Use of Adult T-Cell Leukemia/Lymphoma Cell Lines in a Novel Proteomic Approach for Clarifying the Function of Human Proteins of Unknown Function
Abstract
1. Introduction
2. Overview of Human PUFs
3. ATL Cell Line Is Good Tool for Basic Biology
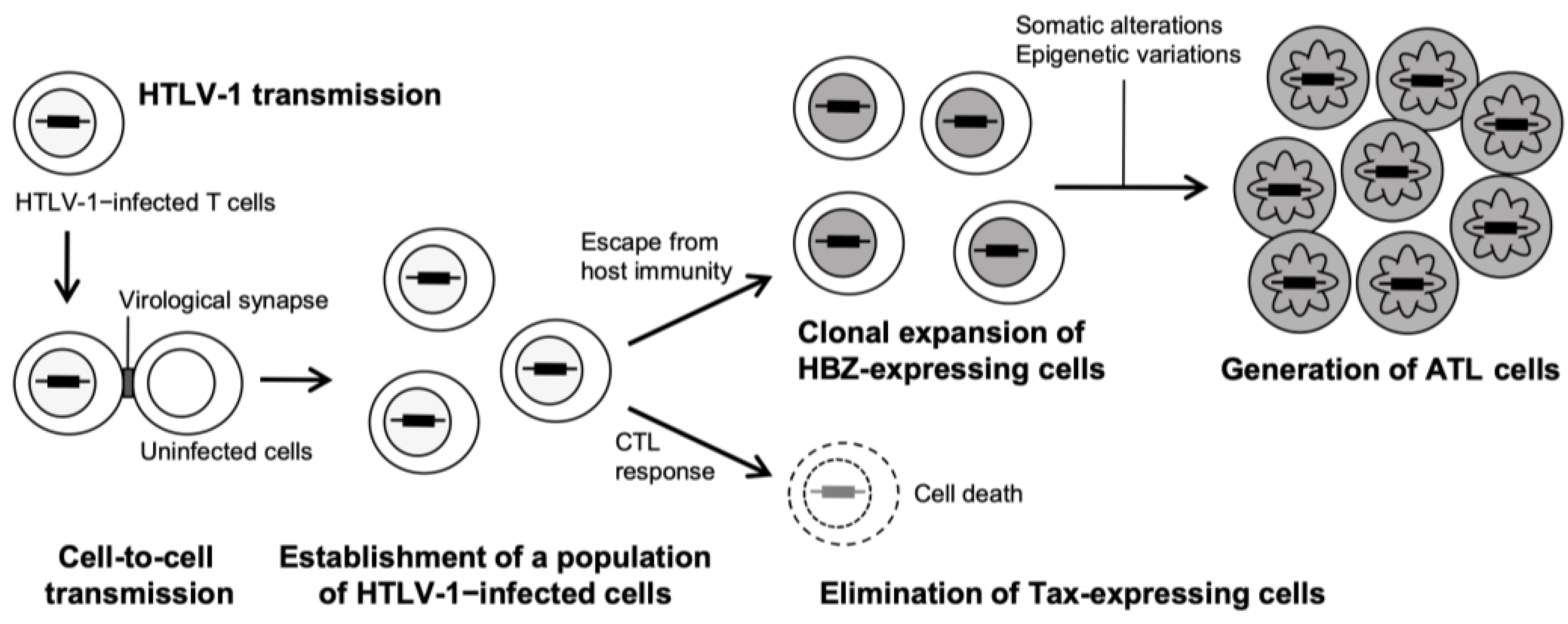
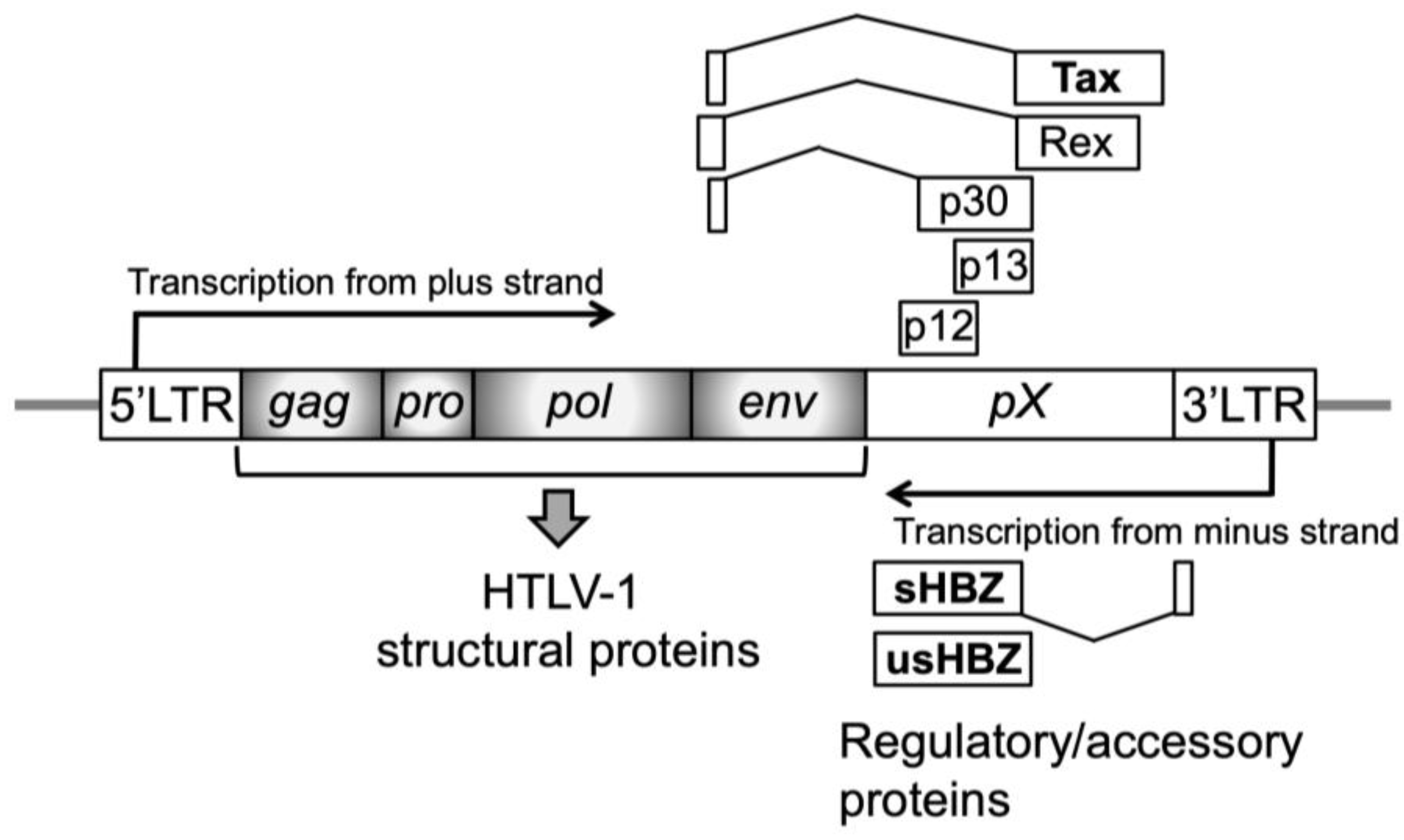
3.1. Leukemogenesis Process and Cellular Characteristics of ATL
3.2. Contribution of ATL Cell to Basic Research
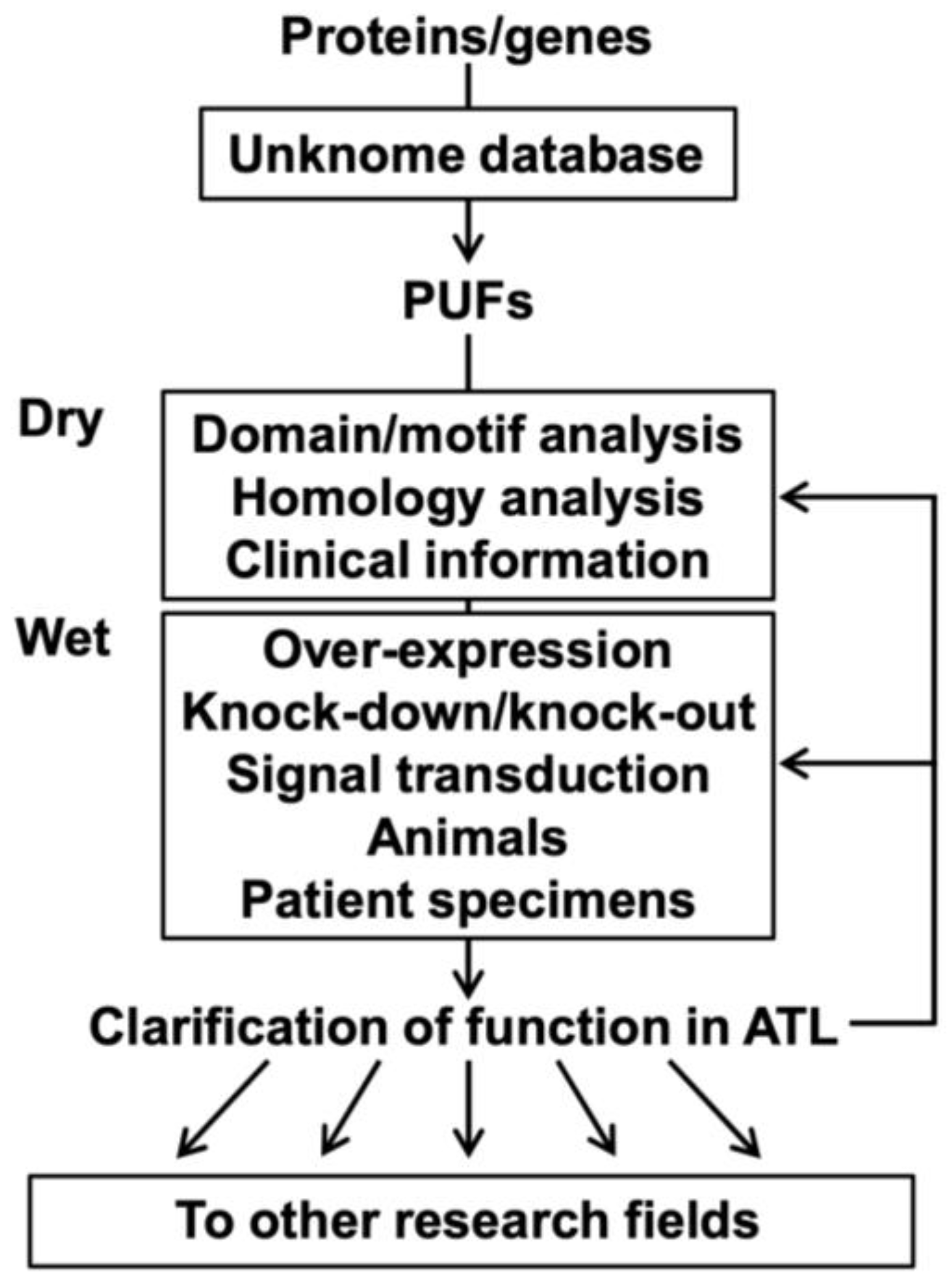
4. Challenges to PUFs in ATL Research
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PUFs | Proteins of unknown function |
| ATL | Adult T-cell leukemia/lymphoma |
| HTLV-1 | Human T-cell leukemia virus type 1 |
| IL-2 | Interleukin-2 |
| IL-2Rα | IL-2 receptor α chain |
| Treg | Regulatory T |
| GO | Gene Ontology |
| PANTHER | Protein analysis through evolutionary relationships |
| HCC | Hepatocellular carcinoma |
| HCV | Hepatitis C virus |
| CTL | Cytotoxic T lymphocyte |
| LTR | Long terminal region |
| HBZ | HTLV-1 bZIP factor |
| sHBZ | Spliced HBZ |
| usHBZ | Unspliced HBZ |
| ADF | ATL-derived factor |
| CCL | Chemokine (C-C motif) ligand |
| CCR | Chemokine (C-C motif) receptor |
| PBMC | Peripheral blood mononuclear cell |
| ADCC | Antibody-dependent cellular cytotoxicity |
References
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef]
- Venter, J.C.; Adams, M.D.; Myers, E.W.; Li, P.W.; Mural, R.J.; Sutton, G.G.; Smith, H.O.; Yandell, M.; Evans, C.A.; Holt, R.A.; et al. The sequence of the human genome. Science 2001, 291, 1304–1351. [Google Scholar] [CrossRef]
- Adhikari, S.; Nice, E.C.; Deutsch, E.W.; Lane, L.; Omenn, G.S.; Pennington, S.R.; Paik, Y.K.; Overall, C.M.; Corrales, F.J.; Cristea, I.M.; et al. A high-stringency blueprint of the human proteome. Nat. Commun. 2020, 11, 5301. [Google Scholar] [CrossRef]
- Sinha, S.; Eisenhaber, B.; Jensen, L.J.; Kalbuaji, B.; Eisenhaber, F. Darkness in the Human Gene and Protein Function Space: Widely Modest or Absent Illumination by the Life Science Literature and the Trend for Fewer Protein Function Discoveries Since 2000. Proteomics 2018, 18, e1800093. [Google Scholar] [CrossRef]
- Wood, V.; Lock, A.; Harris, M.A.; Rutherford, K.; Bahler, J.; Oliver, S.G. Hidden in plain sight: What remains to be discovered in the eukaryotic proteome? Open Biol. 2019, 9, 180241. [Google Scholar] [CrossRef]
- Hinuma, Y.; Nagata, K.; Hanaoka, M.; Nakai, M.; Matsumoto, T.; Kinoshita, K.I.; Shirakawa, S.; Miyoshi, I. Adult T-cell leukemia: Antigen in an ATL cell line and detection of antibodies to the antigen in human sera. Proc. Natl. Acad. Sci. USA 1981, 78, 6476–6480. [Google Scholar] [CrossRef] [PubMed]
- Gessain, A.; Cassar, O. Epidemiological Aspects and World Distribution of HTLV-1 Infection. Front. Microbiol. 2012, 3, 388. [Google Scholar] [CrossRef] [PubMed]
- Shimoyama, M. Diagnostic criteria and classification of clinical subtypes of adult T-cell leukaemia-lymphoma. A report from the Lymphoma Study Group (1984–1987). Br. J. Haematol. 1991, 79, 428–437. [Google Scholar] [CrossRef] [PubMed]
- Shimoyama, M. Chemotherapy of ATL; Oxford University Press: Oxford, UK, 1994. [Google Scholar]
- Katsuya, H.; Ishitsuka, K.; Utsunomiya, A.; Hanada, S.; Eto, T.; Moriuchi, Y.; Saburi, Y.; Miyahara, M.; Sueoka, E.; Uike, N.; et al. Treatment and survival among 1594 patients with ATL. Blood 2015, 126, 2570–2577. [Google Scholar] [CrossRef]
- Sekine, M.; Kameda, T.; Shide, K.; Maeda, K.; Toyama, T.; Kawano, N.; Takeuchi, M.; Kawano, H.; Sato, S.; Ishizaki, J.; et al. Higher average chemotherapy dose intensity improves prognosis in patients with aggressive adult T-cell leukemia/lymphoma. Eur. J. Haematol. 2021, 106, 398–407. [Google Scholar] [CrossRef]
