Abstract
Natural products, including traditional herbal medicine (THM), are known to exert their therapeutic effects by acting on multiple targets, so researchers have employed network pharmacology methods to decipher the potential mechanisms of THM. To conduct THM-network pharmacology (THM-NP) studies, researchers have employed different tools and databases for constructing and analyzing herb–compound–target networks. In this study, we attempted to capture the methodological trends in THM-NP research. We identified the tools and databases employed to conduct THM-NP studies and visualized their combinatorial patterns. We also constructed co-author and affiliation networks to further understand how the methodologies are employed among researchers. The results showed that the number of THM-NP studies and employed databases/tools have been dramatically increased in the last decade, and there are characteristic patterns in combining methods of each analysis step in THM-NP studies. Overall, the Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP) was the most frequently employed network pharmacology database in THM-NP studies. Among the processes involved in THM-NP research, the methodology for constructing a compound–target network has shown the greatest change over time. In summary, our analysis describes comprehensive methodological trends and current ideas in research design for network pharmacology researchers.
1. Introduction
Traditional herbal medicine (THM) has maintained the health of Asian people for thousands of years and built a unique medical system based on empirically accumulated knowledge. Billions of people around the world are taking THM daily, and the drug development field considers THM to be a source of inspiration [1,2]. The research indicates that THM’s therapeutic effects may involve various biomolecules [3]. However, due to the complexity of THM and the limitations of experimental applications, specific mechanisms of action have been fully elucidated for only a few THMs [4]. This is a major obstacle to THM’s modernization and wider application to modern healthcare.
Network pharmacology has emerged as a promising approach to accelerate drug development and elucidate the mechanisms of action of multiple target components [5]. It understands disease as a perturbation of interconnected complex biological networks and identifies the mechanisms of drug action by network topology [6,7]. The conceptual elements of network pharmacology were derived from systems biology, which can address both the connectivity and the interdependence of individual components [8]. The core idea of network pharmacology is well suited for analyzing the multi-targeted agents, so network pharmacology methods may be appropriate for identifying the complex mechanisms of THM.
In the last decade, researchers have employed network pharmacology methods to elucidate the potential targets and toxicity of THMs [9]. THM-network pharmacology (THM-NP) studies are conducted by constructing an herb–compound–target (H-C-T) network by integrating the herbal constituent data and drug-target interactions (DTIs) information. Then, the target network is analyzed to interpret related biological functions, pathways, and diseases (Figure 1). Since there are no gold standard methods for THM-NP studies yet, researchers have developed and applied various tools and databases for each step.
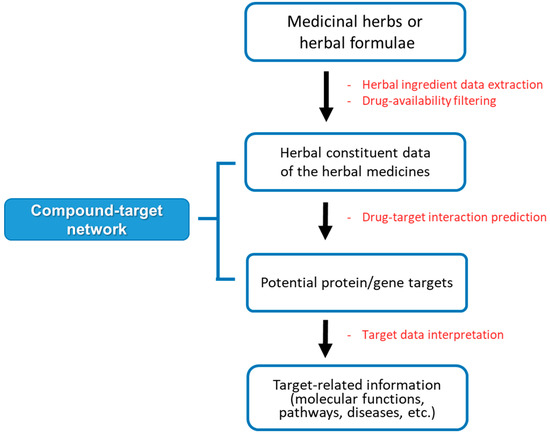
Figure 1.
The general framework of network pharmacology analysis of herbal medicine.
Several studies have described THM-NP researches by summarizing network pharmacology databases for THM and illustrating several representative applications [10,11,12,13,14,15]. Although these studies contributed to a better understanding of THM-NP studies, they were limited in providing quantitative information on the frequencies, variations, and combinatorial patterns of the employed methods in THM-NP studies. In this study, we systematically attempted to capture the methodological trends of the THM-NP research field. We identified the tools and databases employed to conduct THM-NP studies and visualized their frequency, variation, and combinatorial pattern in a step-by-step manner. The THM-NP studies were identified by searching PubMed and then preprocessed. We also constructed and analyzed the co-author and affiliation networks to identify how the diverse methods for THM-NP studies are employed and shared among researchers in the field. We believe that analyzing the methodological trends will provide a comprehensive understanding and valuable insights into the THM-NP research field.
2. Materials and Methods
2.1. Search Strategy
The literature search was performed in PubMed (https://www.ncbi.nlm.nih.gov/pubmed/) from January 2000 to December 2018. The search language was restricted to English. The search terms used were NP-related terms (“network pharmacology” OR “network analysis” OR “system-level” OR “systems-level” OR “systems pharmacology” OR “systems biology” OR “bioinformatics”) in [title] AND THM-related terms (“oriental medicine” OR “traditional medicine” OR “traditional Asian medicine” OR “Chinese medicine” OR “Kampo medicine” OR “Korean medicine”) in [title/abstract]. The search range of THM-related terms was extended to [title/abstract] since the titles of THM studies generally contain only the name of herbs or herbal formulae that are difficult to search.
