of major cerebroside species in seven edible mushrooms. J. Jpn. Oil Chem. Soc. 1996, 45, 51–56. [Google Scholar] [CrossRef][Green Version]
- Fei, T.D.; Zhu, H.R.; Zhen, L.; Fen, Y.Z.; Jing, W.R. Chemical compositions and antimicrobial activity of the volva of Dictyophora echinovolvata (Ⅰ). Mycosystema 2006, 603–610. [Google Scholar]
- Tian, Y.; Zhao, Y.; Huang, J.; Zeng, H.; Zheng, B. Effects of different drying methods on the product quality and volatile compounds of whole Shiitake mushrooms. Food Chem. 2016, 197, 714–722. [Google Scholar] [CrossRef]
- Lyu, Z.Z.; Liu, N.Z.; Qian, Q.G.; Zhou, X.J. Isolation of endophytic fungi from Zanthoxylum simulans and screening of its active strain. China J. Chin. Mater. Medica 2018, 43, 1434–1440. [Google Scholar]
- Morzycki, J.W.; Wawer, I.; Gryszkiewicz, A.; Maj, J.; Siergiejczyk, L.; Zaworska, A. 13C-NMR study of 4-azasteroids in solution and solid state. Steroids 2002, 67, 621–626. [Google Scholar] [CrossRef]
- Shiono, Y.; Tamesada, Y.; Muravayev, Y.D.; Murayama, T.; Ikeda, M. N-phenethylhexadecanamide from the edible mushroom Laetiporus sulphureus. Nat. Prod. Res. 2005, 19, 363–366. [Google Scholar] [CrossRef]
- Jun, T.B.; Long, C.S.; Yu, M.Q.; Li, Y.; Tan, F. Effects of three drying processes on volatile compounds in Lentinus edodes. Food Sci. 2014, 35, 106–110. [Google Scholar]
- Jenkins, K.M.; Jensen, P.R.; Fenical, W. Thraustochytrosides A–C: New glycosphingolipids from a unique marine protist, Thraustochytrium globosum. Tetrahedron Lett. 1999, 40, 7637–7640. [Google Scholar] [CrossRef]
- Chen, J.; Sun, J.; Deering, R.W.; Dasilva, N.; Seeram, N.P.; Wang, H.; Rowley, D.C. Rhizoleucinoside, a rhamnolipid-amino alcohol hybrid from the Rhizobial symbiont bradyrhizobium sp. BTAi1. Org. Lett. 2016, 18, 1490–1493. [Google Scholar] [CrossRef] [PubMed]
- Atolani, O.A.A.E. Chemical composition, antioxidant, anti-lipooxygenase, antimicrobial, anti-parasite and cytotoxic activities of Polyalthia longifolia seed oil. Med. Chem. Res. 2019, 28, 515–527. [Google Scholar] [CrossRef]
- Tsivinska, M.V.; Antonguk, V.O.; Panchak, L.V.; Klguchivska, O.Y.; Stoika, R.S. Biologically active substances of methanol extracts of dried Lactarius quetus and Lactarius volemus basidiomes mushrooms: Identification and potential functions. Biotechnol. Acta 2015, 8, 58–68. [Google Scholar] [CrossRef][Green Version]
- Bi, W.; Ma, Q.; Yin, Y.; Li, Q.; Lei, H.M. GC-MS analysis of the low-polarity components of Rhizoma pinelliae preparation. Northwest. Pharm. J. 2008, 144–145. [Google Scholar]
- Morohashi, A.; Satake, M.; Nagai, H.; Oshima, Y.; Yasumoto, T. The absolute configuration of gambieric acids A–D, potent antifungal polyethers, isolated from the marine dinoflagellate Gambierdiscus toxicus. Tetrahedron 2000, 56, 8995–9001. [Google Scholar] [CrossRef]
- Siddiqi, M.Z.; Muhammad Shafi, S.; Choi, K.D.; Im, W. Compostibacter hankyongensis gen. nov., sp. nov., isolated from compost. Int. J. Syst. Evol. Micr. 2016, 66, 3681–3687. [Google Scholar] [CrossRef]