- Weterings, D.A.; Rowan, A.G.; Cook, L.B. Immunological aspects of HTLV-1 persistence; for the prevention and treatment of Adult T-cell leukaemia-lymphoma (ATL). Leuk. Res. 2025, 148, 107635. [Google Scholar] [CrossRef] [PubMed]
- Epstein-Peterson, Z.D.; Gurumurthi, A.; Horwitz, S.M. New treatments for adult T-cell leukemia/lymphoma. Leuk. Res. 2025, 149, 107642. [Google Scholar] [CrossRef]
- Kanmura, S.; Uto, H.; Kusumoto, K.; Ishida, Y.; Hasuike, S.; Nagata, K.; Hayashi, K.; Ido, A.; Stuver, S.O.; Tsubouchi, H. Early diagnostic potential for hepatocellular carcinoma using the SELDI ProteinChip system. Hepatology 2007, 45, 948–956. [Google Scholar] [CrossRef] [PubMed]
- Ishida, Y.; Yamashita, K.; Sasaki, H.; Takajou, I.; Kubuki, Y.; Morishita, K.; Tsubouchi, H.; Okayama, A. Activation of complement system in adult T-cell leukemia (ATL) occurs mainly through lectin pathway: A serum proteomic approach using mass spectrometry. Cancer Lett. 2008, 271, 167–177. [Google Scholar] [CrossRef]
- Takeshita, M.; Ishida, Y.; Akamatsu, E.; Ohmori, Y.; Sudoh, M.; Uto, H.; Tsubouchi, H.; Kataoka, H. Proanthocyanidin from blueberry leaves suppresses expression of subgenomic hepatitis C virus RNA. J. Biol. Chem. 2009, 284, 21165–21176. [Google Scholar] [CrossRef] [PubMed]
- Ishida, Y.; Yamasaki, M.; Yukizaki, C.; Nishiyama, K.; Tsubouchi, H.; Okayama, A.; Kataoka, H. Carnosol, rosemary ingredient, induces apoptosis in adult T-cell leukemia/lymphoma cells via glutathione depletion: Proteomic approach using fluorescent two-dimensional differential gel electrophoresis. Hum. Cell 2014, 27, 68–77. [Google Scholar] [CrossRef]
- Hiraishi, N.; Ishida, Y.; Nagahama, M. AAA-ATPase NVL2 acts on MTR4-exosome complex to dissociate the nucleolar protein WDR74. Biochem. Biophys. Res. Commun. 2015, 467, 534–540. [Google Scholar] [CrossRef]
- Hiraishi, N.; Ishida, Y.I.; Sudo, H.; Nagahama, M. WDR74 participates in an early cleavage of the pre-rRNA processing pathway in cooperation with the nucleolar AAA-ATPase NVL2. Biochem. Biophys. Res. Commun. 2018, 495, 116–123. [Google Scholar] [CrossRef]
- Ishida, Y.I.; Miyao, S.; Saito, M.; Hiraishi, N.; Nagahama, M. Interactome analysis of the Tudor domain-containing protein SPF30 which associates with the MTR4-exosome RNA-decay machinery under the regulation of AAA-ATPase NVL2. Int. J. Biochem. Cell Biol. 2021, 132, 105919. [Google Scholar] [CrossRef]
- Kang, H.; Lee, C.J. Transmembrane proteins with unknown function (TMEMs) as ion channels: Electrophysiological properties, structure, and pathophysiological roles. Exp. Mol. Med. 2024, 56, 850–860. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Thomas, P.D.; Campbell, M.J.; Kejariwal, A.; Mi, H.; Karlak, B.; Daverman, R.; Diemer, K.; Muruganujan, A.; Narechania, A. PANTHER: A library of protein families and subfamilies indexed by function. Genome Res. 2003, 13, 2129–2141. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef]
- Rouillard, A.D.; Gundersen, G.W.; Fernandez, N.F.; Wang, Z.; Monteiro, C.D.; McDermott, M.G.; Ma’ayan, A. The harmonizome: A collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database 2016, 2016, baw100. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.T.; Mathias, S.; Bologa, C.; Brunak, S.; Fernandez, N.; Gaulton, A.; Hersey, A.; Holmes, J.; Jensen, L.J.; Karlsson, A.; et al. Pharos: Collating protein information to shed light on the druggable genome. Nucleic Acids Res. 2017, 45, D995–D1002. [Google Scholar] [CrossRef] [PubMed]