2.2. Inclusion Criteria
We considered a THM-NP study as the original article that analyzed a THM’s mode of action through the construction of a compound-target network. Full-text articles from the literature search were checked to determine their eligibility. There was no restriction regarding in vivo, in vitro, and in silico studies. THM was considered as (1) extract(s) from a single herb; (2) preparation(s) containing multiple herbs; (3) proprietary herbal product(s); and (4) molecule(s) derived from a single herb.
2.3. Study Selection and Data Extraction
Two authors (W.Y. Lee and C.E. Kim) independently examined titles, abstracts, and journals to select eligible THM-NP studies. When articles were duplicated, only the most recent information was included. Then the full text of potentially relevant studies was retrieved. Two authors (W.Y. Lee and C.E. Kim) independently examined the full-text records to determine which studies met the inclusion criteria. Disagreements about the study selection were resolved by rechecking whether the studies met our criteria for inclusion.
Authors extracted the following data from the included THM-NP studies: authors, affiliations, publication years, tools, and databases. Synonyms for tools and databases were merged and counted under a single keyword. DTpre and SysDT [16] were considered to be the same method as Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP, http://lsp.nwu.edu.cn/tcmsp.php) [17] since these methods were originally developed and implemented in TCMSP.
2.4. Categorizing Drug-Target Interaction Methods
To capture the trends in the methods for constructing compound-target networks, we categorized DTI methods into four groups by their hypothesis and which information was used: the chemogenomic approach, docking simulation approach, ligand-based approach, and others [18,19,20]. The chemogenomic approach predicts potential compound–target pairs similar to validated compound-target pairs. This method is based on the assumption that a compound–target pair is likely to interact with high similarity to a validated compound-target interaction in terms of chemo–physical properties [21]. The docking simulation approach predicts the binding conformation of small-molecule ligands to the appropriate binding site of the target using 3D structural information on the compounds and protein targets [22]. The key hypothesis of this approach is that compounds with a high binding affinity at the binding site are likely to interact with the target [23]. The ligand-based approach predicts interactions by comparing a new ligand to known proteins’ ligands based on the hypothesis that similar molecules usually bind to similar proteins [24]. DTI methods that do not belong to the above categories were assigned to the category “others”, such as data mining techniques, high-throughput screening, and databases that integrate drug-target interaction information from heterogeneous sources.
2.5. Construction of the Co-Author Network and Affiliation Network
The author network and affiliation network were constructed to identify the methodological characteristics of corresponding authors and affiliations. The nodes in each network represent authors or affiliations, and the edges represent co-occurrences of authors or affiliations in THM-NP studies. The frequencies of employed DTI and drug availability methods were counted for each corresponding author or affiliation. These methodologies were mapped to the author network and the affiliation network. Cytoscape 3.7.1 (http://www.cytoscape.org/) was used to visualize the networks [25].
3. Results
3.1. Description of the Search
We initially found 233 potentially relevant articles from PubMed. The search was conducted using combined keywords consisting of THM-related terms and NP-related terms. Another 15 potentially relevant articles were included by searching references in other THM-NP studies or review articles. Titles, abstracts, and journal names were screened, and 167 studies were considered potentially eligible for inclusion. Of these, 20 articles were excluded after screening the full texts. Finally, a total of 147 THM-NP studies were included in our study (Figure 2). The included THM-NP studies are listed in Supplementary Table S1.

Figure 2.
The flowchart of the study selection process.
We identified the number of published THM-NP studies over time, along with the affiliations involved in the studies. THM-NP studies that met our criteria for inclusion have appeared since 2011. In 2011, only three affiliations published two THM-NP studies, but in 2018, the number of affiliations and studies increased to 83 and 52, respectively (Figure 3). The increased number of papers and of affiliations involved indicates that the THM-NP research fields have continuously grown.
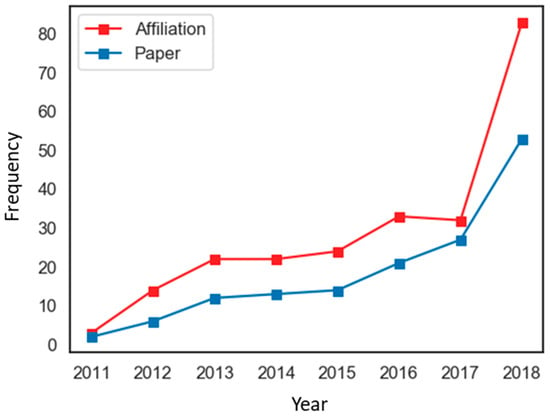
Figure 3.