- Senior, N.M.; Brocklehurst, K.; Cooper, J.B.; Wood, S.P.; Erskine, P.; Shoolingin-Jordan, P.M.; Thomas, P.G.; Warren, M.J. Comparative studies on the 5-aminolaevulinic acid dehydratases from Pisum sativum, Escherichia coli and Saccharomyces cerevisiae. Biochem. J. 1996, 320, 401–412. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.S.; Lee, J.H.; Jeong, W.; Kim, N.; Jin, H.Z.; Hwang, B.Y.; Lee, H.; Lee, S.; Jang, D.S.; Lee, D. Acetylenic acid analogues from the edible mushroom Chanterelle (Cantharellus cibarius) and their effects on the gene expression of peroxisome proliferator-activated receptor-gamma target genes. Bioorg. Med. Chem. Lett. 2012, 22, 2347–2349. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Povedano, M.D.M.; de Medina, V.S.; Bautista, J.; Priego-Capote, F.; de Castro, M.D.L. Tentative identification of the composition of Agaricus bisporus aqueous enzymatic extracts with antiviral activity against HCV: A study by liquid chromatography–tandem mass spectrometry in high resolution mode. J. Funct. Foods 2016, 24, 403–419. [Google Scholar] [CrossRef]
- Shymala Gowri, S. Fatty acid composition of cultivated edible mushroom Lentinus tuberregium VKJM24 (HM060586). World J. Pharm. Res. 2017, 827–832. [Google Scholar] [CrossRef][Green Version]
- Sica, V.P.; Raja, H.A.; El-Elimat, T.; Kertesz, V.; Van Berkel, G.J.; Pearce, C.J.; Oberlies, N.H. Dereplicating and spatial mapping of secondary metabolites from Fungal cultures in situ. J. Nat. Prod. 2015, 78, 1926–1936. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, T.; Ishibashi, M.; Takata, T.; Takano, F.; Ohta, T. Cytotoxic fatty acid from Pleurocybella porrigens. Chem. Pharm. Bull. 2007, 55, 1748–1749. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Pan, X.; Li, F.; Dong, W. UPLC-QTOF-MS metabolomics analysis revealed the contributions of metabolites to the pathogenesis of Rhizoctonia solani strain AG-1-IA. PLoS ONE 2018, 13, e192486. [Google Scholar] [CrossRef]
- Hannemann, K.A.P.V. The common occurrence of furan fatty acids in plants. Lipids 1989, 24, 296–298. [Google Scholar] [CrossRef]
- Du Plessis, L.M.; Vladar, S. Isolation and identification of α-eleostearic acid in the kernels of sandapple (Parinarium capense) and mobolaplum (Parinarium curatellfolium). S. Afr. J. Sci. 1974, 70, 183–184. [Google Scholar]
- Wu, J.; Tsujimori, M.; Hirai, H.; Kawagishi, H. Novel compounds from the mycelia and fruiting bodies of Climacodon septentrionalis. Biosci. Biotechnol. Biochem. 2011, 75, 783–785. [Google Scholar] [CrossRef]
- Mohri, S.; Takahashi, H.; Sakai, M.; Takahashi, S.; Waki, N.; Aizawa, K.; Suganuma, H.; Ara, T.; Matsumura, Y.; Shibata, D.; et al. Wide-range screening of anti-inflammatory compounds in tomato using LC-MS and elucidating the mechanism of their functions. PLoS ONE 2018, 13, e191203. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Wang, Y.; Li, Y.; Ma, S.; Qu, J.; Liu, Y.; Niu, C.; Tang, Z.; Li, Y.; Li, L.; et al. Triterpenoids from the twigs and leaves of Rhododendron latoucheae by HPLC-MS-SPE-NMR. Tetrahedron 2019, 75, 296–307. [Google Scholar] [CrossRef]
- Reis, F.S.; Heleno, S.A.; Barros, L.; Sousa, M.J.; Martins, A.; Santos-Buelga, C.; Ferreira, I.C.F.R. Toward the antioxidant and chemical characterization of mycorrhizal mushrooms from northeast Portugal. J. Food Sci. 2011, 76, C824–C830. [Google Scholar] [CrossRef] [PubMed]
- Das, S.A.D.M. Metabolomic and chemometric study of Achras sapota L. fruit extracts for identification of metabolites contributing to the inhibition of α-amylase and α-glucosidase. Eur. Food Res. Technol. 2016, 242, 733–743. [Google Scholar] [CrossRef]
- Tao, L.; Yan, Y.H.; Xing, L.T.; Hong, X.Z. The study of antioxidant components from oil producing Fungi PJX-29. J. Chin. Cereals Oils Assoc. 2012, 27, 53–59. [Google Scholar]
