Abstract
The recycling of metabolic products is a major way to reduce the energy cost of de novo biosynthesis. The salvage pathways involved not only regain a metabolic product but also generate additional molecules that might serve specific physiological, developmental and/or defensive functions. The isoprenoid pathway is a perfect example of a fine-regulated biosynthetic pathway, by virtue of the large number of molecules with different functions that must be synthesized simultaneously. Additionally, isoprenoid salvage pathways have been characterized. Thus, to produce isoprenoid precursors such as farnesyl diphosphate or phytyl diphosphate, short-chain isoprenols recovered from end-chain metabolites are phosphorylated. In the first instance, the so-called FPP-salvage machinery recycles farnesyl diphosphate from proteolyzed farnesylated proteins. In a second example, phytyl diphosphate is recycled from degraded chlorophyll, to be used for the biosynthesis of vitamin E. Both compounds are recovered as alcohols and require two phosphorylation events to be reactivated and reintegrated into the isoprenoid biosynthetic pathway. This review covers current knowledge of isoprenol biosynthesis, metabolism and function, as well as potential benefits of recycling pathways for plants, with a particular focus on stress responses.
1. Generalities
In response to intrinsic and environmental signals, the entire metabolism must be constantly readjusted to ensure the development and sustainability of a plant. This is achieved by regulating key enzymes but also cycling, recycling and redistributing nutrients such as minerals [1], sugars [2], nucleic acids [3], proteins [4], lipids [4], vitamins [5], etc. Compared to de novo syntheses, such salvage pathways can be energetically beneficial for plant cells. They also have the added benefit of leaving metabolic resources available for the synthesis of other molecules. Finally, it is also a way to connect metabolic pathways together, as is the case for amino acids, lipids and isoprenoids via the mevalonic acid (MVA) shunt involving mitochondrial and peroxisomal hydroxymethylglutaryl-CoA lyase [6]. The senescence of plant organs is a striking example of cellular reprogramming. Thus, during the senescence of leaves, while chlorophyll is catabolized, primary metabolites (proteins, nucleic acids, lipids) are in turn degraded. However, not only can primary metabolites be recovered, specialized metabolites (also called secondary metabolites) are also recyclable. In addition, catabolic routes control phytohormone levels. For example, (+)-S-abscisic acid (ABA), a phytohormone that plays a role in development and stress response, is not only a catabolic product of xanthophyll carotenoids but is further inactivated into catabolites such as (-)-dihydrophaseic acid [7]. Another example is natural rubber, a plant-produced isoprenoid biopolymer containing poly(cis-1,4-isoprene) units. It can be degraded by bacteria or actinomycetes to products entering the β-oxidation pathway [8]. All these recycling routes have an initial catabolic phase in common, possibly followed by the reactivation of small molecules into chemically active metabolites that can enter a metabolic pathway. Phosphorylation is one way to reactivate metabolites. With NADH and acetyl-CoA, ATP is considered as central in energetic metabolism with its ability to act as phosphoryl donor, which thermodynamically promotes group transfer potential [9]. This review will discuss the function of isoprenol phosphorylation in isoprenoid salvage pathways and biosynthesis.
2. Isoprenoid Biosynthesis Pathways
A large proportion of natural products are of plant origin, with terpenoids/isoprenoids being the most widely represented chemical class [10]. It is estimated that over 80,000 isoprenoid compounds have been identified so far, making the isoprenoid pathway the most complex and structurally diversified metabolic pathway. To manage such a sophisticated network, an elaborate regulatory scheme is needed to ensure the biosynthesis of all the compounds required for each cellular function [11]. A multitude of organisms, and in particular plants, rapidly redirect metabolic fluxes to specific isoprenoid subfamilies in response to ad hoc needs such as development, defense against pathogens or adaptation to environmental or climatic changes [12,13,14]. However, despite two centuries of research efforts [15], deciphering how this pathway is regulated remains a work in progress with many question marks. For instance, the estimation of isoprenoid precursors concentrations is a challenging task, because the biosynthetic pathway is compartmentalized in cells but also within plant tissues [16]. This makes the unravelling of metabolic regulations complex and the quantification of metabolic fluxes tricky [17].
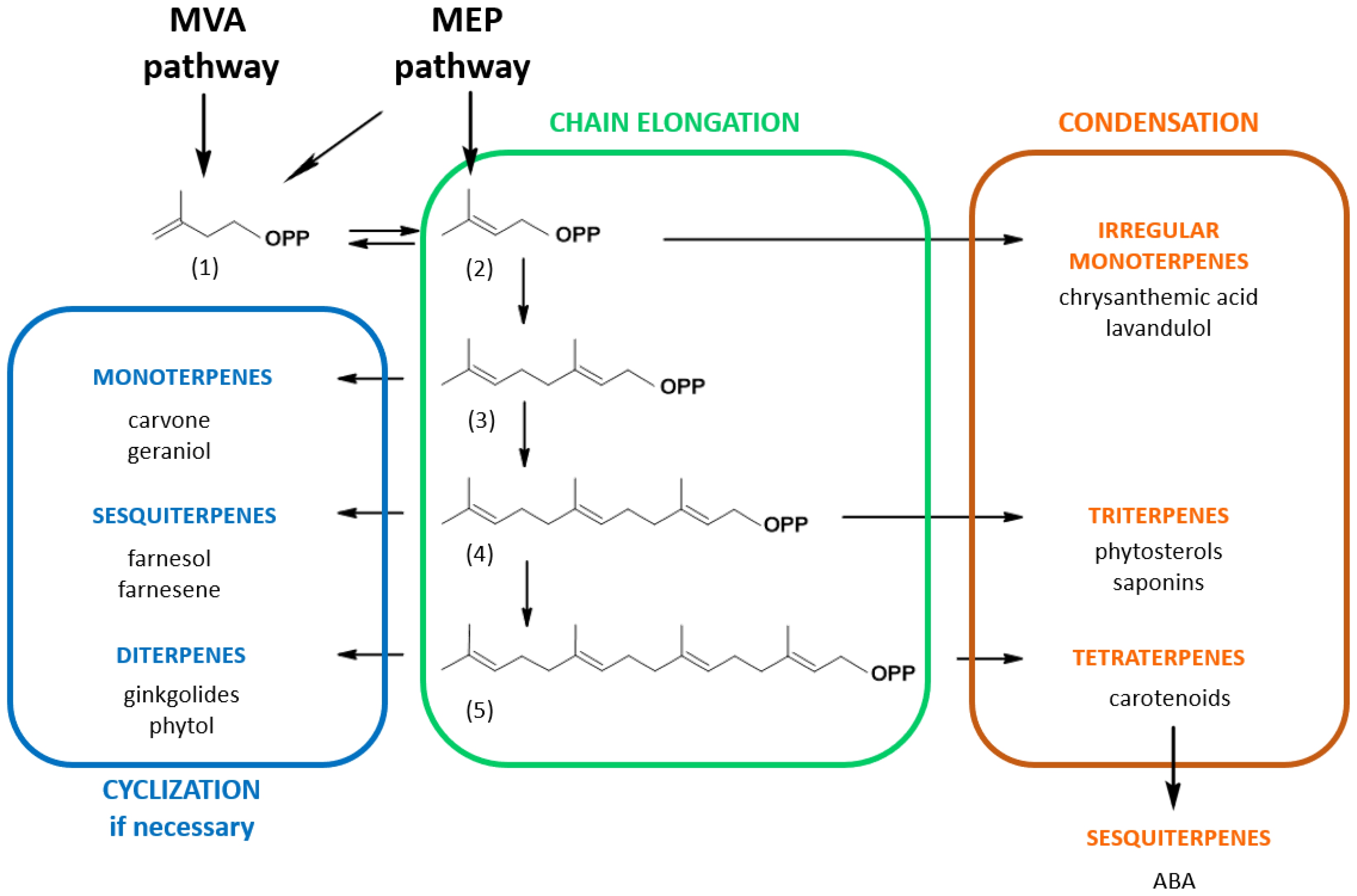
The common thread of every isoprenoid is that at least part of the molecule is derived from a common C5H8 precursor, present as a homoallylic isoprene unit, isopentenyl diphosphate (IPP, 1) or its allylic isomer, dimethylallyl diphosphate (DMAPP, 2) (Figure 1). These building blocks are possibly the most widely used, all metabolisms considered, making IPP/DMAPP one of the eight key metabolites, kinetically stable but thermodynamically activated, that power cell metabolism [9]. Depending on the organism, IPP and DMAPP can either be synthesized by the mevalonic acid/mevalonate (MVA) pathway and/or by the 2C-methylerythritol 4-phosphate (MEP) pathway, plants using both [15]. These precursors undergo condensation (Figure 1) with elimination of the pyrophosphate to yield geranyl diphosphate (C10-GPP, 3), farnesyl diphosphate (C15-FPP, 4) and geranylgeranyl diphosphate (C20-GGPP, 5) that are central molecules embroiled in the biosynthesis of specific branches of compounds classified by the number of isoprene units contained in each molecule. For instance, the GPP-derived branch corresponds to monoterpenes (e.g., carvone), FPP-derived compounds to sesquiterpenes (e.g., farnesol) or triterpenes (e.g., phytosterols) and GGPP-derived molecules to diterpenes (e.g., phytol) or tetraterpenes (e.g., carotenoids) [18]. In addition, isoprenoid precursors (such as C5-DMAPP, C10-GPP, C15-FPP or C20-GGPP) can also be used to modify other types of molecules, such as proteins, tRNA, secondary metabolites, etc., through the action of specific prenyltransferases [19]. Plants exhibit unique metabolic dynamics because they use two distinct and compartmentalized metabolic pathways to synthesize isoprenoid precursors: the MVA pathway in the cytosol/endoplasmic reticulum and the MEP pathway in the plastids (Figure 1). Both pathways can, to some extent, exchange common metabolic precursors such as prenyl diphosphates [15]. In this matter, it has been demonstrated that cytosolic pyruvate is primarily incorporated into plastidial isoprenoids such as phytol via glyceraldehyde 3-phosphate and that plastidial isoprenoid intermediates are incorporated into cytosolic isoprenoids such as phytosterols [20]. HMG-CoA reductase (HMGR), the enzyme catalyzing the biosynthesis of MVA, is recognized as the key enzyme in this pathway. This enzyme is multilevel-regulated and retro-regulated by isoprenoid end-products such as phytosterol precursors [13,16]. The situation of the plastidial MEP pathway is more complex with four enzymes controlling metabolic fluxes (1-deoxy-D-xylulose 5-phosphate synthase, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, (E)-4-hydroxy-3-methyl-but-2-enyl diphosphate synthase and/or (E)-4-hydroxy-3-methyl-but-2-enyl diphosphate reductase) and even appears to be species-specific at some point. In addition, the MEP pathway is subject to retrograde signal regulation [21].
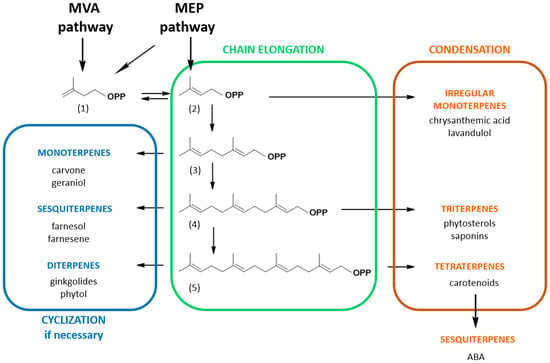
Figure 1.