Annual publication trends of traditional herbal medicine-network pharmacology (THM-NP) studies.
3.2. Methodological Trends in Constructing the Herb-Compound Network
We next investigated the trends in employed methods in THM-NP studies. Commonly used databases and tools are described in Table 1 (see Supplementary Table S2 for complete lists). The construction of the herb-compound network is the first step of a THM-NP study. Among the databases for herbal medicines, TCMSP was most commonly used to construct an herb-compound network. Additionally, some THM-NP researchers used their own experimental results (e.g., Ultra Performance Liquid Chromatography (UPLC) or High-performance liquid chromatography) to identify ingredients of the herbal medicines in their studies (Figure 4A).

Table 1.
The public databases related to traditional herbal medicine-network pharmacology (THM-NP) studies.
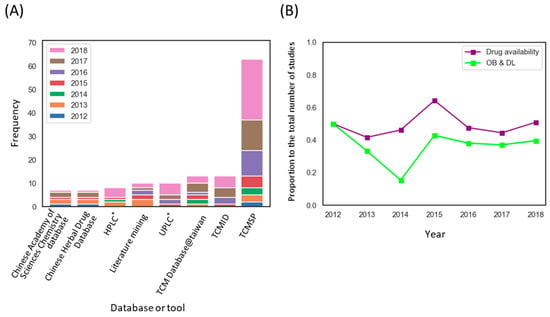
Figure 4.
Methodological trends in the construction of the herb–compound network. (A) Frequency of databases and tools used to identify constituents of herbal medicine. * Analytical techniques to identify the ingredients of herbal medicines; Ultra Performance Liquid Chromatography (UPLC), High-performance liquid chromatography (HPLC) (B) The application rate of the drug availability method by year. Note that Obioavail (OB) and drug-likeness (DL) are among the most commonly used drug availability assessment methods in THM-NP studies. THM-NP studies in 2011 were excluded from the visualization due to the low frequency (n = 2). TCMSP, Traditional Chinese Medicine Systems Pharmacology Database; TCMID, Traditional Chinese Medicine Integrated Database.
Because information on the absorption, distribution, metabolism, and excretion (ADME) properties of herbal medicines in humans are lacking, researchers have employed evaluation methods or machine learning tools to predict those properties. We counted the number of THM-NP studies that evaluated the drug availability of herbal ingredients. We found that approximately half of (72/147, 49.0%) THM-NP studies evaluated the drug availability of herbal ingredients, and the majority of the studies (54/72, 75.0%) employed Obioavail and drug-likeness in combination (Figure 4B). Obioavail is an in silico model that predicts the fraction of an administered dose of a drug that reaches the systemic circulation unchanged [39]. Drug-likeness measures the structural similarity between herbal ingredients and the drugs in the Drugbank database (http://www.drugbank.ca/) using the Tanimoto coefficient [40]. They are applied to screen ADME-favorable compounds and pharmacologically suitable compounds in herbal medicines, respectively.
3.3. Methodological Trends for Constructing Compound-Target Networks
We next attempted to determine the frequency of each DTI method for constructing compound–target (C-T) networks (Note that some of the THM-NP studies combined several methods to identify DTIs. Therefore, the total frequency of the DTI method is greater than the total number of THM-NP studies). The results showed that TCMSP (47/222, 21.1%) and molecular docking (44/222, 19.8%) were the most frequently used. In addition, experimental methods, such as microarrays, were also applied (Figure 5A). It is noteworthy that DTI methods of TCMSP have existed for less than 10 years since its development but have been used most frequently in THM-NP studies [16]. More than one-third of THM-NP studies (54/147, 36.7%) combined several DTI methods for constructing C-T networks, and most of them included TCMSP (e.g., TCMSP-molecular docking and TCMSP-STITCH) (Figure 5B).
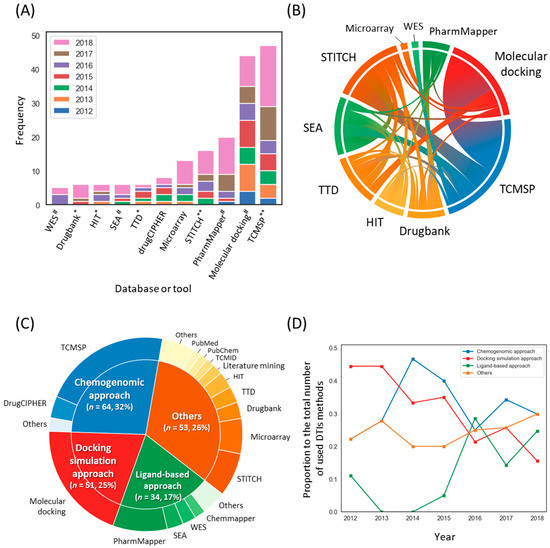
Figure 5.