- Tao, Q.; Ma, K.; Bao, L.; Wang, K.; Han, J.; Zhang, J.; Huang, C.; Liu, H. New sesquiterpenoids from the edible mushroom Pleurotus cystidiosus and their inhibitory activity against α-glucosidase and PTP1B. Fitoterapia 2016, 111, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Jiangbo, H.; Tao, J.; Miao, X.; Bu, W.; Zhang, S.; Dong, Z.; Li, Z.; Feng, T.; Liu, J. Seven new drimane-type sesquiterpenoids from cultures of fungus Laetiporus sulphureus. Fitoterapia 2015, 102, 1–6. [Google Scholar]
- Lu, Z.; Wang, Y.; Miao, C.; Liu, P.; Hong, K.; Zhu, W. Sesquiterpenoids and benzofuranoids from the marine-derived fungus Aspergillus ustus 094102. J. Nat. Prod. 2009, 72, 1761–1767. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, M.L.; Jin, H.Z.; Fu, J.J.; Hu, X.J.; Qin, J.J.; Yan, S.K.; Shen, Y.H.; Zhang, W.D. A new nor-sesquiterpene lactone from Ainsliaea fulvioides. ChemInform 2009, 40, 586–588. [Google Scholar] [CrossRef]
- Wu, J.; Tokuyama, S.; Nagai, K.; Yasuda, N.; Noguchi, K.; Matsumoto, T.; Hirai, H.; Kawagishi, H. Strophasterols A to D with an unprecedented steroid skeleton: From the mushroom Stropharia rugosoannulata. Angew. Chem. Int. Ed. 2012, 51, 10820–10822. [Google Scholar] [CrossRef]
- Choi, J.; Abe, N.; Kodani, S.; Masuda, K.; Koyama, T.; Yazawa, K.; Takahashi, M.; Kawagishi, H. Osteoclast-forming suppressing compounds from the medicinal mushroom Agrocybe chaxingu Huang (Agaricomycetideae). Int. J. Med. Mushrooms 2010, 12, 151–155. [Google Scholar] [CrossRef]
- Nieto, I.J.; Ávila, I.M. Determination of fatty acids and triterpenoid compounds from the fruiting body of Suillus luteus. Rev. Colomb. Quim 2008, 37, 297–304. [Google Scholar]
- Ting, Z.K.; Ying, Y.; Shao, H.J.; Du, X.X.; Wen, Y.; Feng, D.Y. Chemical constituents of fruiting bodies of Lactariuspi peratus. J. Shaanxi Norm. Univ. (Nat. Sci. Ed.) 2013, 41, 104–108. [Google Scholar]
- Liu, K.; Hu, H.G.; Wang, J.L.; Jia, Y.J.; Li, H.X.; Li, J. Chemical constituents from Phellinus robustus. Chin. Pharm. J. 2014, 49, 180–183. [Google Scholar]
- Ridwan, A.Y.; Matoba, R.; Wu, J.; Choi, J.; Hirai, H.; Kawagishi, H. A novel plant growth regulator from Pholiota lubrica. Tetrahedron Lett. 2018, 59, 2559–2561. [Google Scholar] [CrossRef]
- Regerat, F.; Pourrat, H. New sterol isolation, portensterol, from fruit bodies of Tricholoma portenstosum, Rhodopaxillus nudus and Clitocybe nebularis. Ann. Pharm. Fr. 1976, 34, 323–328. [Google Scholar] [PubMed]
- Ma, B.Y.; Zhao, C.A.; Han, L.; Hou, B.; Lu, Z. Anti-phytopathogenic fungi in aqueous extract from cortex dictamni. Agrochemicals 2015, 54, 69–72. [Google Scholar]
- Silva, A.P.D.; Rocha, R.; Silva, C.M.L.; Mira, L.; Duarte, M.F.; Florêncio, M.H. Antioxidants in medicinal plant extracts. A research study of the antioxidant capacity of Crataegus, Hamamelis and Hydrastis. Phytother. Res. 2000, 14, 612–616. [Google Scholar] [CrossRef]
- Liang, C.; Tsai, S.; Huang, S.; Liang, Z.; Mau, J. Taste quality and antioxidant properties of medicinal mushrooms Phellinus linteus and Sparassis crispa mycelia. Int. J. Med. Mushrooms 2010, 12, 141–150. [Google Scholar] [CrossRef]
- Tang, C.L.; Wang, L.; Liu, X.X.; Cheng, M.C.; Xiao, H.B. Chemical fingerprint and metabolic profile analysis of ethyl acetate fraction of Gastrodia elata by ultra performance liquid chromatography/quadrupole-time of flight mass spectrometry. J. Chromatogr. B 2016, 1011, 233–239. [Google Scholar] [CrossRef]
