Computational and Experimental Investigation of the Combined Effect of Various 3D Scaffolds and Bioreactor Stimulation on Human Cells’ Feedback
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Fabrication Methods
2.1.1. Electrospinning
2.1.2. 3D Printing
2.1.3. Sterilization and Coating
2.2. Mechanical Testing
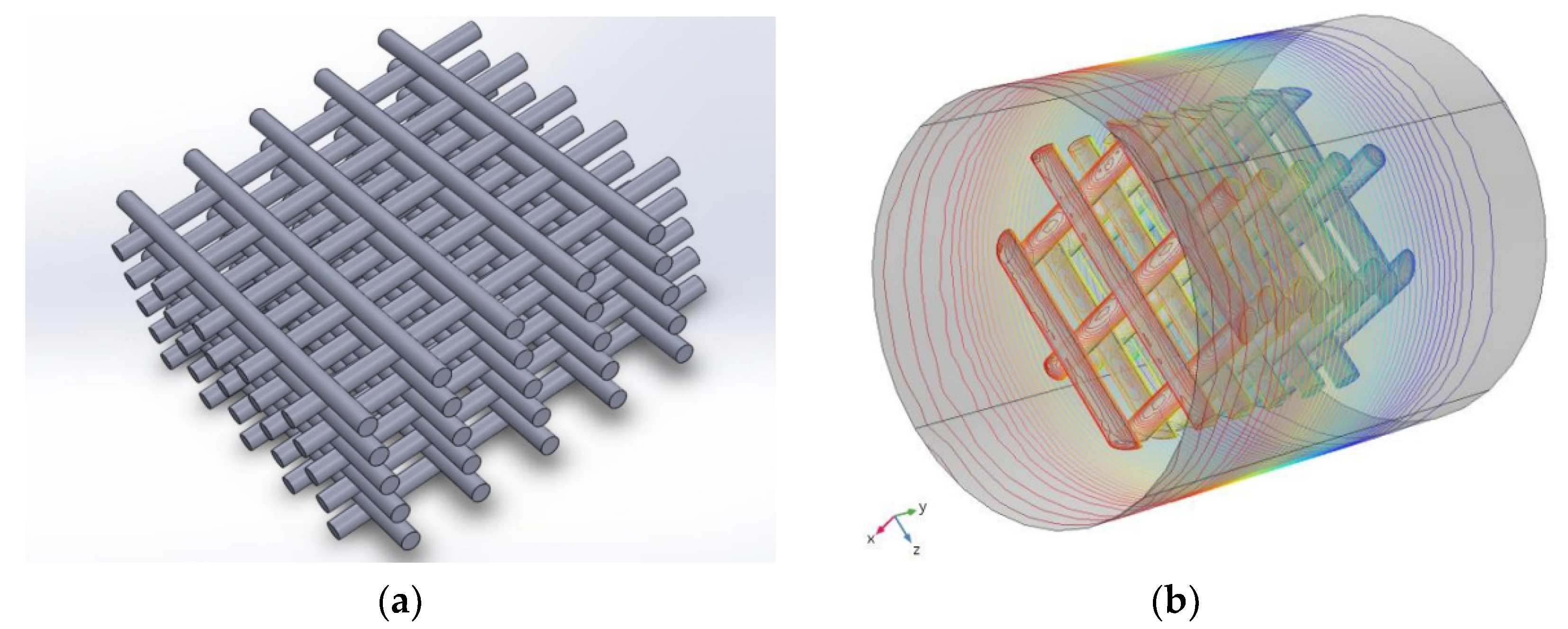
2.3. Computational Analysis
Theory Background for Computational Simulation
- Import of the geometry designed in Solidworks.
- Design of the bounding cylinder.
- Selection of laminar flow and stationary study.
- Insertion of the appropriate parameters for fluid properties, inlet velocity, zero outlet pressure, and non-slip conditions for the tube walls and the scaffold’s walls.