Methodological trends in the construction of the compound–target network. (A) The frequencies of databases and tools used to identify targets of herbal ingredients. * databases that provide validated drug-target interactions (DTIs); ** databases that provide validated databases that provide both validated and predicted DTIs; # Computational tools to predict DTIs. (B) A co-occurrence pattern of the DTI method. (C) Categories of DTI methods and their composition. The outer circle and inner circle represent the DTI methods and their categories, respectively. (D) The annual rate of the groups of DTI methods. Note that THM-NP studies in 2011 were excluded from the visualization due to their low frequency (n = 2). TCMSP, Traditional Chinese Medicine Systems Pharmacology Database; TTD, Therapeutic Target Database; SEA, Similarity Ensemble Approach; HIT, Herb Ingredients’ Targets; WES, Weighted Ensemble Similarity.
We categorized the methods into four groups: the chemogenomic approach, docking simulation approach, ligand-based approach, and others (Figure 5C, see Materials and Methods for details). To identify trends in DTI methods, we counted the frequency of each DTI group each year (Figure 5D). In the early stage, approximately half of THM-NP studies (4/9, 44.4% and 8/18, 44.4% in 2012 and 2013, respectively) employed molecular docking simulation, but the proportion of molecular docking simulations decreased gradually and was the lowest (12/77, 15.9%) in 2018.
3.4. Methodological Trends for Target Interpretation
We identified the frequency of biomedical databases employed to analyze the biological processes, pathways, and diseases from the targets of herbal medicines (Figure 6). Most THM-NP studies (94/96, 98.0%) employed single databases to analyze biological functions and pathways, such as Gene Ontology (GO, http://geneontology.org/) [35] for biological processes or the Kyoto Encyclopedia of Genes and Genomes database (KEGG, https://www.genome.jp/kegg/) [41] for pathways. On the other hand, more than half of the studies (44/76, 57.9%) integrated several databases to analyze target-related diseases, such as the Therapeutic Target Database (TTD, http://bidd.nus.edu.sg/group/cjttd/) [30], Online Mendelian Inheritance in Man (OMIM, https://www.omim.org/) [36], and Drugbank [42].
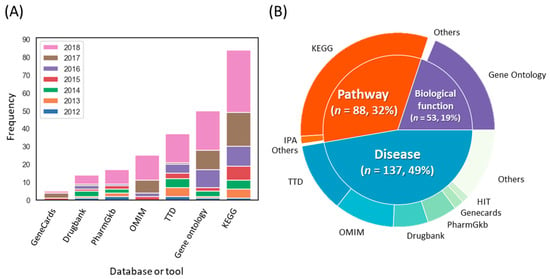
Figure 6.
Trends in the biomedical databases used for target interpretation. (A) The frequencies of databases used for target interpretation. (B) Categories of databases and their composition. The outer circle and inner circle represent the databases used for target interpretation and their categories, respectively. Note that THM-NP studies in 2011 were excluded from the visualization due to their low frequency (n = 2). KEGG, Kyoto Encyclopedia of Genes and Genomes; TTD, Therapeutic Target Database; OMIM, Online Mendelian Inheritance in Man; IPA, Ingenuity Pathway Analysis.
3.5. Combinatorial Patterns in Methodologies of THM-NP Studies
We identified the combinatorial patterns of each step in THM-NP studies by a Sankey diagram-like representation (Figure 7). The Sankey diagram is a visualization tool used to depict quantitative information about flows from one set to another within a network [43]. The nodes in each layer (vertical lines) represent the methods of herb–compound (H-C) network construction, compound–target (C-T) network construction, and target interpretation, respectively. The edges (connected lines) between layers indicate that these methods are used together in the same THM-NP studies.

Figure 7.
The combinatorial pattern of tools and databases employed in THM-NP studies. Each layer represents the process of network pharmacology analysis of herbal medicines, and the components of each layer represent the employed tools and databases. A connecting line between components indicates that the connected tools and databases are used together in the same THM-NP studies. The thickness of each connecting line indicates the frequency with which the two methodologies are used together in the THM-NP studies. TCMSP, Traditional Chinese Medicine Systems Pharmacology Database; TCMID, Traditional Chinese Medicine Integrated Database; HIT, Herb Ingredients’ Targets; KEGG, Kyoto Encyclopedia of Genes and Genomes; TTD, Therapeutic Target Database; OMIM, Online Mendelian Inheritance in Man.