- Duek, P.; Gateau, A.; Bairoch, A.; Lane, L. Exploring the Uncharacterized Human Proteome Using neXtProt. J. Proteome Res. 2018, 17, 4211–4226. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Milacic, M.; Beavers, D.; Conley, P.; Gong, C.; Gillespie, M.; Griss, J.; Haw, R.; Jassal, B.; Matthews, L.; May, B.; et al. The Reactome Pathway Knowledgebase 2024. Nucleic Acids Res. 2024, 52, D672–D678. [Google Scholar] [CrossRef]
- Feuermann, M.; Mi, H.; Gaudet, P.; Muruganujan, A.; Lewis, S.E.; Ebert, D.; Mushayahama, T.; Gene Ontology, C.; Thomas, P.D. A compendium of human gene functions derived from evolutionary modelling. Nature 2025, 640, 146–154. [Google Scholar] [CrossRef]
- Pena-Castillo, L.; Hughes, T.R. Why are there still over 1000 uncharacterized yeast genes? Genetics 2007, 176, 7–14. [Google Scholar] [CrossRef]
- Edwards, A.M.; Isserlin, R.; Bader, G.D.; Frye, S.V.; Willson, T.M.; Yu, F.H. Too many roads not taken. Nature 2011, 470, 163–165. [Google Scholar] [CrossRef] [PubMed]
- Haynes, W.A.; Tomczak, A.; Khatri, P. Gene annotation bias impedes biomedical research. Sci. Rep. 2018, 8, 1362. [Google Scholar] [CrossRef]
- Stoeger, T.; Gerlach, M.; Morimoto, R.I.; Nunes Amaral, L.A. Large-scale investigation of the reasons why potentially important genes are ignored. PLoS Biol. 2018, 16, e2006643. [Google Scholar] [CrossRef]
- Oprea, T.I.; Bologa, C.G.; Brunak, S.; Campbell, A.; Gan, G.N.; Gaulton, A.; Gomez, S.M.; Guha, R.; Hersey, A.; Holmes, J.; et al. Unexplored therapeutic opportunities in the human genome. Nat. Rev. Drug Discov. 2018, 17, 317–332. [Google Scholar] [CrossRef]
- Rocha, J.J.; Jayaram, S.A.; Stevens, T.J.; Muschalik, N.; Shah, R.D.; Emran, S.; Robles, C.; Freeman, M.; Munro, S. Functional unknomics: Systematic screening of conserved genes of unknown function. PLoS Biol. 2023, 21, e3002222. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, M.; Jeang, K.T. Human T-cell leukaemia virus type 1 (HTLV-1) infectivity and cellular transformation. Nat. Rev. Cancer 2007, 7, 270–280. [Google Scholar] [CrossRef]
- Satou, Y.; Matsuoka, M. Molecular and Cellular Mechanism of Leukemogenesis of ATL: Emergent Evidence of a Significant Role for HBZ in HTLV-1-Induced Pathogenesis. Leuk. Res. Treat. 2012, 2012, 213653. [Google Scholar] [CrossRef] [PubMed]
- Poiesz, B.J.; Ruscetti, F.W.; Gazdar, A.F.; Bunn, P.A.; Minna, J.D.; Gallo, R.C. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc. Natl. Acad. Sci. USA 1980, 77, 7415–7419. [Google Scholar] [CrossRef]
- Yoshida, M.; Miyoshi, I.; Hinuma, Y. Isolation and characterization of retrovirus from cell lines of human adult T-cell leukemia and its implication in the disease. Proc. Natl. Acad. Sci. USA 1982, 79, 2031–2035. [Google Scholar] [CrossRef]
- Barbeau, B.; Mesnard, J.M. Does the HBZ gene represent a new potential target for the treatment of adult T-cell leukemia? Int. Rev. Immunol. 2007, 26, 283–304. [Google Scholar] [CrossRef]
- Gaudray, G.; Gachon, F.; Basbous, J.; Biard-Piechaczyk, M.; Devaux, C.; Mesnard, J.M. The complementary strand of the human T-cell leukemia virus type 1 RNA genome encodes a bZIP transcription factor that down-regulates viral transcription. J. Virol. 2002, 76, 12813–12822. [Google Scholar] [CrossRef] [PubMed]
- Satou, Y.; Yasunaga, J.; Zhao, T.; Yoshida, M.; Miyazato, P.; Takai, K.; Shimizu, K.; Ohshima, K.; Green, P.L.; Ohkura, N.; et al. HTLV-1 bZIP factor induces T-cell lymphoma and systemic inflammation in vivo. PLoS Pathog. 2011, 7, e1001274. [Google Scholar] [CrossRef] [PubMed]