Simplified isoprenoid biosynthesis pathway in plants. The non-plastidial mevalonic acid (MVA) pathway and the plastidial 2C-methylerythritol 4-phosphate (MEP) pathway produce C5-isopentenyl diphosphate (1) and C5-dimethylallyl diphosphate (2). Short-chain prenyltransferases catalyze the elongation of C5 molecules into longer chains by successive addition of (1) to the allyl diphosphate molecule to yield C10-geranyl diphosphate (3), farnesyl diphosphate (4) and geranylgeranyl diphosphate (5). These isoprenyl diphosphates are precursors for the synthesis of branch-specific isoprenoid families as indicated. Note that metabolites can be catabolic products as is the case for abscisic acid (ABA), a catabolite of xanthophyll carotenoids.
Recycling is another way to regulate isoprenoid biosynthesis. Thus, isoprenoid salvage pathways have the advantage of producing recycled rather than neo-synthesized precursors. However, the benefits of these processes are debatable because they involve a battery of enzymes including kinases, phosphatases, alcohol dehydrogenases, etc., which consume high-energy molecules. In addition, how far such enzymes are substrate-specific in vivo remains unclear. This review focuses on two well-known salvage pathways, the first one recycling FPP from farnesylated proteins and the second one degrading chlorophyll and recycling the phytyl side chain for tocopherol biosynthesis. Both have in common the use of short chain isoprenols (or oligoprenols) as intermediate substrates necessitating a double phosphorylation to prenyl diphosphates, to be reincorporated into the biosynthesis pathway.
3. Prevalence and Functions of Short-Chain Isoprenols in Plants
GPP (3), (2E,6E)-FPP (4) and (2E,6E,10E)-GGPP (5) play a central role in the metabolism of isoprenoids (Figure 1). They represent branching nodes to different sub-pathways, specific to a family of isoprenic compounds. So, monoterpenes are formed from GPP; sterols, sesquiterpenes and farnesylated proteins from FPP; and carotenoids, gibberellins and chlorophylls are generated from GGPP [16]. Prenyl diphosphates are negatively charged due to their anionic phosphate groups, making them unable to passively cross cell barriers. In contrast, the corresponding alcohols penetrate cell membranes, as seen in tobacco cells labeled with fluorescent analogs [22]. Furthermore, farnesol and geranylgeraniol alter the membrane structure by causing the formation of isoprenoid microdomains [23]. The overexpression of some isoprenyl diphosphate synthases triggers the production of the corresponding alcohols [24]. Additionally, specific enzymes catalyze the production of geraniol (Gol), farnesol (Fol) and geranylgeraniol (GGol) [25,26,27,28]. This reflects well-defined functions of isoprenols in cells. In addition, short-chain isoprenols but also their corresponding diphosphates are naturally present as stereoisomers containing cis- and/or trans-double bonds [26,29,30]. Interestingly, cell-free protein extracts have the capacity to modify the stereochemistry of the double bonds [31,32]. This suggests that specific enzymes are involved in modifying the stereochemistry from the (E) to the (Z) configuration of double bonds, and vice versa. The role of isoprenols is likely to be influenced by stereochemical properties.
For decades, special attention has been given to farnesol, as it was found that this product was active as an antifungal or antibacterial compound [33], also inducing apoptosis of cancer cells [34]. At the same time, farnesol would be a non-sterol metabolite capable of down-regulating the activity of HMGR in mammals and hence the accumulation of cholesterol [35,36]. In nature, this isoprenol is generally found in essential oils, where it is thought that it can relieve oxidative stress. It also serves as a defense compound against abiotic and biotic insults and as a synomone. Its function in plant ecology is undeniable. For example, farnesol functions as an insect attractor [37], has fungistatic effects on Monilinia laxa, a fungal nectarine pathogen [38], and insecticidal effects have been demonstrated against green peach aphids [39]. In plants, farnesol was described as a co-product of terpene synthases [40], but specific farnesol synthases were also characterized in both animals [41] and plants [42,43,44]. However, the metabolite is not just toxic for pathogens, it is also toxic for plant cells. Its concentration must therefore be strictly controlled to avoid exceeding a certain toxicity threshold. While farnesol is cytotoxic at 100 µM, at lower concentrations it interferes with both isoprenoid metabolism and the physiology of the plant. It should be noted that in kiwi flowers (Actinidia chinensis), an accumulation of high concentrations of farnesol, higher than toxic concentrations in plant cells, has been observed possibly sequestered as farnesyl-glycosides [45]. In the 1970s, a correlation was established between farnesol and the regulation of the stomatal opening in Sorghum bicolor [46]. Mansfield and colleagues [47] proposed farnesol as a chemical that acts in synergy with ABA, a phytohormone that induces the closure of stomata and reduces transpiration in water-deficient conditions. In addition, they proposed that farnesol alters the permeability of chloroplast envelope membranes, allowing the release of ABA. However, this explanation is not a convincing one. At this time, the possibility to phosphorylate farnesol into FPP had not been considered. However, more recently, a link between farnesol metabolism, abiotic stress and floral development was established through phenotype analysis of an Arabidopsis farnesol kinase loss-of-function mutant [48].
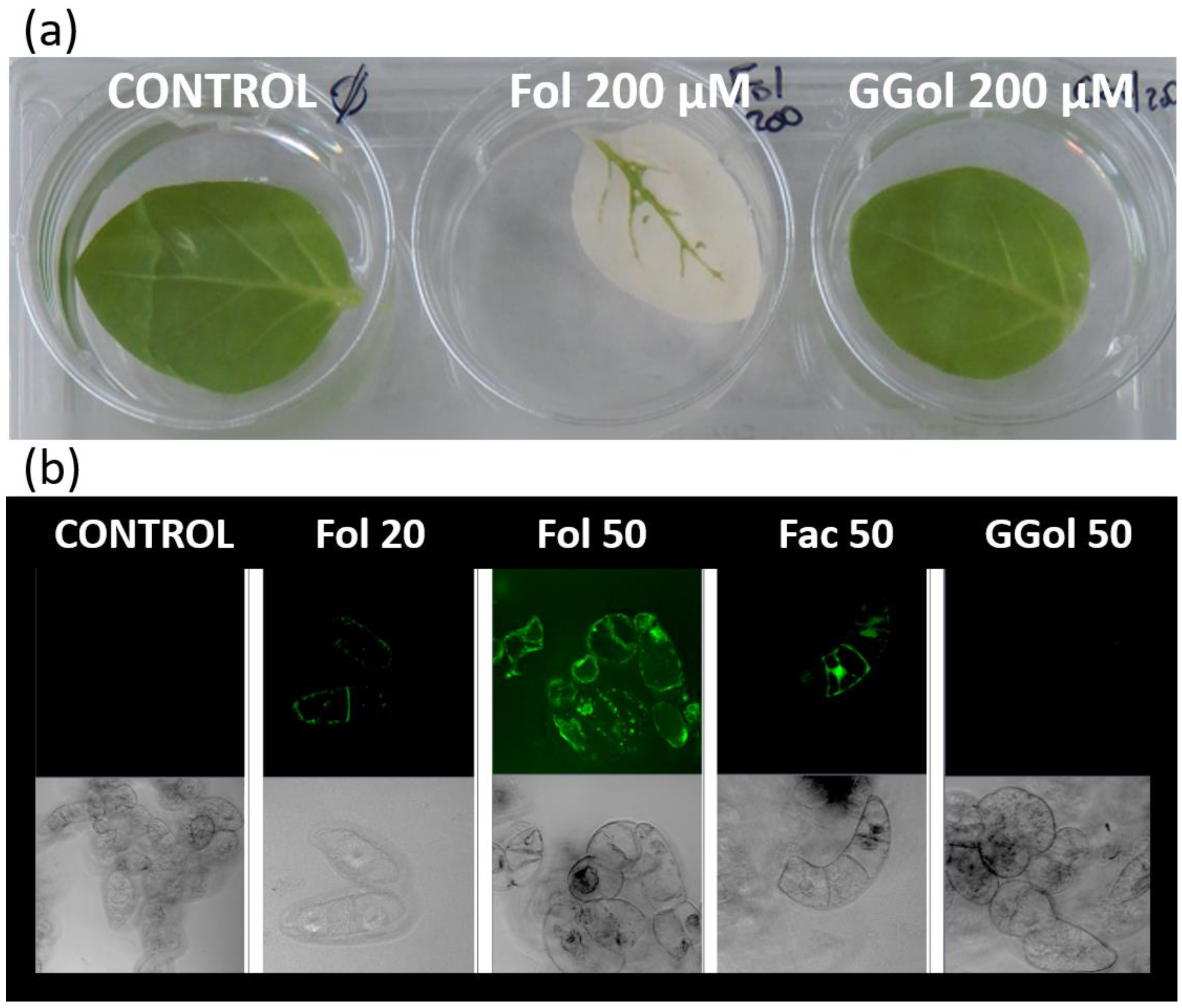
In another context, farnesol was also found to switch the metabolic origin for geranylgeranylation of proteins from the MEP pathway to that of the MVA pathway [49]. Basically, the metabolite acts like an abiotic elicitor by stimulating the production of MVA as a consequence of increased gene expression of HMGR2 in tobacco cells [50,51]. It is important to emphasize that HMGR is encoded by multiple gene families in plants, each of which has a defined function and is expressed in specific tissues. Tobacco HMGR2 is known as a so-called stress isoform: its expression is induced when tobacco cells are challenged with paraquat, an oxidative stress-inducing herbicide [52]. Although it is unlikely, for the moment it cannot be excluded that in plants, the activity of a dedicated HMGR isoform is inhibited in the presence of farnesol, as is the case in animal cells [53]. Confocal microscopy observations of tobacco cells labeled with a fluorescent analog of farnesol, in which an isoprene unit is replaced by 7-nitrobenz-2-oxa-1,3-diazol-4-yl (NBD), showed colocalization with membranes of the endoplasmic reticulum and tonoplast, Golgi apparatus, lipid bodies and mitochondria [22]. These results are consistent with an integration of farnesol into a variety of membrane structures. Further investigation revealed that the fluorescent analog was also incorporated into plastid envelopes [49] and to some extent appeared internalized inside these organelles. At the same time, Fol triggers an apparent proliferation of plastids in tobacco cells [49]. It is interesting to note that tobacco leaves treated with fairly high concentrations of farnesol (200 µM) develop bleaching reminiscent of oxidative stress within 3 days (Figure 2a). This response is specific to the C15-compound, as can be seen from the leaves that remain green when geranylgeraniol is used (Figure 2a). An induction of oxidative stress can be confirmed by applying the fluorogenic probe H2-DC-FDA (2′,7′-dichlorofluorescin diacetate) for detection of reactive oxygen intermediates (Figure 2b). As a result, C15-farnesol, but not C20-geranylgeraniol, induces oxidative stress in plants. Similar findings have also been found in fungi, with interesting applications for treating candidosis [54]. Overall, the usefulness of prenyl alcohols, a fortiori that of farnesol and geraniol [25,27] which are broadly covered in the literature, has been demonstrated.