The Sankey diagram-like representation shows the diversity of databases and tools used in THM-NP studies and their combination patterns (Figure 7). We found that the nodes in the first layer (H-C network construction) tend to be connected to specific nodes in the second layer (C-T network construction), which indicates that the combinatorial pattern between the first and second layer is biased by the methods for H-C network construction. For example, TCMSP in the first layer is mainly connected to TCMSP and molecular docking in the second layer, and Traditional Chinese Medicine Integrated Database (TCMID), UPLC, and literature mining in the first layer are not linked to molecular docking in the second layer. On the other hand, the nodes in the second layer tended to be evenly connected to the nodes in the third layer (target interpretation), which indicates that the combinatorial pattern between the second layer and third layer are relatively independent of the methods for C-T network construction.
3.6. Co-Author Network and Affiliation Network
To further understand how the methodologies of THM-NP studies are employed among researchers, we constructed a co-author network and an affiliation network that were mapped with drug availability and DTI methods. The nodes in each network denote the author and affiliation, and the edges indicate that two of them appear on the same paper. The methods of DTI and drug-availability used by the corresponding author and affiliation are represented by the pie chart and the outline, respectively.
In the author network, Yonghua Wang (n = 18) and Shao Li (n = 8) appeared most frequently as the corresponding author (Figure 8). More than a third of corresponding authors (52/147) combined DTI methods, such as the chemogenomic approach, docking simulation approach, and ligand-based approach. Approximately half of the corresponding authors (69/147) employed evaluation tools to screen for compounds with favorable pharmacokinetic properties, and most of them (50/69) used Obioavail and drug-likeness in combination.

Figure 8.
The co-author network of THM-NP studies. Circles represent corresponding authors, and squares represent non-corresponding authors. The size of the circles and squares reflect the number of occurrences in the THM-NP studies. Nodes that appeared fewer than three times were removed. The box to the right of the network represents the index for the pie chart and the outline of the circle.
We also constructed and visualized the affiliation network (Supplementary Figure S1). Northwest A&F University (n = 22) and China Academy of Chinese Medical Sciences (n = 17) appeared most frequently. Most affiliations combined various DTI methods (68/143) and employed drug-availability methods (86/143).
4. Discussion
In this study, we successfully identified the complex methodological trends of THM-NP research fields by analyzing the frequency of the employed methods in THM-NP studies over time and visualizing the combinatorial patterns between them. Our results showed that the number of THM-NP studies and employed databases/tools have been dramatically increased in the last decade. We also found characteristic patterns exist in combining methods of each analysis step in THM-NP studies. Finally, we showed how the diverse methods for THM-NP studies are employed and shared among researchers in the field by analyzing the co-authorship and affiliation networks.
Among the network pharmacology databases, TCMSP was the most frequently employed database for constructing herb-compound-target networks. This database was developed in 2014 and has been predominantly employed among THM-NP studies [17]. TCMSP provides a network pharmacological analysis of 499 medicinal herbs registered in the Chinese pharmacopeia along with information on ADME properties, such as bioavailability, drug-likeness, and P450, in a one-step manner. Recently, other network pharmacology databases, such as BATMAN-TCM (http://bionet.ncpsb.org/batman-tcm) and TCM-Mesh (http://mesh.tcm.microbioinformatics.org/), were developed [44,45]. They are expected to further facilitate THM-NP research fields by providing network pharmacological analysis for more than 5000 medicinal herbs.
Among the processes used in THM-NP research, the methodology for constructing a C-T network has shown the greatest change over time. In the early stages of THM-NP research, DTI methods for identifying targets of herbal ingredients relied on molecular docking simulations, which require high computational resources (Figure 5D). With the advancement of DTI prediction methods, several methodologies have been applied to THM-NP research fields that can efficiently identify the multiple targets of multiple ingredients in herbal medicines. First, the development and application of machine learning techniques and network-based methods enabled large-scale prediction of the targets of herbal medicines in terms of efficient computational costs [17,44,46,47]. Second, increased computational power made it possible to comprehensively explore potential targets of the compounds using the pharmacophore model [48]. Last, the development of databases that integrate disparate data sources provides comprehensive and high-quality information on DTIs [49]. Furthermore, recently developed DTI prediction models based on deep learning showed higher performance than other state-of-the-art models [50,51]. Such innovation in the machine learning field is expected to facilitate the development of the THM-NP research field.