- Romerio, F. Origin and functional role of antisense transcription in endogenous and exogenous retroviruses. Retrovirology 2023, 20, 6. [Google Scholar] [CrossRef]
- Matsuoka, M.; Yasunaga, J. Human T-cell leukemia virus type 1: Replication, proliferation and propagation by Tax and HTLV-1 bZIP factor. Curr. Opin. Virol. 2013, 3, 684–691. [Google Scholar] [CrossRef] [PubMed]
- Lemasson, I.; Lewis, M.R.; Polakowski, N.; Hivin, P.; Cavanagh, M.H.; Thebault, S.; Barbeau, B.; Nyborg, J.K.; Mesnard, J.M. Human T-cell leukemia virus type 1 (HTLV-1) bZIP protein interacts with the cellular transcription factor CREB to inhibit HTLV-1 transcription. J. Virol. 2007, 81, 1543–1553. [Google Scholar] [CrossRef]
- Zhao, T.; Yasunaga, J.; Satou, Y.; Nakao, M.; Takahashi, M.; Fujii, M.; Matsuoka, M. Human T-cell leukemia virus type 1 bZIP factor selectively suppresses the classical pathway of NF-kappaB. Blood 2009, 113, 2755–2764. [Google Scholar] [CrossRef]
- Kannagi, M.; Harada, S.; Maruyama, I.; Inoko, H.; Igarashi, H.; Kuwashima, G.; Sato, S.; Morita, M.; Kidokoro, M.; Sugimoto, M.; et al. Predominant recognition of human T cell leukemia virus type I (HTLV-I) pX gene products by human CD8+ cytotoxic T cells directed against HTLV-I-infected cells. Int. Immunol. 1991, 3, 761–767. [Google Scholar] [CrossRef]
- Goon, P.K.; Biancardi, A.; Fast, N.; Igakura, T.; Hanon, E.; Mosley, A.J.; Asquith, B.; Gould, K.G.; Marshall, S.; Taylor, G.P.; et al. Human T cell lymphotropic virus (HTLV) type-1-specific CD8+ T cells: Frequency and immunodominance hierarchy. J. Infect. Dis. 2004, 189, 2294–2298. [Google Scholar] [CrossRef]
- Suemori, K.; Fujiwara, H.; Ochi, T.; Ogawa, T.; Matsuoka, M.; Matsumoto, T.; Mesnard, J.M.; Yasukawa, M. HBZ is an immunogenic protein, but not a target antigen for human T-cell leukemia virus type 1-specific cytotoxic T lymphocytes. J. Gen. Virol. 2009, 90, 1806–1811. [Google Scholar] [CrossRef]
- Gazon, H.; Chauhan, P.; Hamaidia, M.; Hoyos, C.; Li, L.; Safari, R.; Willems, L. How Does HTLV-1 Undergo Oncogene-Dependent Replication Despite a Strong Immune Response? Front. Microbiol. 2017, 8, 2684. [Google Scholar] [CrossRef]
- El Hajj, H.; Bazarbachi, A. Interplay between innate immunity and the viral oncoproteins Tax and HBZ in the pathogenesis and therapeutic response of HTLV-1 associated adult T cell leukemia. Front. Immunol. 2022, 13, 957535. [Google Scholar] [CrossRef]
- Raval, G.U.; Bidoia, C.; Forlani, G.; Tosi, G.; Gessain, A.; Accolla, R.S. Localization, quantification and interaction with host factors of endogenous HTLV-1 HBZ protein in infected cells and ATL. Retrovirology 2015, 12, 59. [Google Scholar] [CrossRef]
- Accolla, R.S. The Road to HTLV-1-Induced Leukemia by Following the Subcellular Localization of HTLV-1-Encoded HBZ Protein. Front. Immunol. 2022, 13, 940131. [Google Scholar] [CrossRef]
- Kataoka, K.; Nagata, Y.; Kitanaka, A.; Shiraishi, Y.; Shimamura, T.; Yasunaga, J.; Totoki, Y.; Chiba, K.; Sato-Otsubo, A.; Nagae, G.; et al. Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat. Genet. 2015, 47, 1304–1315. [Google Scholar] [CrossRef] [PubMed]
- Fujikawa, D.; Nakagawa, S.; Hori, M.; Kurokawa, N.; Soejima, A.; Nakano, K.; Yamochi, T.; Nakashima, M.; Kobayashi, S.; Tanaka, Y.; et al. Polycomb-dependent epigenetic landscape in adult T-cell leukemia. Blood 2016, 127, 1790–1802. [Google Scholar] [CrossRef] [PubMed]
- Katsuya, H.; Ishitsuka, K. Treatment advances and prognosis for patients with adult T-cell leukemia-lymphoma. J. Clin. Exp. Hematop. 2017, 57, 87–97. [Google Scholar] [CrossRef]