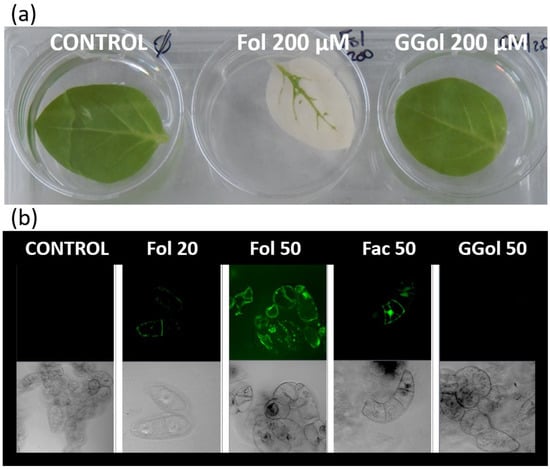
Figure 2.
Farnesol, an inducer of oxidative stress in plants. (a) Tobacco leaf assay. Tobacco leaves from one-month-old plantlets were placed for 3 days in a KH2PO4 (100 µM, pH 7.5) solution containing no additive (CONTROL), containing farnesol (200 µM) (Fol) or geranylgeraniol (200 µM) (GGol). (b) One-week-old stationary tobacco BY-2 cells were diluted (1:5) in fresh Murashige and Skoog medium [50] and treated for 48 h with farnesol (Fol), farnesol acetate (Fac) which cannot be metabolized or geranylgeraniol (GGol). The final concentrations were adjusted to either 20 µM or 50 µM. The green fluorescence observed in the upper series of images corresponds to fluorescein emission. The lower series corresponds to DIC (Differential Interference Contrast) images.
4. Enzyme-Catalyzed Transformation of Isoprenols
Short-chain isoprenols (or oligoprenols) can be further metabolized. Some metabolites are used as scaffolding molecules, others are intended to neutralize the likely cytotoxic effects of isoprenol accumulation. In this section, we will discuss phosphorylations, oxidations and esterifications.
4.1. Oligoprenol Phosphorylations
The phosphorylation/dephosphorylation procedures for the synthesis of isoprenols and their reactivation into isoprenyl diphosphates have recently been reviewed [28]; it is encouraged to refer to this comprehensive review for more details. Here, we will focus on the phosphorylation process required to recycle farnesol into farnesyl diphosphate and phytol into phytyl diphosphate. We will see that isoprenols, as intermediates, will be reactivated by a two-step phosphorylation process to isoprenyl diphosphates, which can then reintegrate the isoprenoid biosynthetic pathway.
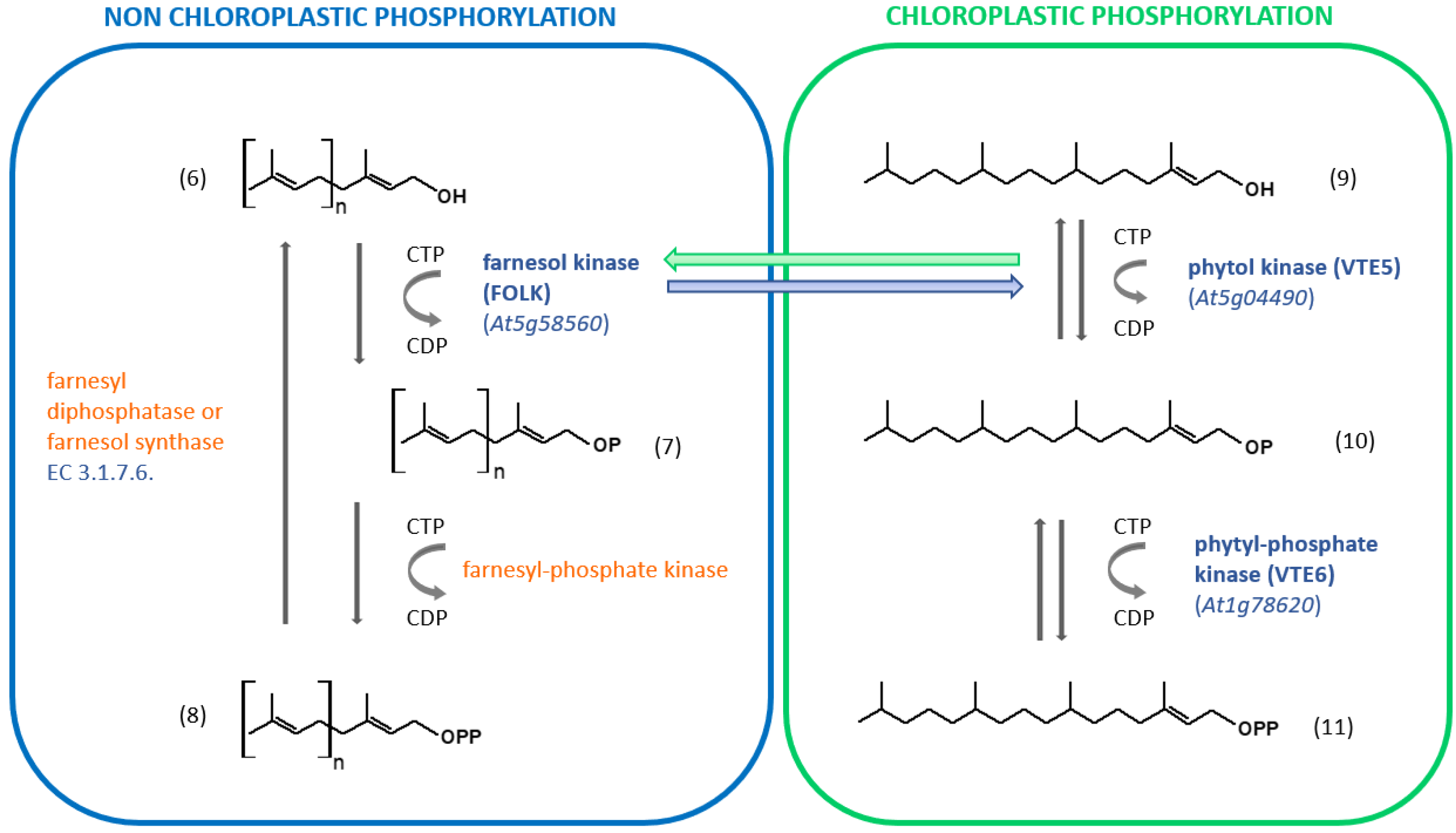
The phosphorylation of short isoprenol chains is a widespread mechanism, but the specific substrate used by dedicated kinases will differ with the type of organism [28]. There is no real evidence that C5-isopentenol or C5-dimethylalcohol is phosphorylated in non-engineered plants. Enzyme-catalyzed phosphorylation of farnesol (and geranylgeraniol) has been evidenced in several organisms including animals, humans, algae and higher plants [55,56]. When tobacco microsomal membranes were used as a source of kinase activity, farnesol was phosphorylated to farnesyl-phosphate (FP) when ATP, CTP, GTP or UTP was present as a phosphoryl donor [56]. Phosphorylation of FP to FPP occurred only in the presence of CTP as a phosphoryl donor [56]. The results suggest that two distinct enzymes catalyze the phosphorylation of farnesol and FP (Figure 3).
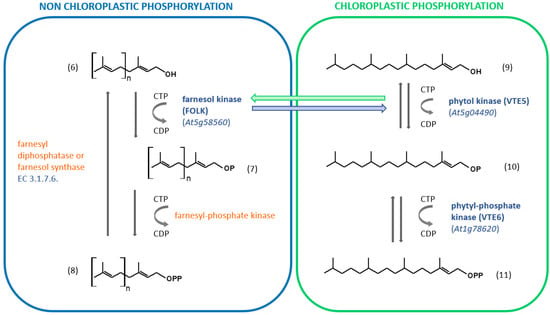
Figure 3.
Phosphorylation/dephosphorylation of short-chain isoprenols in plants. When identified, the Arabidopsis Genome Initiative (AGI) locus identifier code is indicated in blue. When the gene is unidentified, the enzyme is indicated in orange. (6): Isoprenol corresponding to geraniol if n = 1, farnesol if n = 2 or geranylgeraniol if n = 3; (7) monophosphorylated molecules described in (6); (8) diphosphorylated molecules described in (6). (9): phytol; (10) phytyl-phosphate; (11) phytyl diphosphate.
Farnesol kinase from Arabidopsis thaliana (FOLK; Arabidopsis Genome Initiative/AGI locus code At5g58560) was later isolated and characterized. The enzyme uses all NTP as a phosphoryl donor substrate, but CTP is preferred [57]. Interestingly, the tobacco farnesol kinase does not accept ATP as a phosphoryl donor substrate [56]. Furthermore, the enzyme also recognizes geraniol and geranylgeraniol as isoprenol substrates [57]. VTE5 (AGI locus code At5g04490) shares similarities with FOLK [58,59]. FOLK exhibits 38.7% identity at the deduced amino acid sequence level to VTE5. Similar to FOLK, VTE5 catalyzes the phosphorylation of phytol by using CTP as the preferred phosphoryl donor substrate. Thus, it was hypothetized that this enzyme may also phosphorylate Fol, but this assumption turned out to be wrong [57]. More recently, the reverse hypothesis has been assessed and it appears that FOLK catalyzes the phosphorylation of phytol [60], but cellular compartimentation might be a problem for such cross-activities (Figure 3). The identity of the second enzyme (farnesyl-phosphate kinase) remains unclear. Instead, an enzyme that phosphorylates isopentenyl phosphate (isopentenyl phosphate kinase: IPK; AGI locus code At1g26640) using ATP as a cofactor has been identified and characterized. It is involved in the alternate MVA pathway for IPP biosynthesis, where MVA 5-phosphate is decarboxylated into isopentenyl-phosphate, instead of being phosphorylated into MVA 5-diphosphate [61]. IPK can to some extent phosphorylate geranyl phosphate, but with a catalytical efficiency (kcat/Km) about 6500-fold lower. It therefore appears to be specific for C5 isoprenoid precursors. For its part, the enzyme catalyzing the formation of phytyl phosphate has been characterized in Arabidopsis [62]. VTE6 (AGI locus code At1g78620) catalyzes the formation of phytyl diphosphate using CTP as a cosubstrate (Figure 3).
It can be concluded that the phosphorylation of short-chain isoprenyl alcohols from chloroplasts and that which operates outside the organelle have a notable parallelism (Figure 3). Interactions between these two pathways can reasonably be expected to lead to better adaptation to changing environmental conditions.
4.2. Isoprenol Oxidations and Reductions
Oxidation of isoprenols is well documented in plants. All isoprenols are prone to chemical oxidation, which might contribute to the metabolic regulation [63]. The oxidation of the alcohol group into aldehyde and corresponding acids has been characterized. In addition, microorganisms can also break down plant secondary metabolites such as monoterpenes by oxidation reactions [64]. Here we will provide a few pertinent examples of oxidized products that have been characterized in plants.
Geraniol and farnesol were found to be oxidized into geranial and farnesal by different protein plant extracts [65,66,67]. A soluble dehydrogenase activity found in chicory (Cichorium intybus L.) root protein extracts was able to catalyze the formation of (2E,6E)-farnesal and (2Z,6E)-farnesal using (2E,6E)-farnesol and NAD(P)+ as substrates [65]. The enzyme could also catalyze the hydroxylation of cyclic sesquiterpenes [65]. Later, a gene coding for farnesol dehydrogenase was identified in Arabidopsis thaliana (AGI locus code At4g33360, Figure 4) [68]. The recombinant membrane-associated protein is NAD+-dependent and shows only partial specificity for farnesol as a substrate. In squash (Cucurbita maxima), the alcohols are oxidized into geranylgeranoic acid, farnesoic acid and geranoic acid [69].