- Design of the mesh (triangular for the scaffold walls and tetrahedral for the remaining tube).
- Running of stationary simulation.
- Insertion of a time-dependent particle tracing for fluid flow.
- Insertion of the appropriate parameters for particle properties, inlet number of particles, drag force, freezing particles’ condition for the scaffold’s walls and the outlet.
- Running of a time-dependent simulation.
2.4. Bioreactor Setup
2.5. Cell Culture
3. Results
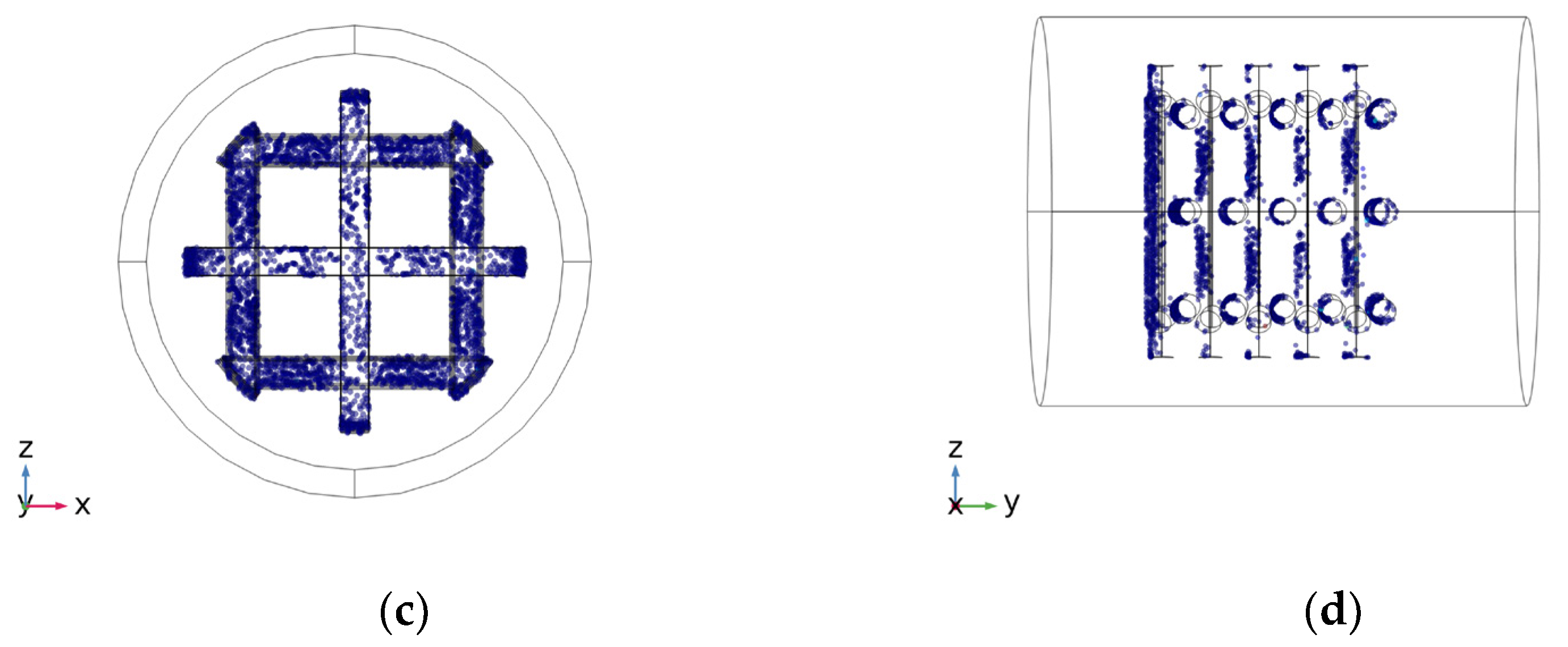