- Letafati, A.; Mehdigholian Chaijani, R.; Edalat, F.; Eslami, N.; Askari, H.; Askari, F.; Shirvani, S.; Talebzadeh, H.; Tarahomi, M.; MirKhani, N.; et al. Advances in epigenetic treatment of adult T-cell leukemia/lymphoma: A comprehensive review. Clin. Epigenetics 2025, 17, 39. [Google Scholar] [CrossRef]
- Liu, M.M.; Furusato, E.; Cao, X.; Shen, D.; Chan, C.C. Ocular manifestations and pathology of adult T-cell leukemia/lymphoma associated with human T-lymphotropic virus type 1. Rare Tumors 2010, 2, e63. [Google Scholar] [CrossRef]
- Tezuka, K.; Xun, R.; Tei, M.; Ueno, T.; Tanaka, M.; Takenouchi, N.; Fujisawa, J. An animal model of adult T-cell leukemia: Humanized mice with HTLV-1-specific immunity. Blood 2014, 123, 346–355. [Google Scholar] [CrossRef]
- Fuji, S.; Muta, M.; Hisakata, T.; Kawano, N.; Ikeda, E.; Kouno, H.; Tanabe, Y.; Nakanishi, K. Comprehensive review of morphological diagnosis of adult T-cell leukemia-lymphoma. Expert. Rev. Hematol. 2025, 18, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Shimoyama, M.; Minato, K.; Tobinai, K.; Nagai, M.; Setoya, T.; Takenaka, T.; Ishihara, K.; Watanabe, S.; Hoshino, H.; Miwa, M.; et al. Atypical adult T-cell leukemia-lymphoma: Diverse clinical manifestations of adult T-cell leukemia-lymphoma. Jpn. J. Clin. Oncol. 1983, 13 (Suppl. S2), 165–187. [Google Scholar] [PubMed]
- Okamoto, T.; Ohno, Y.; Tsugane, S.; Watanabe, S.; Shimoyama, M.; Tajima, K.; Miwa, M.; Shimotohno, K. Multi-step carcinogenesis model for adult T-cell leukemia. Jpn. J. Cancer Res. 1989, 80, 191–195. [Google Scholar] [CrossRef]
- Koya, J.; Saito, Y.; Kameda, T.; Kogure, Y.; Yuasa, M.; Nagasaki, J.; McClure, M.B.; Shingaki, S.; Tabata, M.; Tahira, Y.; et al. Single-Cell Analysis of the Multicellular Ecosystem in Viral Carcinogenesis by HTLV-1. Blood Cancer Discov. 2021, 2, 450–467. [Google Scholar] [CrossRef]
- Hattori, T.; Uchiyama, T.; Toibana, T.; Takatsuki, K.; Uchino, H. Surface phenotype of Japanese adult T-cell leukemia cells characterized by monoclonal antibodies. Blood 1981, 58, 645–647. [Google Scholar] [CrossRef]
- Nikaido, T.; Shimizu, A.; Ishida, N.; Sabe, H.; Teshigawara, K.; Maeda, M.; Uchiyama, T.; Yodoi, J.; Honjo, T. Molecular cloning of cDNA encoding human interleukin-2 receptor. Nature 1984, 311, 631–635. [Google Scholar] [CrossRef]
- Teshigawara, K.; Maeda, M.; Nishino, K.; Nikaido, T.; Uchiyama, T.; Tsudo, M.; Wano, Y.; Yodoi, J. Adult T leukemia cells produce a lymphokine that augments interleukin 2 receptor expression. J. Mol. Cell Immunol. 1985, 2, 17–26. [Google Scholar] [PubMed]
- Tagaya, Y.; Maeda, Y.; Mitsui, A.; Kondo, N.; Matsui, H.; Hamuro, J.; Brown, N.; Arai, K.; Yokota, T.; Wakasugi, H.; et al. ATL-derived factor (ADF), an IL-2 receptor/Tac inducer homologous to thioredoxin; possible involvement of dithiol-reduction in the IL-2 receptor induction. EMBO J. 1989, 8, 757–764. [Google Scholar] [CrossRef]
- Wakasugi, N.; Tagaya, Y.; Wakasugi, H.; Mitsui, A.; Maeda, M.; Yodoi, J.; Tursz, T. Adult T-cell leukemia-derived factor/thioredoxin, produced by both human T-lymphotropic virus type I- and Epstein-Barr virus-transformed lymphocytes, acts as an autocrine growth factor and synergizes with interleukin 1 and interleukin 2. Proc. Natl. Acad. Sci. USA 1990, 87, 8282–8286. [Google Scholar] [CrossRef]