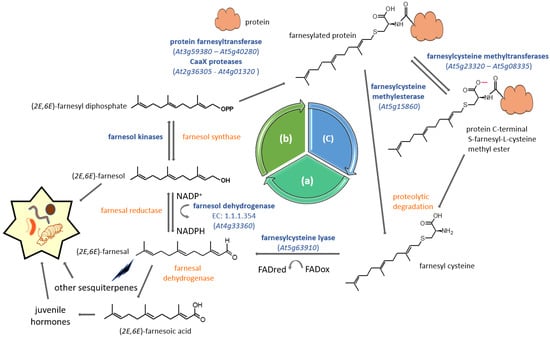
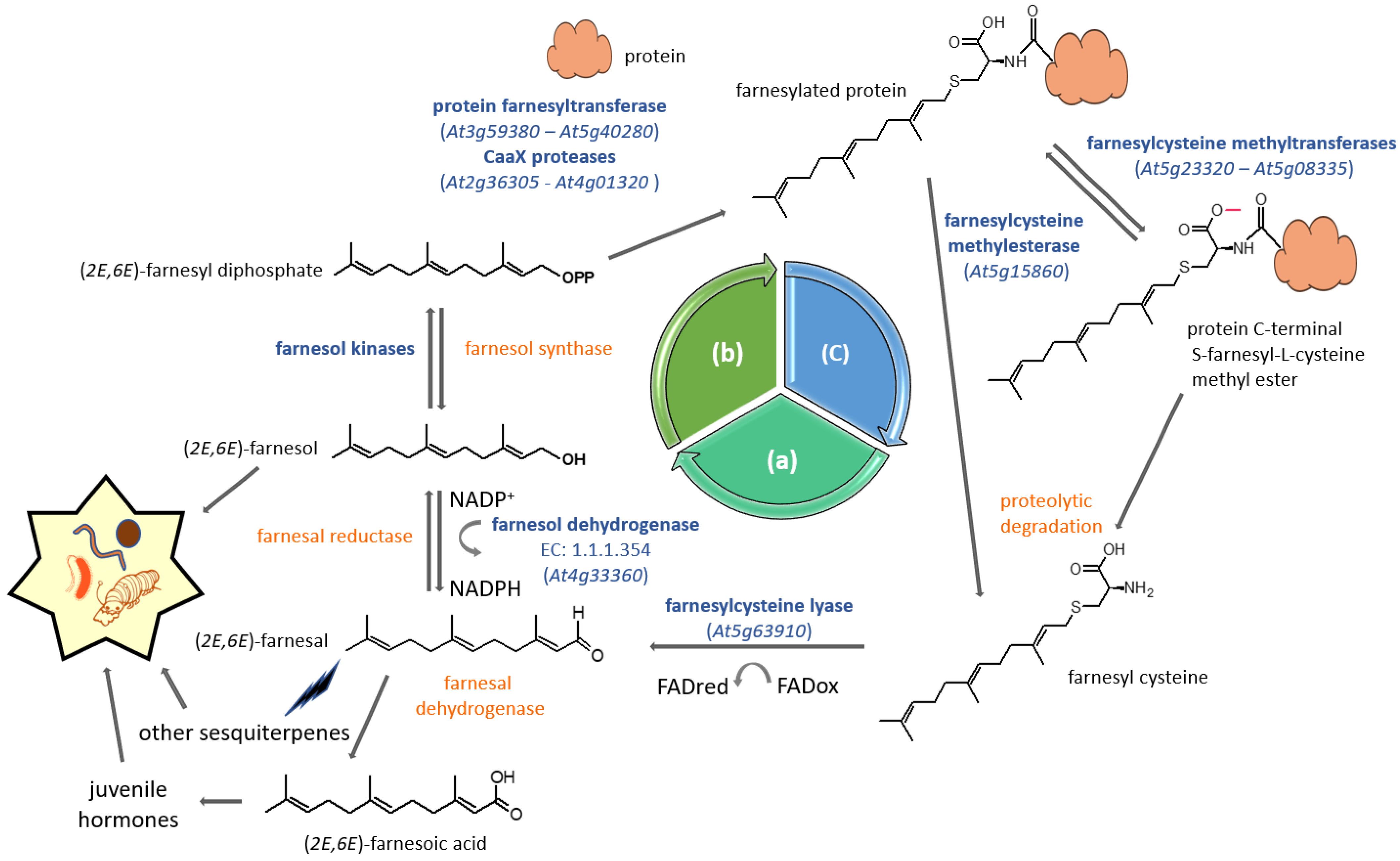
Figure 4.
FPP salvage pathway for protein prenylation. The cyclic pathway undergoes three steps including (a) the degradation of prenylated (farnesylated) proteins and the recovery of farnesol, (b) a phosphorylation process leading to the reactivation into farnesyl diphosphate and finally (c) the prenylation and maturation of the protein. Enzyme reactions catalyzed by farnesol kinases have been described in Figure 3. Characterized genes and proteins are indicated in blue, as is the corresponding AGI locus code. When the gene is unidentified, the enzyme is indicated in orange.
4.3. Isoprenol Esterifications
Short-chain isoprenyl alcohols can be present as glucoside or fatty acid esters. Farnesyl glycosides (cupanioidesosides) have been isolated in Lecaniodiscus cupanioides roots [70], while Eucalyptus perriniana, Strophanthus gratus and Phytolacca americana plant cell cultures produce glucosylated farnesol and geraniol [71] and that of Vitis vinifera produces geraniol [66]. Such molecular combinations reduce the hydrophobicity of the prenyl alcohol and can facilitate a possible vacuolar storage. Other farnesyl glycosides have a cytotoxic effect, which may suggest a possible role for plant defense mechanisms [72]. In addition, fatty acid farnesyl esters, geranylgeranyl esters and phytyl ester can be accumulated in Arabidopsis thaliana loss-of-function mutants deficient in isoprenol kinase activities [58,59,60,62].
5. Farnesyl Diphosphate Salvage Pathway for Protein Prenylation
For some time now, isoprenol phosphorylation has been proposed as an alternative to FPP neo-biosynthesis generated by the MVA pathway [73]. Dring Crowell and his team later proposed a comprehensive way to recycle FPP, the so-called FPP salvage machinery that recycles the farnesyl group used to modify CaaX-motif proteins [74]. Many of the enzymes involved in this process have been characterized in plants, starting with type I protein prenyltransferases. Type I protein prenylation is a post-translational process associating CaaX motif (C = cyst; a = aliphatic; X = C-terminus amino acid) proteins to prenyl moieties via thioether linkages to cysteines located in the fourth position of the carboxyl terminus [75]. This eukaryotic process involves type I protein prenyltransferases being used as co-substrates to the CaaX protein, either farnesyl diphosphate or geranylgeranyl diphosphate [76]. Once modified, a number of proteins undergo further post-prenylation maturation events, including aaX peptide proteolysis and cysteinyl carboxymethylation [77] (Figure 4).
The FPP salvage machinery is proposed as a cycle based on three successive phases: protein prenylation, protein degradation and prenyl diphosphate recycling (Figure 4). Basically, prenylated proteins are prone to proteolysis, which would ultimately liberate farnesylcysteine as a metabolite (Figure 4). A FAD-dependent farnesylcysteine lyase (EC 1.8.3.6; AGI locus code At5g63910) catalyzes the cleavage of the strong thioether bond and releases farnesal [74,78], which would then subsequently be reduced into farnesol by an uncharacterized farnesal reductase. However, such an enzyme has been isolated in humans, but its substrate specificity is rather broad, because besides farnesal, it uses several aldehydes, some ketones and also quinones [79]. As for farnesylcysteine lyase, in contrast to the bovine enzyme, the Arabidopsis one is highly specific to farnesylcysteine and seems to make poor use of geranylgeranylcysteine as a substrate [74]. This suggests that plants have either developed an alternative way to oxidize geranylgeranylcysteine or that there is no real need to recover geranylgeranyl diphosphate from proteins. In addition, the plant enzyme recognizes N-acetyl-farnesylcysteine, which provides an additional argument for its function in the deprenylation of farnesylated proteins [74]. Although described as a membrane-associated enzyme in Arabidopsis thaliana [74], this protein has also been identified as part of the proteome of aging leaf lipid droplets [80]. In the next step, the two membrane-localized kinases (Figure 3) phosphorylate farnesol into farnesyl-phosphate and farnesyl diphosphate, respectively (Figure 4). At this stage, the prenyl diphosphate can again be used as a substrate, for instance by the hetorodimeric protein farnesyltransferase to modify a CaaX-protein prenylation substrate (Figure 4). It has been decribed as a salvage pathway for protein prenylation, but it cannot be excluded that recycled FPP could be used for other purposes, such as phytosterol biosynthesis or quinone side chain biosynthesis, for example [81].
The significance of such a recycling process is still unclear. The membrane localization of prenylated proteins makes them efficient transducers in signaling pathways [82,83]. This is only possible if their function is ephemeral, which could be attained by protein degradation. On the other hand, two defense-related metabolites are synthesized during this process: (2E,6E)-farnesal and (2E,6E)-farnesol. (2E,6E)-Farnesal is oxidized to (2E,6E)-farnesoic acid, which is a precursor of plant juvenile hormones used by plants to fight against insect attacks [84]. Likewise, as mentioned above, farnesol is a well-known compound, which triggers pathogen resistance. Very recently, in nectarine (Prunus persica), an analysis of the relative expression of three genes that code for enzymes involved in the FPP salvage pathway (β-subunit of farnesyltransferase, farnesylcysteine methyltransferase and farnesol kinase) was investigated [38]. It could be demonstrated that in resistant tissues infected with Monilinia laxa, a fungal pathogen, the expression remained unchanged, while in susceptible tissues, the general expression of those genes significantly decreased over time. These findings could indicate that an active pathway is helping the plant fight the pathogen. It is therefore reasonable to think that such a salvage pathway could promote plant defense against pest attacks and strengthen their “immunity”. In addition, all characterized enzymes are subject to ABA regulation and appear to participate in the negative regulation of ABA signaling, i.e., its sensitivity [48,57,68,74,85,86]. It may be possible to find a relationship between the negative regulation of ABA signaling and the initial observations made with farnesol on stomatal closure (Section 3 Occurrence and functions of short chain isoprenols in plants), but the direct link remains purely speculative. It is important to remind that ABA plays multiple roles in abiotic stress, but also biotic, by interacting with other stress-response hormones such as jasmonates, salicylic acid or ethylene [82]. Different farnesylated proteins have been identified as being associated with ABA hypersensitivity. In a first example, the defective farnesylation of the heat-shock protein 40 (HSP40), encoded by J2 and J3 genes, is sufficient to make the plant able to withstand water stress [87]. Arabidopsis plants inactivated in a second gene encoding the farnesylated WD40 repeat protein ALTERED SEED GERMINATION2 (ASG2) display an ABA-hypersensitive phenotype correlated to the inhibition of germination [88]. However, resistance to drought could not be observed in those plants [88]. Only a few CaaX proteins have been characterized so far, and more will be expected in the future. This would provide a more accurate response to this interference.