- Meng, Y.J.; Ling, Z.; Ze, J.D.; Ji, K.L. Two new metabolites from basidiomycete Sparassis crispa. Nat. Prod. 2009, 7, 1087–1089. [Google Scholar]
- Kodani, S.; Hayashi, K.; Tokuyama, S.; Hashimoto, M.; Kimura, T.; Dombo, M.; Kawagishi, H. Occurrence and identification of chalcones from the culinary-medicinal cauliflower mushroom Sparassis crispa (Wulf.) Fr. (Aphyllophoromycetideae). Int. J. Med. Mushrooms 2008, 10, 331–336. [Google Scholar] [CrossRef]
- Yim, H.S.; Akowuah, G.A.; Chye, F.Y.; Sia, C.M.; Okechukwu, P.N.; Ho, C.W. Identification of apigenin-7-glucoside and luteolin-7-glucoside in Pleurotus porrigens and Schizophyllum commune mushrooms by liquid chromatography-ion trap tandem mass spectrometry. Curr. Bioact. Compd. 2015, 11, 202–208. [Google Scholar] [CrossRef]
- König, S.; Romp, E.; Krauth, V.; Rühl, M.; Dörfer, M.; Liening, S.; Hofmann, B.; Häfner, A.; Steinhilber, D.; Karas, M.; et al. Melleolides from honey mushroom inhibit 5-lipoxygenase via CYS159. Cell Chem. Biol. 2019, 26, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, E.; Torres, F.R.; Martínez, M.; Pérez-Castorena, A.L. Sucrose Esters from the fruits of Physalis nicandroides var. attenuata. J. Nat. Prod. 2006, 69, 1511–1513. [Google Scholar] [CrossRef] [PubMed]
- Cho, D.; Seo, H.; Kim, K. Analysis of the volatile flavor compounds produced during the growth stages of the shiitake mushrooms (Lentinus edodes). Prev. Nutr. Food Sci. 2003, 8, 306–314. [Google Scholar] [CrossRef]
- Son, B.W.; Cho, Y.J.; Choi, J.S.; Lee, W.K.; Kim, D.; Choi, H.D.; Choi, J.S.; Jung, J.H.; Im, K.S.; Choi, W.C. New galactolipids from the marine bacillariophycean microalga Nitzschia sp. Nat. Prod. Lett. 2001, 15, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Naquvi, K.J.A.A. Two new aliphatic lactones from the fruits of Coriandrum sativum L. Org. Med. Chem. Lett. 2012, 2, 28. [Google Scholar] [CrossRef] [PubMed]
- Fo, E.R.; Fernandes, J.B.; Vieira, P.C.; Silva, M.F.D.G. Isolation of secoisolariciresinol diesters from stems of Simaba cuneata. Phytochemistry 1992, 31, 2115–2116. [Google Scholar] [CrossRef]
- Li, Q.; Hai, Y.; Shi, H.; Du, Y.G.; Xi, Y.; Guo, L.Y.; Zhu, D.H. Analysis of volatile aroma components of Pleurotus ostreatus and Lentinus edodes with gas chromatography-mass spectrometry. Chem. Bioeng. 2010, 27, 87–89. [Google Scholar]
- Pan, F.; Su, T.; Cai, S.; Wu, W. Fungal endophyte-derived Fritillaria unibracteata var. wabuensis: Diversity, antioxidant capacities in vitro and relations to phenolic, flavonoid or saponin compounds. Sci. Rep.-UK 2017, 7, 42008. [Google Scholar] [CrossRef] [PubMed]
- Spitzer, R.; Jain, A.N. Surflex-Dock: Docking benchmarks and real-world application. J. Comput. Aid. Mol. Des. 2012, 26, 687–699. [Google Scholar] [CrossRef] [PubMed]
- Yuki, H.; Kikuzato, K.; Koda, Y.; Mikuni, J.; Tomabechi, Y.; Kukimoto-Niino, M.; Tanaka, A.; Shirai, F.; Shirouzu, M.; Koyama, H.; et al. Activity cliff for 7-substituted pyrrolo-pyrimidine inhibitors of Hck explained in terms of predicted basicity of the amine nitrogen. Bioorg. Med. Chem. 2017, 25, 4259–4264. [Google Scholar] [CrossRef] [PubMed]
- Williams, N.K.; Lucet, I.S.; Peter, K.S.; Evan, I.; Jamie, R. Crystal structures of the Lyn protein tyrosine kinase domain in its Apo and inhibitor-bound state. J. Biol. Chem. 2009, 284, 284–291. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, T.; Matsubara, M.; Ishiguro, H.; Okita, K.; Tada, T. Structure of human Fyn kinase domain complexed with staurosporine. Biochem. Biophys. Res. Commun. 2006, 346, 840–844. [Google Scholar] [CrossRef] [PubMed]