- Yoshie, O.; Fujisawa, R.; Nakayama, T.; Harasawa, H.; Tago, H.; Izawa, D.; Hieshima, K.; Tatsumi, Y.; Matsushima, K.; Hasegawa, H.; et al. Frequent expression of CCR4 in adult T-cell leukemia and human T-cell leukemia virus type 1-transformed T cells. Blood 2002, 99, 1505–1511. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, T.; Yodoi, J.; Sagawa, K.; Takatsuki, K.; Uchino, H. Adult T-cell leukemia: Clinical and hematologic features of 16 cases. Blood 1977, 50, 481–492. [Google Scholar] [CrossRef]
- Watanabe, T.; Seiki, M.; Yoshida, M. Retrovirus terminology. Science 1983, 222, 1178. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Seiki, M.; Yoshida, M. HTLV type I (U. S. isolate) and ATLV (Japanese isolate) are the same species of human retrovirus. Virology 1984, 133, 238–241. [Google Scholar] [CrossRef]
- Gallo, R.C.; Montagnier, L. The discovery of HIV as the cause of AIDS. N. Engl. J. Med. 2003, 349, 2283–2285. [Google Scholar] [CrossRef]
- Hinuma, Y. Retrovirus etiology of adult T-cell leukemia. Leuk. Res. 1993, 17, 379–381. [Google Scholar] [CrossRef]
- de The, G. The history of HTLV-I. Leuk. Res. 1993, 17, 383–384. [Google Scholar] [CrossRef]
- Blattner, W.A. HTLV-I and adult T-cell leukemia. Leuk. Res. 1993, 17, 385–386. [Google Scholar] [CrossRef]
- Aoki, T. Discovery of HTLV-1. Leuk. Res. 1993, 17, 387–388. [Google Scholar] [CrossRef] [PubMed]
- Hatano, Y.; Ideta, T.; Hirata, A.; Hatano, K.; Tomita, H.; Okada, H.; Shimizu, M.; Tanaka, T.; Hara, A. Virus-Driven Carcinogenesis. Cancers 2021, 13, 2625. [Google Scholar] [CrossRef] [PubMed]
- Leonard, W.J.; Depper, J.M.; Crabtree, G.R.; Rudikoff, S.; Pumphrey, J.; Robb, R.J.; Kronke, M.; Svetlik, P.B.; Peffer, N.J.; Waldmann, T.A.; et al. Molecular cloning and expression of cDNAs for the human interleukin-2 receptor. Nature 1984, 311, 626–631. [Google Scholar] [CrossRef]
- Sakaguchi, S.; Sakaguchi, N.; Asano, M.; Itoh, M.; Toda, M. Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J. Immunol. 1995, 155, 1151–1164. [Google Scholar] [CrossRef]
- Wood, K.J.; Sakaguchi, S. Regulatory T cells in transplantation tolerance. Nat. Rev. Immunol. 2003, 3, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Bien, E.; Balcerska, A. Serum soluble interleukin 2 receptor alpha in human cancer of adults and children: A review. Biomarkers 2008, 13, 1–26. [Google Scholar] [CrossRef]
- Yasuda, N.; Lai, P.K.; Ip, S.H.; Kung, P.C.; Hinuma, Y.; Matsuoka, M.; Hattori, T.; Takatsuki, K.; Purtilo, D.T. Soluble interleukin 2 receptors in sera of Japanese patients with adult T cell leukemia mark activity of disease. Blood 1988, 71, 1021–1026. [Google Scholar] [CrossRef] [PubMed]
- Lokau, J.; Petasch, L.M.; Garbers, C. The soluble IL-2 receptor alpha/CD25 as a modulator of IL-2 function. Immunology 2024, 171, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Brandl, N.; Seitz, R.; Sendtner, N.; Muller, M.; Gulow, K. Living on the Edge: ROS Homeostasis in Cancer Cells and Its Potential as a Therapeutic Target. Antioxidants 2025, 14, 1002. [Google Scholar] [CrossRef]
- Li, B.; Ming, H.; Qin, S.; Nice, E.C.; Dong, J.; Du, Z.; Huang, C. Redox regulation: Mechanisms, biology and therapeutic targets in diseases. Signal Transduct. Target. Ther. 2025, 10, 72. [Google Scholar] [CrossRef]
- Jones, D.P.; Sies, H. The Redox Code. Antioxid. Redox Signal 2015, 23, 734–746. [Google Scholar] [CrossRef]
- Silic-Benussi, M.; Cavallari, I.; Vajente, N.; Vidali, S.; Chieco-Bianchi, L.; Di Lisa, F.; Saggioro, D.; D’Agostino, D.M.; Ciminale, V. Redox regulation of T-cell turnover by the p13 protein of human T-cell leukemia virus type 1: Distinct effects in primary versus transformed cells. Blood 2010, 116, 54–62. [Google Scholar] [CrossRef]