6. Phytyl Diphosphate Salvage Pathway for Vitamin E Biosynthesis
The second salvage pathway recycles phytol in plants. Like the previous one, the salvage pathway is based on the breakdown of an isoprenoid compound, in this case chlorophyll. Basically, chlorophyll is an iron porphyrin complex anchored to chloroplast thylakoid membranes by a C20-phytyl group. Dephytylation is an enzymatic step required for the recycling of chlorophyll [89]. Dephytylation of chlorophyll is also necessary for tocopherol (vitamin E) biosynthesis, a liposoluble antioxidant [62]. A large majority of the phytyl diphosphate used to prenylate HBA to obtain, for instance, α-tocopherol (Figure 5) is not originated from de novo synthesis of MEP-derived GGPP, but is recovered from degraded chlorophyll [90]. Vitamin E can only be synthesized in organisms with plastids. Tocopherols are made up of a 6-chromanol ring and an isoprenoid side chain of 16 carbon atoms. These side chains derive from C20-phytyl, which is necessary for membrane anchoring [91,92]. Tocopherols accumulate in Arabidopsis of senescent leaves or in plants subjected to abiotic or biotic stress [91,92]. At the same time, the ultrastructure of chloroplasts is altered, their number and size decreases and the storage of metabolites such as starch declines. However, the number and size of plastoglobules increases [93]. These lipoprotein particules, linked to thylakoids, participate actively in tocopherol biosynthesis [94]. However, more careful work using proteomics and protein subcellular localizations showed that only the last steps of tocopherol biosynthesis happen in plastoglobules [95]. Furthermore, enzymes involved in phytol esterification have also been found to be associated with plastoglobules [95]. Besides being involved in tocopherol biosynthesis, chromoplast plastoglobules are also implicated in carotenoid metabolism, specifically recruiting biosynthetic enzymes for carotenoid accumulation in tomato fruits, including enzymes of the MEP pathway [96].
The identification of enzymes involved in the salvage pathway and their corresponding genes has recently been reviewed [97]. How the metabolism of phytol is involved in tocopherol biosynthesis has been carefully described [58,59,60,90]. Interestingly, the salvage pathway is dependent on the nature of the tissue and the plant model used [98]. For the sake of clarity, we will focus on Arabidopsis thaliana: Figure 5 illustrates a simplified view of the current knowledge.
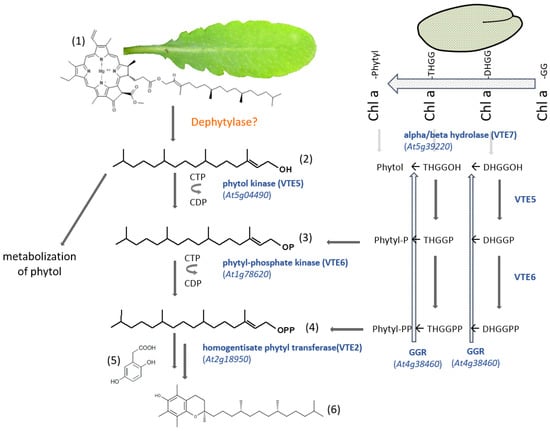
Figure 5.
Phytyl diphosphate salvage pathway for tocopherol biosynthesis in Arabidopsis thaliana. There are two biosynthetic pathways that have been described, depending on whether the synthesis takes place in seeds [98] or in leaves [58]. (1) Chlorophyll a; (2) phytol; (3) phytyl-phosphate; (4) phytyl diphosphate; (5) homogentisic acid -HGA; (6) α-tocopherol. Chl a, chlorophyll a; GGR, geranylgeranyl reductase; DHGG, dihydrogeranylgeranyl; DHGGP, dihydrogeranylgeranyl-phosphate; DHGGPP, dihydrogeranylgeranyl diphosphate; THGG, tetrahydrogeranylgeranyl; THGGP, tetrahydrogeranylgeranyl-phophate; THGGPP, tetrahydrogeranylgeranyl diphosphate. Alternatively, phytol can also be metabolized [90].
Figure 5.
Phytyl diphosphate salvage pathway for tocopherol biosynthesis in Arabidopsis thaliana. There are two biosynthetic pathways that have been described, depending on whether the synthesis takes place in seeds [98] or in leaves [58]. (1) Chlorophyll a; (2) phytol; (3) phytyl-phosphate; (4) phytyl diphosphate; (5) homogentisic acid -HGA; (6) α-tocopherol. Chl a, chlorophyll a; GGR, geranylgeranyl reductase; DHGG, dihydrogeranylgeranyl; DHGGP, dihydrogeranylgeranyl-phosphate; DHGGPP, dihydrogeranylgeranyl diphosphate; THGG, tetrahydrogeranylgeranyl; THGGP, tetrahydrogeranylgeranyl-phophate; THGGPP, tetrahydrogeranylgeranyl diphosphate. Alternatively, phytol can also be metabolized [90].
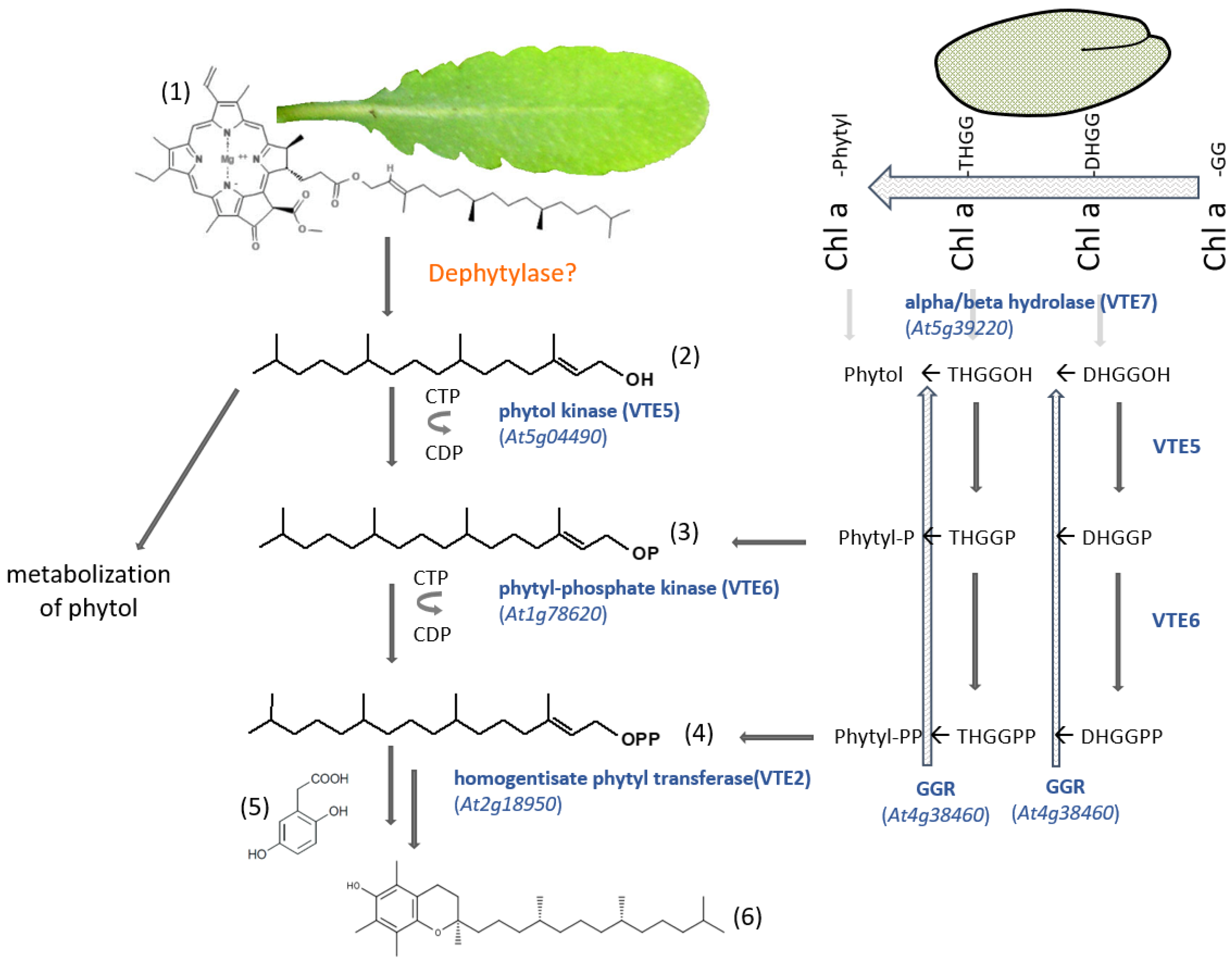
A so far unknown dephytylase in Arabidopsis leaves releases phytol, which will be used as a substrate for VTE5 (phytol kinase). Phytyl phosphate will further be phosphorylated into phytyl diphosphate by VTE6 (phytyl phosphate kinase). A homogentisate phytyltransferase will transfer the activated phytyl group to homogentisic acid (HGA), which will ultimately rise to tocopherol (Figure 5). A different way of obtaining tocopherol has been found in Arabidopsis seeds [98]. VTE7 has been identified as a seed-specific alpha/beta hydrolase being capable of synthesizing phytol (Figure 5). However, the enzyme, instead of recognizing chlorophyll a as a substrate, uses metabolic intermediates and releases dihydrogeranylgeraniol or tetrahydrogeranylgeraniol. It has been proposed that those prenyl alcohols can reenter the tocopherol biosynthesis pathway through the action of geranylgeranyl reductase and phosphorylation by VTE5 and VTE6 according to combinations described in Figure 5. The organization of this pathway shows how plants can achieve their development and defense goals in a very flexible way.
7. Conclusions and Perspectives
There are only two isoprenoid salvage pathways that have been documented, so conclusions can only be partial and speculative. They not only make prenyl diphosphates available, but they also enable the synthesis of metabolites (tocopherol, farnesol, farnesal and farnesoic acid) that can contribute to stress responses (Figure 6).
In the end, over and above the economic incentive to recycle metabolites, there is room to improve stress tolerance with protective roles against environmental constraints. Indeed, as sessile organisms, plants must continuously maintain a metabolic balance that allows them to both grow and respond quickly to attacks [99]. Access to and availability of metabolic precursors are essential to control metabolic pathways and carbohydrates generated by photosynthesis are an important source of those precursors. Physiological and molecular reprogramming controls the entire process, with phytohormones as major players. In this regard, ABA functions as a signal molecule and plays an essential role in many physiological processes such as seed dormancy and germination, seedling growth, aging and responses to abiotic and biotic stresses [100]. On a molecular level, ABA induces the expression of a battery of transcription factors belonging to different families such as MYB, NAC, bHLH, bZIP, WRKY, etc. [101]. Many of them have been identified as regulators of isoprenoid biosynthesis genes, causing the accumulation of specific metabolites [14,18,102,103]. The link between the gene products involved in the FPP salvage pathway to ABA signaling was discussed in this review. In particular, ABA represses the expression of FOLK permitting the accumulation of Fol [57]. Many ABA-responsive elements are also present in the promoter region of tocopherol biosynthesis genes, suggesting an ABA-dependent regulation of the production of tocopherol [104]. The biosynthesis of ABA is promoted by drought stress in roots, which, once transported to leaves, favors stomatal closure to reduce water loss. However, stomatal closure has both advantages and disadvantages. It has a negative impact on gas exchange, resulting in a reduction in CO2 taken up, decreasing photosynthetic processes and triggering oxidative damages. In addition, ABA induces metabolic changes that lead to the senescence of old leaves, which is necessary for the survival of sprouting leaves [105]. These new constraints caused by stomatal closure could improve the performance of salvage pathways by boosting chlorophyll and protein degradation accompanied by an increase in lipid particles (Figure 6). It should be noted that proteomic analyses have shown that enzymes from both salvage pathways have been identified as associated with lipid-containing particles [80,94,95]. Whether these particles are central to isoprenoid salvage pathways remains to be determined.