- Barker, M.D.; Liddle, J.; Atkinson, F.L.; Wilson, D.M.; Dickson, M.C.; Ramirez-Molina, C.; Lewis, H.; Davis, R.P.; Somers, D.O.; Neu, M.; et al. Discovery of potent and selective spleen tyrosine kinase inhibitors for the topical treatment of inflammatory skin disease. Bioorg. Med. Chem. Lett. 2018, 28, 3458–3462. [Google Scholar] [CrossRef]
- Han, S.W.; Sun, L.; He, F.; Che, H.L. Anti-allergic activity of glycyrrhizic acid on IgE-mediated allergic reaction by regulation of allergy-related immune cells. Sci. Rep.-UK 2017, 7, 7222. [Google Scholar] [CrossRef]
- Passante, E.; Frankish, N. The RBL-2H3 cell line: Its provenance and suitability as a model for the mast cell. Inflamm. Res. 2009, 58, 737–745. [Google Scholar] [CrossRef]
Sample Availability: Samples of the reference compounds riboflavin, citric acid, ergosterol, fraxinellone, mannitol, sparoside A, linoleic acid, ainsliatone A and the S. crispa pileus are available from the authors. |

| Category | Compound Name | tR (min) | Formula | Exact Mass | Adduct Ion m/z | Mass Error (ppm) | Fragment Ion m/z |
|---|---|---|---|---|---|---|---|
| alkaloid | riboflavin (1) a | 3.97 | C17H20O6N4 | 376.1383 | 377.1459 [M + H]+ | 0.820 | 359(35.6) b, 341(20.3), 243(100), 99(16.7) |
| organic acid | citric acid (2) | 0.80 | C6H8O7 | 192.0270 | −191.0195 [M − H]- | −0.920 | 173(41.9), 129(35.0), 111(100), 85(31.8) |
| sesquiterpene | ainsliatone A (3) | 4.57 | C14H18O4 | 250.1200 | 251.1278 [M + H]+ 273.1095 [M + Na]+ | 0.257 −0.953 | 233(32.2), 215(65.0), 205(48.5), 197(7.3), 187(100),169(28.1), 159(20.5), 145(33.9) |
| sterol | ergosterol (4) | 18.48 | C28H44O | 396.3392 | 397.3463 [M + H]- | −0.610 | 379(100), 271(58.7), 253(31.2), 231(40.0), 213(53.1) |
| phthalide | fraxinellone (5) | 11.34 | C14H16O3 | 232.1094 | 233.1171 [M + H]+ | −0.390 | 215(100), 197(9.9), 187(73.5), 169(13.9), 159(18.0), 95(22.3) |
| other | mannitol (6) | 0.76 | C6H14O6 | 182.0790 | 183.0864 [M + H]+ 205.0683 [M + Na]+ | 0.521 0.198 | 165(70.0), 147(44.1), 129(32.2), 111(100) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Liu, J.; Zhong, X.; Li, J.; Wang, X.; Ji, L.; Shang, X. Rapid Characterization of Chemical Components in Edible Mushroom Sparassis crispa by UPLC-Orbitrap MS Analysis and Potential Inhibitory Effects on Allergic Rhinitis. Molecules 2019, 24, 3014. https://doi.org/10.3390/molecules24163014
Wang Z, Liu J, Zhong X, Li J, Wang X, Ji L, Shang X. Rapid Characterization of Chemical Components in Edible Mushroom Sparassis crispa by UPLC-Orbitrap MS Analysis and Potential Inhibitory Effects on Allergic Rhinitis. Molecules. 2019; 24(16):3014. https://doi.org/10.3390/molecules24163014
Chicago/Turabian StyleWang, Zhixin, Jingyu Liu, Xiangjian Zhong, Jinjie Li, Xin Wang, Linlin Ji, and Xiaoya Shang. 2019. "Rapid Characterization of Chemical Components in Edible Mushroom Sparassis crispa by UPLC-Orbitrap MS Analysis and Potential Inhibitory Effects on Allergic Rhinitis" Molecules 24, no. 16: 3014. https://doi.org/10.3390/molecules24163014
APA StyleWang, Z., Liu, J., Zhong, X., Li, J., Wang, X., Ji, L., & Shang, X. (2019). Rapid Characterization of Chemical Components in Edible Mushroom Sparassis crispa by UPLC-Orbitrap MS Analysis and Potential Inhibitory Effects on Allergic Rhinitis. Molecules, 24(16), 3014. https://doi.org/10.3390/molecules24163014