- Takahashi, M.; Higuchi, M.; Makokha, G.N.; Matsuki, H.; Yoshita, M.; Tanaka, Y.; Fujii, M. HTLV-1 Tax oncoprotein stimulates ROS production and apoptosis in T cells by interacting with USP10. Blood 2013, 122, 715–725. [Google Scholar] [CrossRef]
- Romeo, M.; Hutchison, T.; Malu, A.; White, A.; Kim, J.; Gardner, R.; Smith, K.; Nelson, K.; Bergeson, R.; McKee, R.; et al. The human T-cell leukemia virus type-1 p30(II) protein activates p53 and induces the TIGAR and suppresses oncogene-induced oxidative stress during viral carcinogenesis. Virology 2018, 518, 103–115. [Google Scholar] [CrossRef]
- Rushing, A.W.; Rushing, B.; Hoang, K.; Sanders, S.V.; Peloponese, J.M., Jr.; Polakowski, N.; Lemasson, I. HTLV-1 basic leucine zipper factor protects cells from oxidative stress by upregulating expression of Heme Oxygenase I. PLoS Pathog. 2019, 15, e1007922. [Google Scholar] [CrossRef]
- Wollman, E.E.; d’Auriol, L.; Rimsky, L.; Shaw, A.; Jacquot, J.P.; Wingfield, P.; Graber, P.; Dessarps, F.; Robin, P.; Galibert, F.; et al. Cloning and expression of a cDNA for human thioredoxin. J. Biol. Chem. 1988, 263, 15506–15512. [Google Scholar] [CrossRef]
- Powis, G.; Mustacich, D.; Coon, A. The role of the redox protein thioredoxin in cell growth and cancer. Free Radic. Biol. Med. 2000, 29, 312–322. [Google Scholar] [CrossRef] [PubMed]
- Oberacker, T.; Kraft, L.; Schanz, M.; Latus, J.; Schricker, S. The Importance of Thioredoxin-1 in Health and Disease. Antioxidants 2023, 12, 1078. [Google Scholar] [CrossRef]
- Ishii, T.; Ishida, T.; Utsunomiya, A.; Inagaki, A.; Yano, H.; Komatsu, H.; Iida, S.; Imada, K.; Uchiyama, T.; Akinaga, S.; et al. Defucosylated humanized anti-CCR4 monoclonal antibody KW-0761 as a novel immunotherapeutic agent for adult T-cell leukemia/lymphoma. Clin. Cancer Res. 2010, 16, 1520–1531. [Google Scholar] [CrossRef] [PubMed]
- Yoshie, O.; Matsushima, K. CCR4 and its ligands: From bench to bedside. Int. Immunol. 2015, 27, 11–20. [Google Scholar] [CrossRef]
- Baggiolini, M.; Moser, B. Blocking chemokine receptors. J. Exp. Med. 1997, 186, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
- Sherry, B.; Cerami, A. Small cytokine superfamily. Curr. Opin. Immunol. 1991, 3, 56–60. [Google Scholar] [CrossRef]
- Imai, T.; Yoshida, T.; Baba, M.; Nishimura, M.; Kakizaki, M.; Yoshie, O. Molecular cloning of a novel T cell-directed CC chemokine expressed in thymus by signal sequence trap using Epstein-Barr virus vector. J. Biol. Chem. 1996, 271, 21514–21521. [Google Scholar] [CrossRef]
- Imai, T.; Baba, M.; Nishimura, M.; Kakizaki, M.; Takagi, S.; Yoshie, O. The T cell-directed CC chemokine TARC is a highly specific biological ligand for CC chemokine receptor 4. J. Biol. Chem. 1997, 272, 15036–15042. [Google Scholar] [CrossRef]
- Matsushima, K. Chemokines. Introduction. Springer Semin. Immunopathol. 2000, 22, 321–328. [Google Scholar] [CrossRef]
- Imai, T.; Nagira, M.; Takagi, S.; Kakizaki, M.; Nishimura, M.; Wang, J.; Gray, P.W.; Matsushima, K.; Yoshie, O. Selective recruitment of CCR4-bearing Th2 cells toward antigen-presenting cells by the CC chemokines thymus and activation-regulated chemokine and macrophage-derived chemokine. Int. Immunol. 1999, 11, 81–88. [Google Scholar] [CrossRef]
- Niwa, R.; Shoji-Hosaka, E.; Sakurada, M.; Shinkawa, T.; Uchida, K.; Nakamura, K.; Matsushima, K.; Ueda, R.; Hanai, N.; Shitara, K. Defucosylated chimeric anti-CC chemokine receptor 4 IgG1 with enhanced antibody-dependent cellular cytotoxicity shows potent therapeutic activity to T-cell leukemia and lymphoma. Cancer Res. 2004, 64, 2127–2133. [Google Scholar] [CrossRef] [PubMed]