Agriculture research faces a significant challenge in developing biofortified plants with enhanced stress tolerance. Clearly, these pathways may make crop plants more resistant to a variety of environmental conditions and help them produce vitamins important for human nutrition (Figure 6). Over the next few years, we will need to figure out which regulatory nodes lead to the activation of these salvage pathways.
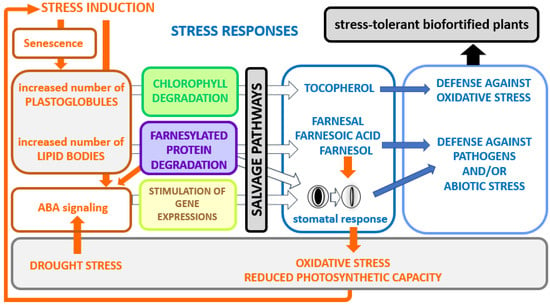
Figure 6.
A model for the implication of isoprenoid salvage pathways on biotic or abiotic stress responses. Stress induction modes are shown in orange, stress responses in blue. Stresses such as heat, cold drought, etc., induce the formation of lipid particles. This is accompanied by plastidial chlorophyll degradation and/or ER/cytosolic farnesylated protein degradation. In this way, defense-related metabolites, such as tocopherol, farnesal, farnesoic acid and farnesol, are produced for improved stress tolerance. Protein farnesylation is involved in negative regulation of abscisic acid (ABA), thus degradation of those proteins causes an enhanced response to ABA [106]. The stomatal closure caused by the hormonal signal triggers oxidative stress and a diminished photosynthesis capacity, which in turn stimulates the entire process.
Figure 6.
A model for the implication of isoprenoid salvage pathways on biotic or abiotic stress responses. Stress induction modes are shown in orange, stress responses in blue. Stresses such as heat, cold drought, etc., induce the formation of lipid particles. This is accompanied by plastidial chlorophyll degradation and/or ER/cytosolic farnesylated protein degradation. In this way, defense-related metabolites, such as tocopherol, farnesal, farnesoic acid and farnesol, are produced for improved stress tolerance. Protein farnesylation is involved in negative regulation of abscisic acid (ABA), thus degradation of those proteins causes an enhanced response to ABA [106]. The stomatal closure caused by the hormonal signal triggers oxidative stress and a diminished photosynthesis capacity, which in turn stimulates the entire process.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
I am thankful to NCBI for their freely accessible chemical information PubChem (https://pubchem.ncbi.nlm.nih.gov/, accessed on 15 March 2023). Chemical structures used in this review were downloaded from there, where chemical structures were recovered from their library then further worked out with ACD/ChemSketch.
Conflicts of Interest
The author declares no conflict of interest.
References
- Sattelmacher, B. The apoplast and its significance for plant mineral nutrition. New Phytol. 2001, 149, 167–192. [Google Scholar] [CrossRef] [PubMed]
- Bar-Peled, M.; O’Neill, M.A. Plant nucleotide sugar formation, interconversion, and salvage by sugar recycling. Annu. Rev. Plant Biol. 2011, 62, 127–155. [Google Scholar] [CrossRef] [PubMed]
- Ashihara, H.; Stasolla, C.; Fujimura, T.; Crozier, A. Purine salvage in plants. Phytochemistry 2018, 147, 89–124. [Google Scholar] [CrossRef]
- McLoughlin, F.; Augustine, R.C.; Marshall, R.S.; Li, F.; Kirkpatrick, L.D.; Otegui, M.S.; Vierstra, R.D. Maize multi-omics reveal roles for autophagic recycling in proteome remodelling and lipid turnover. Nat. Plants 2018, 4, 1056–1070. [Google Scholar] [CrossRef] [PubMed]
- Colinas, M.; Fitzpatrick, T.B. Natures balancing act: Examining biosynthesis de novo, recycling and processing damaged vitamin B metabolites. Curr. Opin. Plant Biol. 2015, 25, 98–106. [Google Scholar] [CrossRef]
- Hemmerlin, A.; Huchelmann, A.; Tritsch, D.; Schaller, H.; Bach, T.J. The specific molecular architecture of plant 3-hydroxy-3-methylglutaryl-CoA lyase. J. Biol. Chem. 2019, 294, 16186–16197. [Google Scholar] [CrossRef]
- Cutler, A.J.; Krochko, J.E. Formation and breakdown of ABA. Trends Plant Sci. 1999, 4, 472–478. [Google Scholar] [CrossRef]
- Kasai, D. Poly(cis-1,4-isoprene)-cleavage enzymes from natural rubber-utilizing bacteria. Biosci. Biotechnol. Biochem. 2020, 84, 1089–1097. [Google Scholar] [CrossRef]
- Walsh, C.T.; Tu, B.P.; Tang, Y. Eight kinetically stable but thermodynamically activated molecules that power cell metabolism. Chem. Rev. 2018, 118, 1460–1494. [Google Scholar] [CrossRef]
- Chassagne, F.; Cabanac, G.; Hubert, G.; David, B.; Marti, G. The landscape of natural product diversity and their pharmacological relevance from a focus on the Dictionary of Natural Products®. Phytochem. Rev. 2019, 18, 601–622. [Google Scholar] [CrossRef]
- Gutensohn, M.; Hartzell, E.; Dudareva, N. Another level of complexity: The role of metabolic channeling and metabolons in plant terpenoid metabolism. Front. Plant Sci. 2022, 13, 954083. [Google Scholar] [CrossRef]
- Bianchetti, R.; De Luca, B.; de Haro, L.A.; Rosado, D.; Demarco, D.; Conte, M.; Bermudez, L.; Freschi, L.; Fernie, A.R.; Michaelson, L.V.; et al. Phytochrome-dependent temperature perception modulates isoprenoid metabolism. Plant Physiol. 2020, 183, 869–882. [Google Scholar] [CrossRef]
- Hemmerlin, A. Post-translational events and modifications regulating plant enzymes involved in isoprenoid precursor biosynthesis. Plant Sci. 2013, 203–204, 41–54. [Google Scholar] [CrossRef] [PubMed]
- Vranová, E.; Coman, D.; Gruissem, W. Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu. Rev. Plant Biol. 2013, 64, 665–700. [Google Scholar] [CrossRef] [PubMed]
- Hemmerlin, A.; Harwood, J.L.; Bach, T.J. A raison d’être for two distinct pathways in the early steps of plant isoprenoid biosynthesis? Prog. Lipid Res. 2012, 51, 95–148. [Google Scholar] [CrossRef] [PubMed]
- Vranová, E.; Coman, D.; Gruissem, W. Structure and dynamics of the isoprenoid pathway network. Mol. Plant 2012, 5, 318–333. [Google Scholar] [CrossRef] [PubMed]
- Shih, M.L.; Morgan, J.A. Metabolic flux analysis of secondary metabolism in plants. Metab. Eng. Commun. 2020, 10, e00123. [Google Scholar] [CrossRef]
- Tholl, D. Biosynthesis and biological functions of terpenoids in plants. Adv. Biochem. Eng. Biotechnol. 2015, 148, 63–106. [Google Scholar]
- Chang, H.Y.; Cheng, T.H.; Wang, A.H.J. Structure, catalysis, and inhibition mechanism of prenyltransferase. IUBMB Life 2021, 73, 40–63. [Google Scholar] [CrossRef]
- Ladd, S.N.; Nelson, D.B.; Bamberger, I.; Daber, L.E.; Kreuzwieser, J.; Kahmen, A.; Werner, C. Metabolic exchange between pathways for isoprenoid synthesis and implications for biosynthetic hydrogen isotope fractionation. New Phytol. 2021, 231, 1708–1719. [Google Scholar] [CrossRef]
- Jiang, J.; Dehesh, K. Plastidial retrograde modulation of light and hormonal signaling: An odyssey. New Phytol. 2021, 230, 931–937. [Google Scholar] [CrossRef] [PubMed]
- Hemmerlin, A.; Reents, R.; Mutterer, J.; Feldtrauer, J.-F.; Waldmann, H.; Bach, T.J. Monitoring farnesol-induced toxicity in tobacco BY-2 cells with a fluorescent analog. Arch. Biochem. Biophys. 2006, 448, 93–103. [Google Scholar] [CrossRef]
- Funari, S.S.; Prades, J.; Escribá, P.V.; Barceló, F. Farnesol and geranylgeraniol modulate the structural properties of phosphatidylethanolamine model membranes. Mol. Membr. Biol. 2005, 22, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A.; Waݶchtler, B.; Temp, U.; Krekling, T.; Seݩguin, A.; Gershenzon, J. A bifunctional geranyl and geranylgeranyl diphosphate synthase is involved in terpene oleoresin formation in Picea abies. Plant Physiol. 2010, 152, 639–655. [Google Scholar] [CrossRef] [PubMed]
- Conart, C.; Saclier, N.; Foucher, F.; Goubert, C.; Rius-Bony, A.; Paramita, S.N.; Moja, S.; Thouroude, T.; Douady, C.; Sun, P.; et al. Duplication and specialization of NUDX1 in Rosaceae led to geraniol production in rose petals. Mol. Biol. Evol. 2022, 39, msac002. [Google Scholar] [CrossRef]
- Gupta, P.; Sharma, M.; Arora, N.; Pruthi, V.; Poluri, K.M. Chemistry and biology of farnesol and its derivatives: Quorum sensing molecules with immense therapeutic potential. Curr. Top. Med. Chem. 2018, 18, 1937–1954. [Google Scholar] [CrossRef]
- Iijima, Y.; Gang, D.R.; Fridman, E.; Lewinsohn, E.; Pichersky, E. Characterization of geraniol synthase from the peltate glands of sweet basil. Plant Physiol. 2004, 134, 370–379. [Google Scholar] [CrossRef]
- Verdaguer, I.B.; Crispim, M.; Hernández, A.; Katzin, A.M. The Biomedical Importance of the missing pathway for farnesol and geranylgeraniol salvage. Molecules 2022, 27, 8691. [Google Scholar] [CrossRef]
- Cornish-Bowden, A. Nomenclature of prenols (Recommendations 1986). Pure Appl. Chem. 1987, 59, 683–689. [Google Scholar] [CrossRef]
- Sallaud, C.; Rontein, D.; Onillon, S.; Jabes, F.; Duffe, P.; Giacalone, C.; Thoraval, S.; Escoffier, C.; Herbette, G.; Leonhardt, N.; et al. A novel pathway for sesquiterpene biosynthesis from Z,Z-farnesyl pyrophosphate in the wild tomato Solanum habrochaites. Plant Cell 2009, 21, 301–317. [Google Scholar] [CrossRef]