To conduct THM-NP research, various tools and databases are combined in each phase of a study (Figure 7). We found that the methods for H-C network construction tend to be linked with specific methods for C-T network construction. This result indicates that there might be a preferred combinatorial pattern when choosing the methods for constructing H-C-T network. On the other hand, the combinatorial patterns between the methods for C-T network construction and target interpretation are relatively independent when compared to the previous step. This result indicates that the databases used for target interpretation tend to be chosen for the purpose of the study, while each method was preferred by different researchers in the previous steps. Further studies are needed to evaluate the reliability of network pharmacological analysis by evaluating the consistency between predicted results according to the methodologies of THM-NP studies.
There are some limitations to our study that should be noted. First, we identified potentially relevant articles using combined keywords consisting of THM-related terms and NP-related terms. Although we carefully selected search terms, we cannot guarantee that our search strategy can fully identify THM-NP studies. Second, we found potentially relevant articles only in PubMed. It is one of the largest electronic database in the world. However, there are other databases which may include other potential THM-NP studies, such as Embase, China Knowledge Resource Integrated Database (CNKI), Research Information Sharing Service (RISS), and Japan Science Technology Information Aggregator (J-stage). Last, we limited the search range of our study to English literature, which might have introduced some bias. In spite of these limitations, our results will help to improve the understanding of the methodological trends of the THM-NP research fields.
5. Conclusions
In conclusion, we investigated the methodological trends in THM-NP studies. Our results provide researchers with the current status of which methodologies are used in THM-NP studies and how they are applied.
Supplementary Materials
The following are available online at https://www.mdpi.com/2218-273X/9/8/362/s1. Supplementary Figure S1. The affiliation network of THM-NP studies; Supplementary Table S1. Selected THM-NP studies; Supplementary Table S2. Providing information and their types of methods employed in THM-NP studies.
Author Contributions
Conceptualization, Y.-S.K. and C.-E.K.; Data curation, W.-Y.L.; Formal analysis, W.-Y.L. and C.-Y.L.; Visualization, W.-Y.L.; Writing—original draft, W.-Y.L.; Writing—review and editing, C.-Y.L., Y.-S.K., and C.-E.K.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP; Ministry of Science, ICT & Future Planning) (No. 2017-0129). This work was also supported by the Gachon University research fund of 2019 (GCU-2019-0025).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Kong, D.X.; Li, X.J.; Zhang, H.Y. Where is the hope for drug discovery? Let history tell the future. Drug Discov. Today 2009, 14, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Verpoorte, R.; Crommelin, D.; Danhof, M.; Gilissen, L.J.; Schuitmaker, H.; van der Greef, J.; Witkamp, R.F. Commentary: “A systems view on the future of medicine: Inspiration from Chinese medicine?”. J. Ethnopharmacol. 2009, 121, 479–481. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Wu, M.; Wang, H.; Li, S.; Wang, X.; Li, Y.; Wang, D.; Li, S. Network Pharmacology to Unveil the Biological Basis of Health-Strengthening Herbal Medicine in Cancer Treatment. Cancers (Basel) 2018, 10, 461. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.H.; Zheng, C.J.; Han, L.Y.; Xie, B.; Jia, J.; Cao, Z.W.; Li, Y.X.; Chen, Y.Z. Synergistic therapeutic actions of herbal ingredients and their mechanisms from molecular interaction and network perspectives. Drug Discov. Today 2009, 14, 579–588. [Google Scholar] [CrossRef] [PubMed]
- Berg, E.L. Systems biology in drug discovery and development. Drug Discov. Today 2014, 19, 113–125. [Google Scholar] [CrossRef] [PubMed]