3.1. Computational Analysis
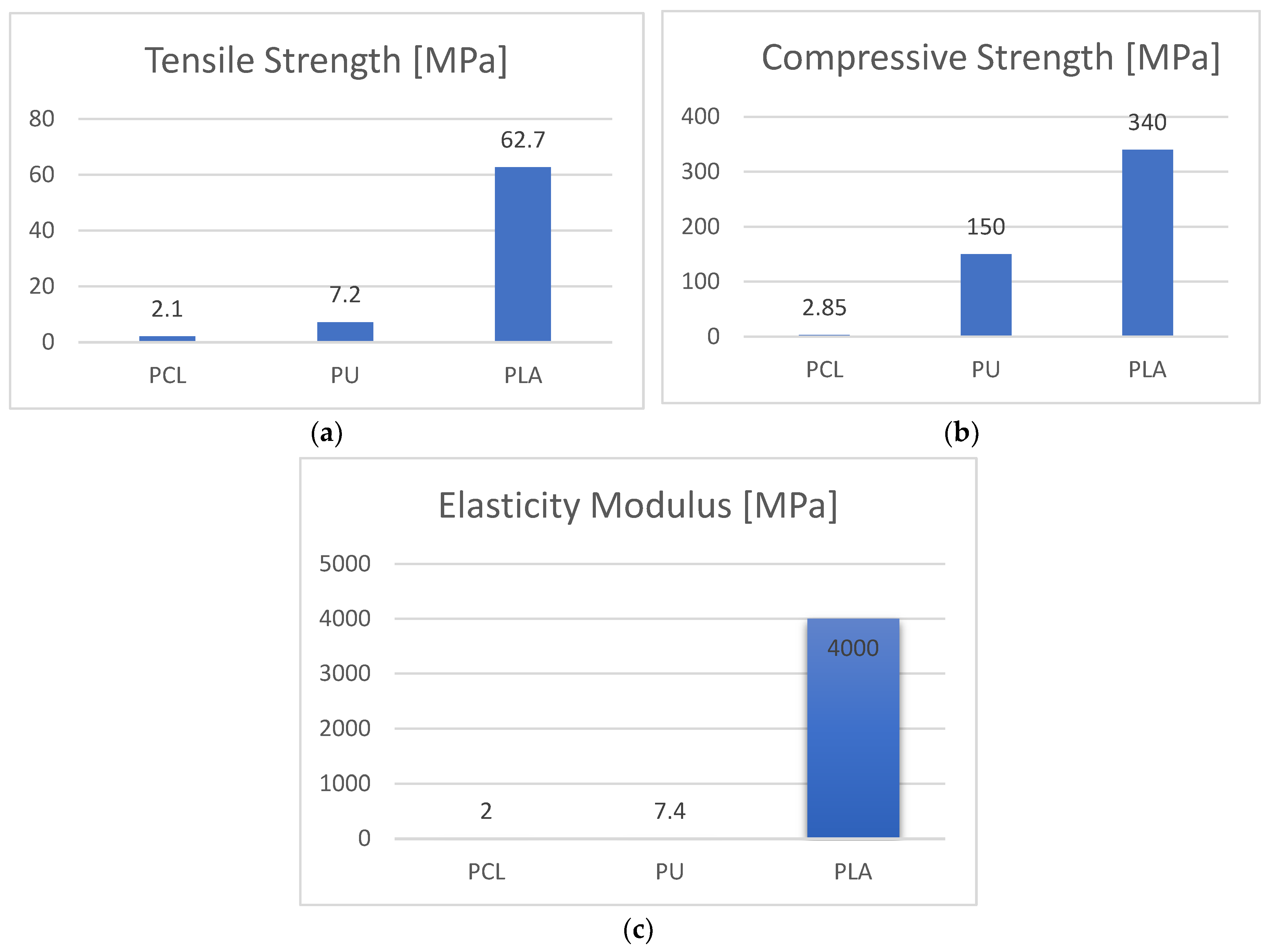
3.2. Mechanical Evaluation of the Scaffolds
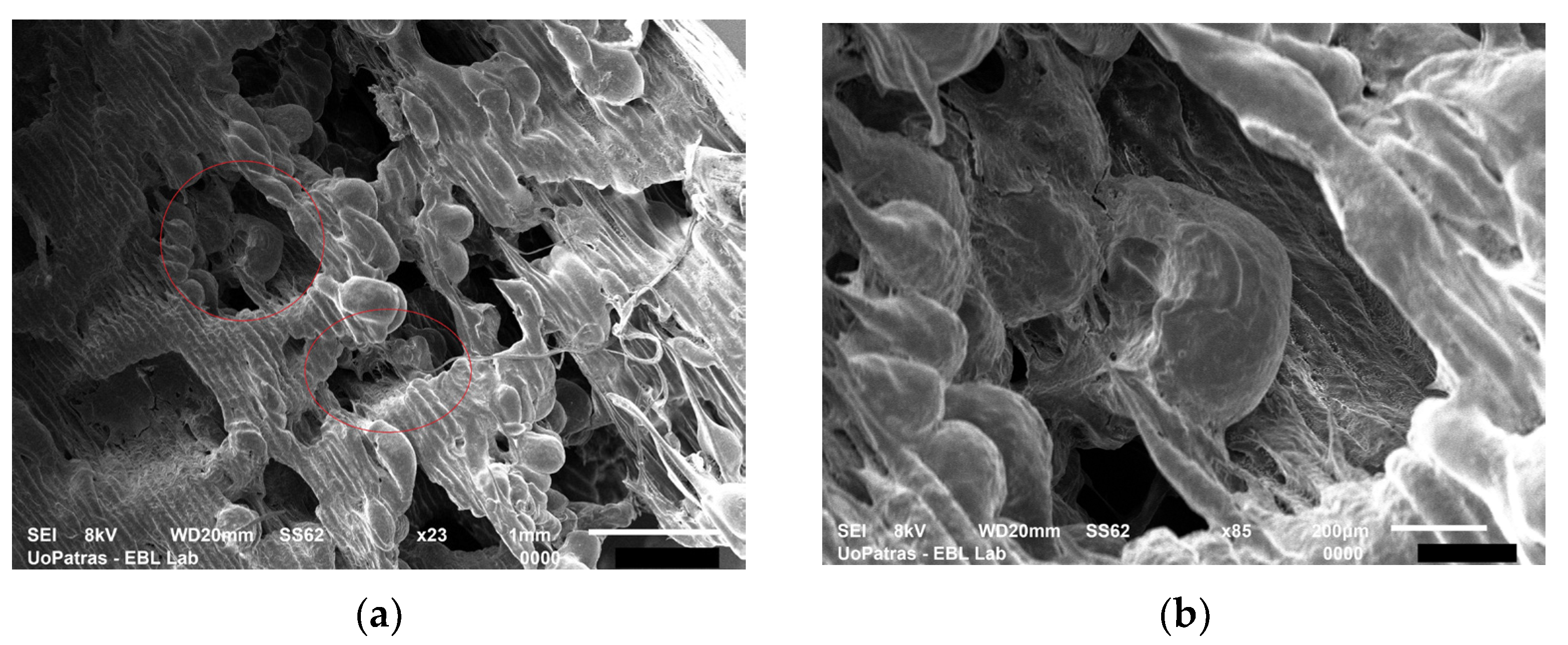