- Overton, K.H.; Roberts, F.M. Interconversion of trans, trans and cis, trans farnesol by enzymes from Andrographis. Phytochemistry 1974, 13, 2741–2743. [Google Scholar] [CrossRef]
- Shine, W.E.; Loomis, W.D. Isomerization of geraniol and geranyl phosphate by enzymes from carrot and peppermint. Phytochemistry 1974, 13, 2095–2101. [Google Scholar] [CrossRef]
- Costa, A.F.; Silva, L.D.C.; Amaral, A.C. Farnesol: An approach on biofilms and nanotechnology. Med. Mycol. 2021, 59, 958–969. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.Y.; Hwang, S.T.; Sethi, G.; Fan, L.; Arfuso, F.; Ahn, K.S. Potential anti-inflammatory and anti-cancer properties of farnesol. Molecules 2018, 23, 2827. [Google Scholar] [CrossRef]
- Bahr, T.; Butler, G.; Rock, C.; Welburn, K.; Allred, K.; Rodriguez, D. Cholesterol-lowering activity of natural mono-and sesquiterpenoid compounds in essential oils: A review and investigation of mechanisms using in silico protein–ligand docking. Phytother. Res. 2021, 35, 4215–4245. [Google Scholar] [CrossRef]
- Meigs, T.E.; Roseman, D.S.; Simoni, R.D. Regulation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase degradation by the nonsterol mevalonate metabolite farnesol in vivo. J. Biol. Chem. 1996, 271, 7916–7922. [Google Scholar] [CrossRef]
- Ma, F.M.; Zheng, L.X.; Gao, Z.Z.; Wu, W.J. Farnesol, a synomone component between lantana (Lamiales: Verbenaceae) and the omnivorous predator, Campylomma chinensis Schuh (Hemiptera: Miridae). Arthropod Plant Interact. 2017, 11, 703–708. [Google Scholar] [CrossRef]
- Balsells-Llauradó, M.; Vall-Llaura, N.; Usall, J.; Silva, C.J.; Blanco-Ulate, B.; Teixidó, N.; Caballol, M.; Torres, R. Transcriptional profiling of the terpenoid biosynthesis pathway and in vitro tests reveal putative roles of linalool and farnesal in nectarine resistance against brown rot. Plant Sci. 2023, 327, 111558. [Google Scholar] [CrossRef]
- Cantó-Tejero, M.; Guirao, P.; Pascual-Villalobos, M.J. Aphicidal activity of farnesol against the green peach aphid–Myzus persicae. Pest Manag. Sci. 2022, 78, 2714–2721. [Google Scholar] [CrossRef]
- Schnee, C.; Köllner, T.G.; Gershenzon, J.; Dengenhardt, J. The maize gene terpene synthase 1 encodes a sesquiterpene synthase catalyzing the formation of (E)-β-farnesene, (E)-nerolidol, and (E,E)-farnesol after herbivore damage. Plant Physiol. 2002, 130, 2049–2060. [Google Scholar] [CrossRef]
- Bansal, V.S.; Vaidya, S. Characterization of two distinct allyl pyrophosphatase activities from rat liver microsomes. Arch. Biochem. Biophys. 1994, 315, 393–399. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, V.; Kumar, S.R.; Shilpashree, H.B.; Krishna, R.; Rao, S.; Shasany, A.K.; Olsson, S.B.; Nagegowda, D.A. An inducible potato (E,E)-farnesol synthase confers tolerance against bacterial pathogens in potato and tobacco. Plant J. 2022, 111, 1308–1323. [Google Scholar] [PubMed]
- Parveen, I.; Wang, M.; Zhao, J.; Chittiboyina, A.G.; Tabanca, N.; Ali, A.; Baerson, S.R.; Techen, N.; Chappell, J.; Khan, I.A.; et al. Investigating sesquiterpene biosynthesis in Ginkgo biloba: Molecular cloning and functional characterization of (E,E)-farnesol and α-bisabolene synthases. Plant Mol. Biol. 2015, 89, 451–462. [Google Scholar] [CrossRef] [PubMed]
- Pérez, L.M.; Taucher, G.; Cori, O. Hydrolysis of allylic phosphates by enzymes from the flavedo of Citrus sinensis. Phytochemistry 1980, 19, 183–197. [Google Scholar] [CrossRef]
- Green, S.A.; Chen, X.; Nieuwenhuizen, N.J.; Matich, A.J.; Wang, M.Y.; Bunn, B.J.; Yauk, Y.K.; Atkinson, R.G. Identification, functional characterization, and regulation of the enzyme responsible for floral (E)-nerolidol biosynthesis in kiwifruit (Actinidia chinensis). J. Exp. Bot. 2012, 63, 1951–1967. [Google Scholar] [CrossRef]
- Fenton, R.; Davies, W.J.; Mansfield, T.A. The role of farnesol as a regulator of stomatal opening in Sorghum. J. Exp. Bot. 1977, 28, 1043–1053. [Google Scholar] [CrossRef]
- Mansfield, T.A.; Wellburn, A.R.; Moreira, T.J. The role of abscisic acid and farnesol in the alleviation of water stress. Philos. Trans. R. Soc. London B Biol. Sci. 1978, 284, 471–482. [Google Scholar]
- Fitzpatrick, A.H.; Shrestha, N.; Bhandari, J.; Crowell, D.N. Roles for farnesol and ABA in Arabidopsis flower development. Plant Signal. Behav. 2011, 6, 1189–1191. [Google Scholar] [CrossRef]
- Huchelmann, A.; Brahim, M.S.; Gerber, E.; Tritsch, D.; Bach, T.J.; Hemmerlin, A. Farnesol-mediated shift in the metabolic origin of prenyl groups used for protein prenylation in plants. Biochimie 2016, 127, 95–102. [Google Scholar] [CrossRef]
- Hemmerlin, A.; Bach, T.J. Farnesol induced cell death and stimulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase activity in tobacco cv. Bright Yellow-2 cells. Plant Physiol. 2000, 123, 1257–1268. [Google Scholar] [CrossRef]
- Hemmerlin, A.; Gerber, E.; Feldtrauer, J.-F.; Wentzinger, L.; Hartmann, M.A.; Tritsch, D.; Hoeffler, J.F.; Rohmer, M.; Bach, T.J. A review of tobacco BY-2 cells as an excellent system to study the biosynthesis and function of sterols and other isoprenoids. Lipids 2004, 39, 723–735. [Google Scholar] [CrossRef]
- Merret, R.; Cirioni, J.R.; Bach, T.J.; Hemmerlin, A. A serine involved in actin-dependent subcellular localization of a stress-induced tobacco BY-2 hydroxymethylglutaryl-CoA reductase isoform. FEBS Lett. 2007, 581, 5295–5299. [Google Scholar] [CrossRef]
- Meigs, T.E.; Simoni, R.D. Farnesol as a regulator of HMG-CoA reductase degradation: Characterization and role of farnesyl pyrophosphatase. Arch. Biochem. Biophys. 1997, 345, 1–9. [Google Scholar] [CrossRef]
- Abe, S.; Tsunashima, R.; Iijima, R.; Yamada, T.; Maruyama, N.; Hisajima, T.; Abe, Y.; Oshima, H.; Yamazaki, M. Suppression of anti-Candida activity of macrophages by a quorum-sensing molecule, farnesol, through induction of oxidative stress. Microbiol. Immunol. 2009, 53, 323–330. [Google Scholar] [CrossRef] [PubMed]
- Inoue, H.; Korenaga, T.; Sagami, H.; Koyama, T.; Ogura, K. Phosphorylation of farnesol by a cell-free system from Botryococcus braunii. Biochem. Biophys. Res. Commun. 1994, 200, 1036–1041. [Google Scholar] [CrossRef] [PubMed]
- Thai, L.; Rush, J.S.; Maul, J.E.; Devarenne, T.; Rodgers, D.L.; Chappell, J.; Waechter, C.J. Farnesol is utilized for isoprenoid biosynthesis in plant cells via farnesyl pyrophosphate formed by successive monophosphorylation reactions. Proc. Natl. Acad. Sci. USA 1999, 96, 13080–13085. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, A.H.; Bhandari, J.; Crowell, D.N. Farnesol kinase is involved in farnesol metabolism, ABA signaling and flower development in Arabidopsis. Plant J. 2011, 66, 1078–1088. [Google Scholar] [CrossRef] [PubMed]
- Ischebeck, T.; Zbierzak, A.M.; Kanwischer, M.; Dörmann, P. A salvage pathway for phytol metabolism in Arabidopsis. J. Biol. Chem. 2006, 281, 2470–2477. [Google Scholar] [CrossRef]
- Valentin, H.E.; Lincoln, K.; Moshiri, F.; Jensen, P.K.; Qi, Q.; Venkatesh, T.V.; Karunanandaa, B.; Baszis, S.R.; Norris, S.R.; Savidge, B.; et al. The Arabidopsis vitamin E pathway gene5-1 mutant reveals a critical role for phytol kinase in seed tocopherol biosynthesis. Plant Cell 2006, 18, 212–224. [Google Scholar] [CrossRef]
- Romer, J. The Role of the Salvage Pathway of Phytol, Geranylgeraniol, and Farnesol Phosphorylation in Arabidopsis thaliana. Doctoral Dissertation, Universitäts-und Landesbibliothek Bonn, Bonn, Germany, 2021. [Google Scholar]
- Henry, L.K.; Thomas, S.T.; Widhalm, J.R.; Lynch, J.H.; Davis, T.C.; Kessler, S.A.; Bohlmann, J.; Noel, J.P.; Dudareva, N. Contribution of isopentenyl phosphate to plant terpenoid metabolism. Nat. Plants 2018, 4, 721–729. [Google Scholar] [CrossRef]
- vom Dorp, K.; Hölzl, G.; Plohmann, C.; Eisenhut, M.; Abraham, M.; Weber, A.P.M.; Hanson, A.D.; Dörmann, P. Remobilization of phytol from chlorophyll degradation is essential for tocopherol synthesis and growth of Arabidopsis. Plant Cell 2015, 27, 2846–2859. [Google Scholar] [CrossRef] [PubMed]
- Molińska, E.; Klimczak, U.; Komaszyło, J.; Derewiaka, D.; Obiedziński, M.; Kania, M.; Danikiewicz, W.; Swiezewska, E. Double bond stereochemistry influences the susceptibility of short-chain isoprenoids and polyprenols to decomposition by thermo-oxidation. Lipids 2015, 50, 359–370. [Google Scholar] [CrossRef] [PubMed]
- Lüddeke, F.; Wülfing, A.; Timke, M.; Germer, F.; Weber, J.; Dikfidan, A.; Rahnfeld, T.; Linder, D.; Meyerdierks, A.; Harder, J. Geraniol and geranial dehydrogenases induced in anaerobic monoterpene degradation by Castellaniella defragrans. Appl. Environ. Microbiol. 2012, 78, 2128–2136. [Google Scholar] [CrossRef] [PubMed]
- de Kraker, J.W.; Schurink, M.; Franssen, M.C.; König, W.A.; de Groot, A.; Bouwmeester, H.J. Hydroxylation of sesquiterpenes by enzymes from chicory (Cichorium intybus L.) roots. Tetrahedron 2003, 59, 409–418. [Google Scholar] [CrossRef]