- Barabási, A.-L.; Gulbahce, N.; Loscalzo, J. Network medicine: A network-based approach to human disease. Nat. Rev. Genet. 2011, 12, 56–68. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, A.L. Network pharmacology: The next paradigm in drug discovery. Nat. Chem. Biol. 2008, 4, 682–690. [Google Scholar] [CrossRef]
- Van der Greef, J. Perspective: All systems go. Nature 2011, 480, S87. [Google Scholar] [CrossRef]
- Li, S.; Fan, T.-P.; Jia, W.; Lu, A.; Zhang, W. Network Pharmacology in Traditional Chinese Medicine. Evid. Based Complement. Altern. Med. 2014, 2014, 138460. [Google Scholar] [CrossRef]
- Li, S.; Zhang, B. Traditional Chinese medicine network pharmacology: Theory, methodology and application. Chin. J. Nat. Med. 2013, 11, 110–120. [Google Scholar] [CrossRef]
- Zhang, R.; Zhu, X.; Bai, H.; Ning, K. Network Pharmacology Databases for Traditional Chinese Medicine: Review and Assessment. Front. Pharmacol. 2019, 10, 123. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Chen, J.-L.; Xu, L.-W.; Ji, G. Navigating Traditional Chinese Medicine Network Pharmacology and Computational Tools. Evid. Based Complement. Altern. Med. 2013, 2013, 731969. [Google Scholar] [CrossRef] [PubMed]
- Kibble, M.; Saarinen, N.; Tang, J.; Wennerberg, K.; Mäkelä, S.; Aittokallio, T. Network pharmacology applications to map the unexplored target space and therapeutic potential of natural products. Nat. Prod. Rep. 2015, 32, 1249–1266. [Google Scholar] [CrossRef] [PubMed]
- Hao, D.C.; Xiao, P.G. Network pharmacology: A rosetta stone for traditional chinese medicine. Drug Dev. Res. 2014, 75, 299–312. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ai, N.; Keys, A.; Fan, X.; Chen, M. Network Pharmacology for Traditional Chinese Medicine Research: Methodologies and Applications. Chin. Herb. Med. 2015, 7, 18–26. [Google Scholar] [CrossRef]
- Yu, H.; Chen, J.; Xu, X.; Li, Y.; Zhao, H.; Fang, Y.; Li, X.; Zhou, W.; Wang, W.; Wang, Y. A systematic prediction of multiple drug-target interactions from chemical, genomic, and pharmacological data. PLoS ONE 2012, 7, e37608. [Google Scholar] [CrossRef] [PubMed]
- Ru, J.; Li, P.; Wang, J.; Zhou, W.; Li, B.; Huang, C.; Li, P.; Guo, Z.; Tao, W.; Yang, Y.; et al. TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. J. Cheminform. 2014, 6, 13. [Google Scholar] [CrossRef]
- Katsila, T.; Spyroulias, G.A.; Patrinos, G.P.; Matsoukas, M.T. Computational approaches in target identification and drug discovery. Comput. Struct. Biotechnol. J. 2016, 14, 177–184. [Google Scholar] [CrossRef]
- Macalino, S.J.Y.; Gosu, V.; Hong, S.; Choi, S. Role of computer-aided drug design in modern drug discovery. Arch. Pharm. Res. 2015, 38, 1686–1701. [Google Scholar] [CrossRef]
- Yang, S.Y. Pharmacophore modeling and applications in drug discovery: Challenges and recent advances. Drug Discov. Today 2010, 15, 444–450. [Google Scholar] [CrossRef]
- Ding, H.; Takigawa, I.; Mamitsuka, H.; Zhu, S. Similarity-basedmachine learning methods for predicting drug-target interactions: A brief review. Brief. Bioinform. 2013, 15, 734–747. [Google Scholar] [CrossRef] [PubMed]
- Lengauer, T.; Rarey, M. Computational methods for biomolecular docking. Curr. Opin. Struct. Biol. 1996, 6, 402–406. [Google Scholar] [CrossRef]
- Cherfils, J.; Duquerroy, S.; Janin, J. Protein-protein recognition analyzed by docking simulation. Proteins Struct. Funct. Bioinform. 1991, 11, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Vidal, D.; Garcia-Serna, R.; Mestres, J. Ligand-Based Approaches to In Silico Pharmacology. Methods Mol. Biol. 2010, 672, 489–502. [Google Scholar]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Xue, R.; Fang, Z.; Zhang, M.; Yi, Z.; Wen, C.; Shi, T. TCMID: Traditional Chinese medicine integrative database for herb molecular mechanism analysis. Nucleic Acids Res. 2013, 41, 1089–1095. [Google Scholar] [CrossRef]
- Chen, C.Y.C. TCM Database@Taiwan: The world’s largest traditional Chinese medicine database for drug screening In Silico. PLoS ONE 2011, 6, e15939. [Google Scholar] [CrossRef]
- Liu, X.; Ouyang, S.; Yu, B.; Liu, Y.; Huang, K.; Gong, J.; Zheng, S.; Li, Z.; Li, H.; Jiang, H. PharmMapper server: A web server for potential drug target identification using pharmacophore mapping approach. Nucleic Acids Res. 2010, 38, 5–7. [Google Scholar] [CrossRef]