- Ogura, M.; Ishida, T.; Hatake, K.; Taniwaki, M.; Ando, K.; Tobinai, K.; Fujimoto, K.; Yamamoto, K.; Miyamoto, T.; Uike, N.; et al. Multicenter phase II study of mogamulizumab (KW-0761), a defucosylated anti-cc chemokine receptor 4 antibody, in patients with relapsed peripheral T-cell lymphoma and cutaneous T-cell lymphoma. J. Clin. Oncol. 2014, 32, 1157–1163. [Google Scholar] [CrossRef] [PubMed]
- Yoshimitsu, M. Targeted antibody therapy as a treatment strategy for aggressive adult T-cell leukemia/lymphoma. Leuk. Res. 2025, 149, 107653. [Google Scholar] [CrossRef]
- Yoshimitsu, M.; Choi, I.; Kusumoto, S.; Shimokawa, M.; Utsunomiya, A.; Suehiro, Y.; Hidaka, T.; Nosaka, K.; Sasaki, H.; Rai, S.; et al. A phase 2 trial of CHOP with anti-CCR4 antibody mogamulizumab for older patients with adult T-cell leukemia/lymphoma. Blood 2025, 146, 1440–1449. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, H.; Nishikata, I.; Shiraga, T.; Akamatsu, E.; Fukami, T.; Hidaka, T.; Kubuki, Y.; Okayama, A.; Hamada, K.; Okabe, H.; et al. Overexpression of a cell adhesion molecule, TSLC1, as a possible molecular marker for acute-type adult T-cell leukemia. Blood 2005, 105, 1204–1213. [Google Scholar] [CrossRef] [PubMed]
- Maes, E.; Mertens, I.; Valkenborg, D.; Pauwels, P.; Rolfo, C.; Baggerman, G. Proteomics in cancer research: Are we ready for clinical practice? Crit. Rev. Oncol. Hematol. 2015, 96, 437–448. [Google Scholar] [CrossRef]
- Sudo, H.; Tonoyama, Y.; Ikebe, E.; Hasegawa, H.; Iha, H.; Ishida, Y.I. Proteomic analysis of adult T-cell leukemia/lymphoma: A biomarker identification strategy based on preparation and in-solution digestion methods of total proteins. Leuk. Res. 2024, 138, 107454. [Google Scholar] [CrossRef]
- Yin, J.; Fu, W.; Dai, L.; Jiang, Z.; Liao, H.; Chen, W.; Pan, L.; Zhao, J. ANKRD22 promotes progression of non-small cell lung cancer through transcriptional up-regulation of E2F1. Sci. Rep. 2017, 7, 4430. [Google Scholar] [CrossRef]
- Utsumi, T.; Hosokawa, T.; Shichita, M.; Nishiue, M.; Iwamoto, N.; Harada, H.; Kiwado, A.; Yano, M.; Otsuka, M.; Moriya, K. ANKRD22 is an N-myristoylated hairpin-like monotopic membrane protein specifically localized to lipid droplets. Sci. Rep. 2021, 11, 19233. [Google Scholar] [CrossRef] [PubMed]
- Abe, M.; Uchihashi, K.; Kazuto, T.; Osaka, A.; Yanagihara, K.; Tsukasaki, K.; Hasegawa, H.; Yamada, Y.; Kamihira, S. Foxp3 expression on normal and leukemic CD4+CD25+ T cells implicated in human T-cell leukemia virus type-1 is inconsistent with Treg cells. Eur. J. Haematol. 2008, 81, 209–217. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tonoyama, Y.; Ishida, Y.-i. Use of Adult T-Cell Leukemia/Lymphoma Cell Lines in a Novel Proteomic Approach for Clarifying the Function of Human Proteins of Unknown Function. Lymphatics 2025, 3, 38. https://doi.org/10.3390/lymphatics3040038
Tonoyama Y, Ishida Y-i. Use of Adult T-Cell Leukemia/Lymphoma Cell Lines in a Novel Proteomic Approach for Clarifying the Function of Human Proteins of Unknown Function. Lymphatics. 2025; 3(4):38. https://doi.org/10.3390/lymphatics3040038
Chicago/Turabian StyleTonoyama, Yasuhiro, and Yo-ichi Ishida. 2025. "Use of Adult T-Cell Leukemia/Lymphoma Cell Lines in a Novel Proteomic Approach for Clarifying the Function of Human Proteins of Unknown Function" Lymphatics 3, no. 4: 38. https://doi.org/10.3390/lymphatics3040038
APA StyleTonoyama, Y., & Ishida, Y.-i. (2025). Use of Adult T-Cell Leukemia/Lymphoma Cell Lines in a Novel Proteomic Approach for Clarifying the Function of Human Proteins of Unknown Function. Lymphatics, 3(4), 38. https://doi.org/10.3390/lymphatics3040038