- Luan, F.; Mosandl, A.; Münch, A.; Wüst, M. Metabolism of geraniol in grape berry mesocarp of Vitis vinifera L. cv. Scheurebe: Demonstration of stereoselective reduction, E/Z-isomerization, oxidation and glycosylation. Phytochemistry 2005, 66, 295–303. [Google Scholar] [CrossRef]
- Potty, V.H.; Bruemmer, J.H. Oxidation of geraniol by an enzyme system from orange. Phytochemistry 1970, 9, 1003–1007. [Google Scholar] [CrossRef]
- Bhandari, J.; Fitzpatrick, A.H.; Crowell, D.N. Identification of a novel abscisic acid-regulated farnesol dehydrogenase from Arabidopsis. Plant Physiol. 2010, 154, 1116–1127. [Google Scholar] [CrossRef]
- Nagaki, M.; Imaruoka, H.; Kawakami, J.; Saga, K.; Kitahara, H.; Sagami, H.; Oba, R.; Ohya, N.; Koyama, T. Biotransformation of prenyl alcohols by cultured cells of Cucurbita maxima. J. Mol. Catal. B Enzym. 2007, 47, 33–36. [Google Scholar] [CrossRef]
- Messi, L.M.; Noté, O.P.; Mbing, J.N.; Vansteelandt, M.; Lavedan, P.; Vedrenne, M.; Pegnyemb, D.E.; Haddad, M. Farnesyl glycosides and one new triterpenoid saponin from the roots of Lecaniodiscus cupanioides Planch. ex Benth. Carbohydr. Res. 2020, 495, 108092. [Google Scholar] [CrossRef]
- Shimoda, K.; Sakamoto, S.; Nakajima, N.; Hamada, H.; Hamada, H. Synthesis of unnatural mono-and oligosaccharides of farnesol, geraniol, and (S)-perillyl alcohol by biocatalytic glycosylations. Chem. Lett. 2008, 37, 556–557. [Google Scholar] [CrossRef]
- Éparvier, V.; Thoison, O.; Bousserouel, H.; Guéritte, F.; Sévenet, T.; Litaudon, M. Cytotoxic farnesyl glycosides from Pittosporum pancheri. Phytochemistry 2007, 68, 604–608. [Google Scholar] [CrossRef] [PubMed]
- Crick, D.C.; Andres, D.A.; Waechter, C.J. Novel salvage pathway utilizing farnesol and geranylgeraniol for protein isoprenylation. Biochem. Biophys. Res. Commun. 1997, 211, 590–599. [Google Scholar] [CrossRef] [PubMed]
- Crowell, D.N.; Huizinga, D.H.; Deem, A.K.; Trobaugh, C.; Denton, R.; Sen, S.E. Arabidopsis thaliana plants possess a specific farnesylcysteine lyase that is involved in detoxification and recycling of farnesylcysteine. Plant J. 2007, 50, 839–847. [Google Scholar] [CrossRef] [PubMed]
- Rilling, H.C.; Bruenger, E.; Epstein, W.W.; Kandutsch, A.A. Prenylated proteins: Demonstration of a thioether linkage to cysteine of proteins. Biochem. Biophys. Res. Commun. 1989, 163, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Hála, M.; Žárský, V. Protein prenylation in plant stress responses. Molecules 2019, 24, 3906. [Google Scholar] [CrossRef]
- Bracha-Drori, K.; Shichrur, K.; Lubetzky, T.C.; Yalovsky, S. Functional analysis of Arabidopsis postprenylation CaaX processing enzymes and their function in subcellular protein targeting. Plant Physiol. 2008, 148, 119–131. [Google Scholar] [CrossRef]
- Eggers, R.; Jammer, A.; Jha, S.; Kerschbaumer, B.; Lahham, M.; Strandback, E.; Toplak, M.; Wallner, S.; Winkler, A.; Macheroux, P. The scope of flavin-dependent reactions and processes in the model plant Arabidopsis thaliana. Phytochemistry 2021, 189, 112822. [Google Scholar] [CrossRef]
- Endo, S.; Matsunaga, T.; Nishinaka, T. The role of AKR1B10 in physiology and pathophysiology. Metabolites 2021, 11, 332. [Google Scholar] [CrossRef]
- Brocard, L.; Immel, F.; Coulon, D.; Esnay, N.; Tuphile, K.; Pascal, S.; Claverol, S.; Fouillen, L.; Bessoule, J.J.; Bréhelin, C. Proteomic analysis of lipid droplets from Arabidopsis aging leaves brings new insight into their biogenesis and functions. Front. Plant Sci. 2017, 8, 894. [Google Scholar] [CrossRef]
- Hartmann, M.-A.; Bach, T.J. Incorporation of all-trans-farnesol into sterols and ubiquinone in Nicotiana tabacum L. cv Bright Yellow-2 cell cultures. Tetrahedron Lett. 2001, 42, 655–657. [Google Scholar] [CrossRef]
- Ku, Y.S.; Sintaha, M.; Cheung, M.Y.; Lam, H.M. Plant hormone signaling crosstalks between biotic and abiotic stress responses. Int. J. Mol. Sci. 2018, 19, 3206. [Google Scholar] [CrossRef] [PubMed]
- McTaggart, S.J. Isoprenylated proteins. Cell. Mol. Life Sci. 2006, 63, 255–267. [Google Scholar] [CrossRef]
- Bede, J.C.; Tobe, S.S. Insect juvenile hormones in plants. Stud. Nat. Prod. Chem. 2000, 22, 369–418. [Google Scholar]
- Cutler, S.; Ghassemian, M.; Bonetta, D.; Cooney, S.; McCourt, P. A protein farnesyl transferase involved in abscisic acid signal transduction in Arabidopsis. Science 1996, 273, 1239–1241. [Google Scholar] [CrossRef] [PubMed]
- Pei, Z.M.; Ghassemian, M.; Kwak, C.M.; McCourt, P.; Schroeder, J.I. Role of farnesyltransferase in ABA regulation of guard cell anion channels and plant water loss. Science 1998, 282, 287–290. [Google Scholar] [CrossRef]
- Barghetti, A.; Sjögren, L.; Floris, M.; Paredes, E.B.; Wenkel, S.; Brodersen, P. Heat-shock protein 40 is the key farnesylation target in meristem size control, abscisic acid signaling, and drought resistance. Genes Dev. 2017, 31, 2282–2295. [Google Scholar] [CrossRef]
- Dutilleul, C.; Ribeiro, I.; Blanc, N.; Nezames, C.D.; Deng, X.W.; Zglobicki, P.; Palacio Barrera, A.M.; Atehortùa, L.; Courtois, M.; Labas, V.; et al. ASG2 is a farnesylated DWD protein that acts as ABA negative regulator in Arabidopsis. Plant Cell Environ. 2016, 39, 185–198. [Google Scholar] [CrossRef]
- Lin, Y.P.; Charng, Y.Y. Chlorophyll dephytylation in chlorophyll metabolism: A simple reaction catalyzed by various enzymes. Plant Sci. 2021, 302, 110682. [Google Scholar] [CrossRef]
- Gutbrod, K.; Romer, J.; Dörmann, P. Phytol metabolism in plants. Prog. Lipid Res. 2019, 74, 1–17. [Google Scholar] [CrossRef]
- Munné-Bosch, S.; Alegre, L. The function of tocopherols and tocotrienols in plants. Crit. Rev. Plant Sci. 2002, 21, 31–57. [Google Scholar] [CrossRef]
- Maeda, H.; DellaPenna, D. Tocopherol functions in photosynthetic organisms. Curr. Opin. Plant Biol. 2007, 10, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Zechmann, B. Ultrastructure of plastids serves as reliable abiotic and biotic stress marker. PLoS ONE 2019, 14, e0214811. [Google Scholar] [CrossRef]
- Bréhélin, C.; Kessler, F. The plastoglobule: A bag full of lipid biochemistry tricks. Photochem. Photobiol. 2008, 84, 1388–1394. [Google Scholar] [CrossRef] [PubMed]
- Nacir, H.; Bréhélin, C. When proteomics reveals unsuspected roles: The plastoglobule example. Front. Plant Sci. 2013, 4, 114. [Google Scholar] [CrossRef]
- Zita, W.; Bressoud, S.; Glauser, G.; Kessler, F.; Shanmugabalaji, V. Chromoplast plastoglobules recruit the carotenoid biosynthetic pathway and contribute to carotenoid accumulation during tomato fruit maturation. PLoS ONE 2022, 17, e0277774. [Google Scholar] [CrossRef]
- Niu, Y.; Zhang, Q.; Wang, J.; Li, Y.; Wang, X.; Bao, Y. Vitamin E synthesis and response in plants. Front. Plant Sci. 2022, 13, 994058. [Google Scholar] [CrossRef]
- Albert, E.; Kim, S.; Magallanes-Lundback, M.; Bao, Y.; Deason, N.; Danilo, B.; Wu, D.; Li, X.; Wood, J.C.; Bornowski, N.; et al. Genome-wide association identifies a missing hydrolase for tocopherol synthesis in plants. Proc. Natl. Acad. Sci. USA 2022, 119, e2113488119. [Google Scholar] [CrossRef]
- Huot, B.; Yao, J.; Montgomery, B.L.; He, S.Y. Growth–defense tradeoffs in plants: A balancing act to optimize fitness. Mol. Plant 2014, 7, 1267–1287. [Google Scholar] [CrossRef]
- Ma, Y.; Cao, J.; He, J.; Chen, Q.; Li, X.; Yang, Y. Molecular mechanism for the regulation of ABA homeostasis during plant development and stress responses. Int. J. Mol. Sci. 2018, 19, 364. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.A.; Li, M.Z.; Wang, S.M.; Yin, H.J. Revisiting the role of plant transcription factors in the battle against abiotic stress. Int. J. Mol. Sci. 2018, 19, 1634. [Google Scholar] [CrossRef]
- Goyal, P.; Devi, R.; Verma, B.; Hussain, S.; Arora, P.; Tabassum, R.; Gupta, S. WRKY transcription factors: Evolution, regulation, and functional diversity in plants. Protoplasma 2023, 260, 331–348. [Google Scholar] [CrossRef] [PubMed]
- Nagegowda, D.A.; Gupta, P. Advances in biosynthesis, regulation, and metabolic engineering of plant specialized terpenoids. Plant Sci. 2020, 294, 110457. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Qiu, D.; Pang, Y.; Gao, H.; Wang, X.; Qin, Y. Diverse roles of tocopherols in responses to abiotic and biotic stresses and strategies for genetic biofortification in plants. Mol. Breed. 2020, 40, 18. [Google Scholar] [CrossRef]
- Asad, M.A.U.; Zakari, S.A.; Zhao, Q.; Zhou, L.; Ye, Y.; Cheng, F. Abiotic stresses intervene with ABA signaling to induce destructive metabolic pathways leading to death: Premature leaf senescence in plants. Int. J. Mol. Sci. 2019, 20, 256. [Google Scholar] [CrossRef]
- Huizinga, D.H.; Denton, R.; Koehler, K.G.; Tomasello, A.; Wood, L.; Sen, S.E.; Crowell, D.N. Farnesylcysteine lyase is involved in negative regulation of abscisic acid signaling in Arabidopsis. Mol. Plant 2010, 3, 143–155. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).