- Kuhn, M.; von Mering, C.; Campillos, M.; Jensen, L.J.; Bork, P. STITCH: Interaction networks of chemicals and proteins. Nucleic Acids Res. 2008, 36, 684–688. [Google Scholar] [CrossRef]
- Chen, X.; Ji, Z.L.; Chen, Y.Z. TTD: Therapeutic Target Database. Nucleic Acids Res. 2002, 30, 412–415. [Google Scholar] [CrossRef]
- Keiser, M.J.; Roth, B.L.; Armbruster, B.N.; Ernsberger, P.; Irwin, J.J.; Shoichet, B.K. Relating protein pharmacology by ligand chemistry. Nat. Biotechnol. 2007, 25, 197–206. [Google Scholar] [CrossRef]
- Ye, H.; Ye, L.; Kang, H.; Zhang, D.; Tao, L.; Tang, K.; Liu, X.; Zhu, R.; Liu, Q.; Chen, Y.Z.; et al. HIT: Linking herbal active ingredients to targets. Nucleic Acids Res. 2011, 39, 1055–1059. [Google Scholar] [CrossRef]
- Wishart, D.S. DrugBank: A comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2005, 34, D668–D672. [Google Scholar] [CrossRef]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999, 28, 27–30. [Google Scholar] [CrossRef]
- Harris, M.A.; Clark, J.; Ireland, A.; Lomax, J.; Ashburner, M.; Foulger, R.; Eilbeck, K.; Lewis, S.; Marshall, B.; Mungall, C.; et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004, 32, D258–D261. [Google Scholar]
- Hamosh, A.; Scott, A.F.; Amberger, J.S.; Bocchini, C.A.; McKusick, V.A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005, 33, D514–D517. [Google Scholar] [CrossRef]
- Hewett, M.; Oliver, D.E.; Rubin, D.L.; Easton, K.L.; Stuart, J.M.; Altman, R.B.; Klein, T.E. PharmGKB: The pharmacogenetics knowledge base. Nucleic Acids Res. 2002, 30, 163–165. [Google Scholar] [CrossRef]
- Safran, M.; Solomon, I.; Shmueli, O.; Lapidot, M.; Shen-Orr, S.; Adato, A.; Ben-Dor, U.; Esterman, N.; Rosen, N.; Peter, I.; et al. GeneCardsTM 2002: Towards a complete, object-oriented, human gene compendium. Bioinformatics 2002, 18, 1542–1543. [Google Scholar] [CrossRef]
- Wang, X.; Xu, X.; Tao, W.; Li, Y.; Wang, Y.; Yang, L. A Systems Biology Approach to Uncovering Pharmacological Synergy in Herbal Medicines with Applications to Cardiovascular Disease. Evid. Based Complement. Altern. Med. 2012, 2012, 519031. [Google Scholar] [CrossRef]
- Vistoli, G.; Pedretti, A.; Testa, B. Assessing drug-likeness—What are we missing? Drug Discov. Today 2008, 13, 285–294. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Soundararajan, K.; Ho, H.K.; Su, B. Sankey diagram framework for energy and exergy flows. Appl. Energy 2014, 136, 1035–1042. [Google Scholar] [CrossRef]
- Liu, Z.; Guo, F.; Wang, Y.; Li, C.; Zhang, X.; Li, H.; Diao, L.; Gu, J.; Wang, W.; Li, D.; et al. BATMAN-TCM: A Bioinformatics Analysis Tool for Molecular mechANism of Traditional Chinese Medicine. Sci. Rep. 2016, 6, 21146. [Google Scholar] [CrossRef]
- Zhang, R.Z.; Yu, S.J.; Bai, H.; Ning, K. TCM-Mesh: The database and analytical system for network pharmacology analysis for TCM preparations. Sci. Rep. 2017, 7, 2821. [Google Scholar] [CrossRef]
- Zhao, S.; Li, S. Network-based relating pharmacological and genomic spaces for drug target identification. PLoS ONE 2010, 5, e11764. [Google Scholar] [CrossRef]
- Wu, Z.; Lu, W.; Wu, D.; Luo, A.; Bian, H.; Li, J.; Li, W.; Liu, G.; Huang, J.; Cheng, F.; et al. In silico prediction of chemical mechanism of action via an improved network-based inference method. Br. J. Pharmacol. 2016, 173, 3372–3385. [Google Scholar] [CrossRef]
- Wang, X.; Shen, Y.; Wang, S.; Li, S.; Zhang, W.; Liu, X.; Lai, L.; Pei, J.; Li, H. PharmMapper 2017 update: A web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res. 2017, 45, W356–W360. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Santos, A.; Von Mering, C.; Jensen, L.J.; Bork, P.; Kuhn, M. STITCH 5: Augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016, 44, D380–D384. [Google Scholar] [CrossRef]
- Bahi, M.; Batouche, M. Deep semi-supervised learning for DTI prediction using large datasets and H2O-spark platform. In Proceedings of the 2018 International Conference on Intelligent Systems and Computer Vision (ISCV), Fez, Morocco, 2–4 April 2018; pp. 1–7. [Google Scholar]
- Wen, M.; Zhang, Z.; Niu, S.; Sha, H.; Yang, R.; Yun, Y.; Lu, H. Deep-Learning-Based Drug-Target Interaction Prediction. J. Proteome Res. 2017, 16, 1401–1409. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).