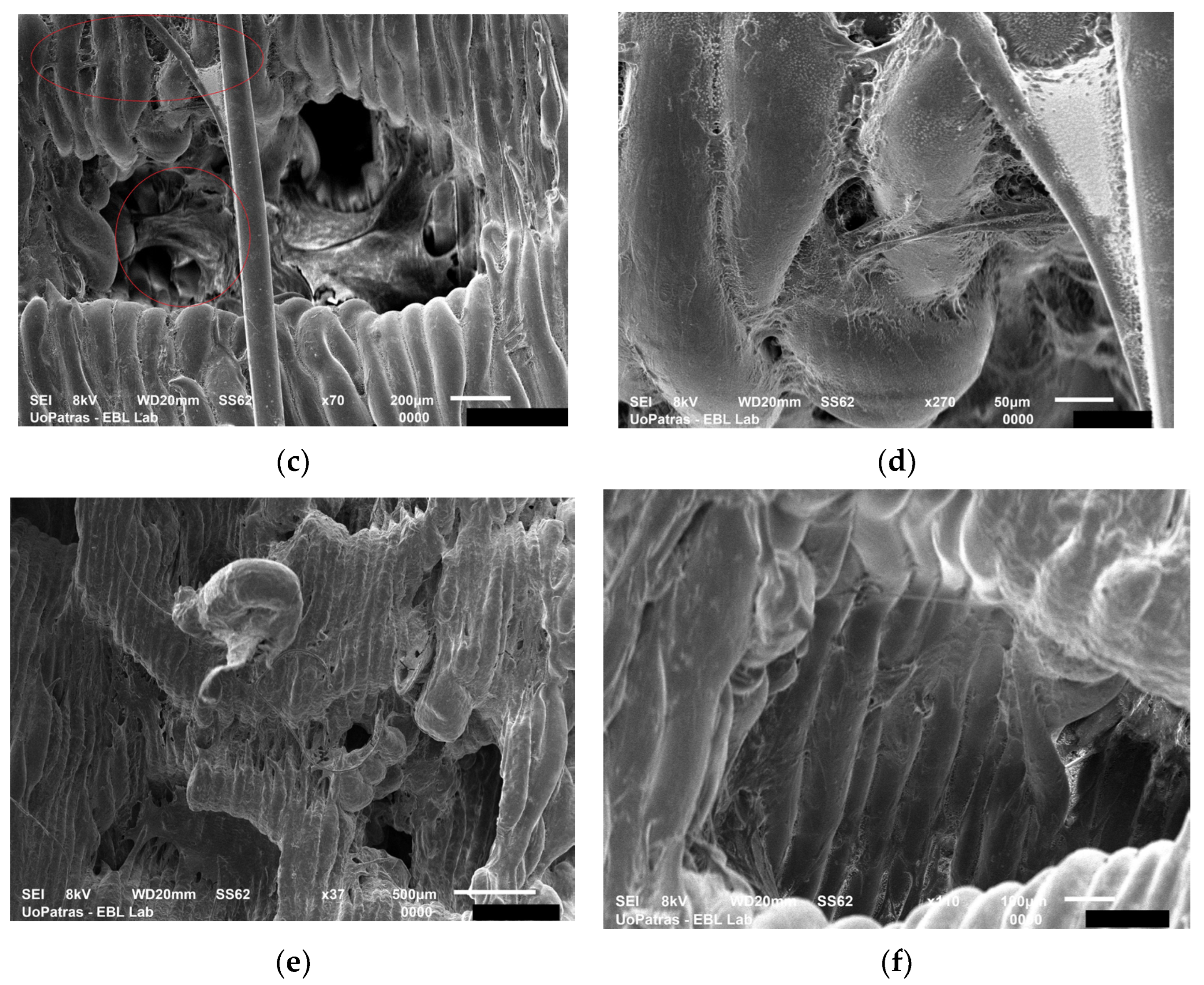
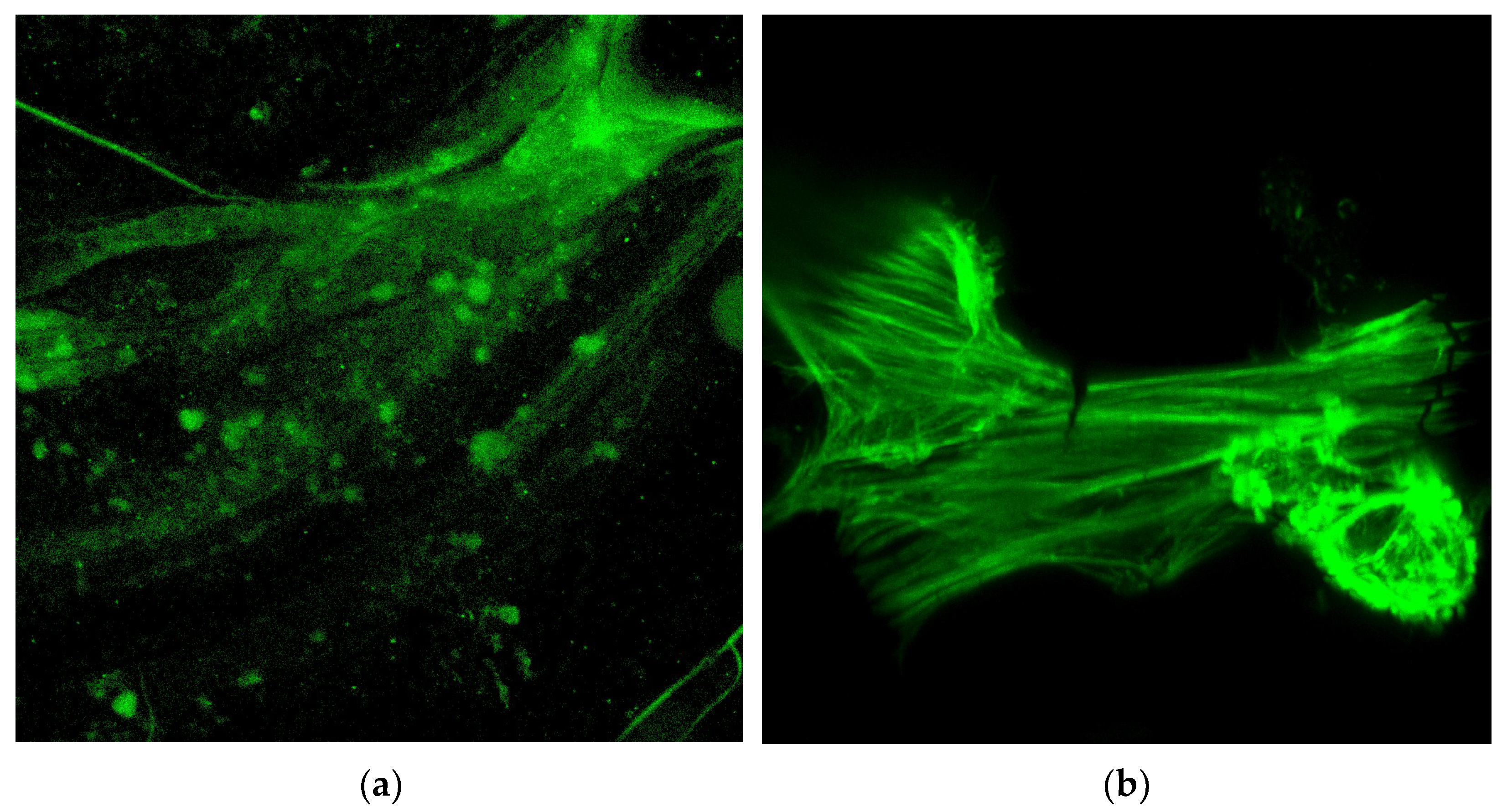
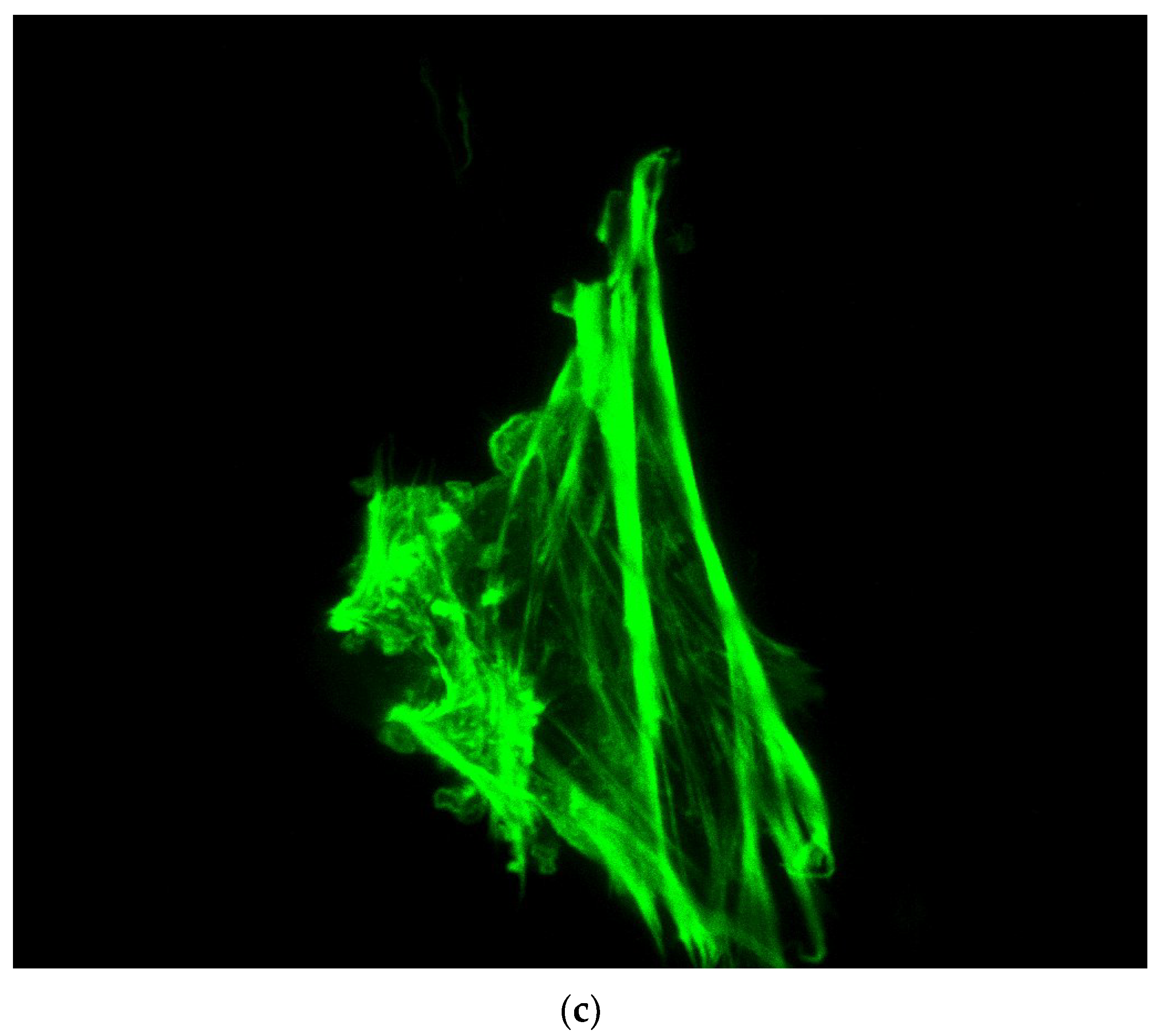
3.3. Stem Cells Feedback to Scaffolds in a Bioreactor
3.3.1. Experiments with PCL Electrospun Scaffolds
3.3.2. Experiments with PLA and PU Printed Scaffolds
4. Discussion
5. Conclusions
5.1. Theoretical Conclusions
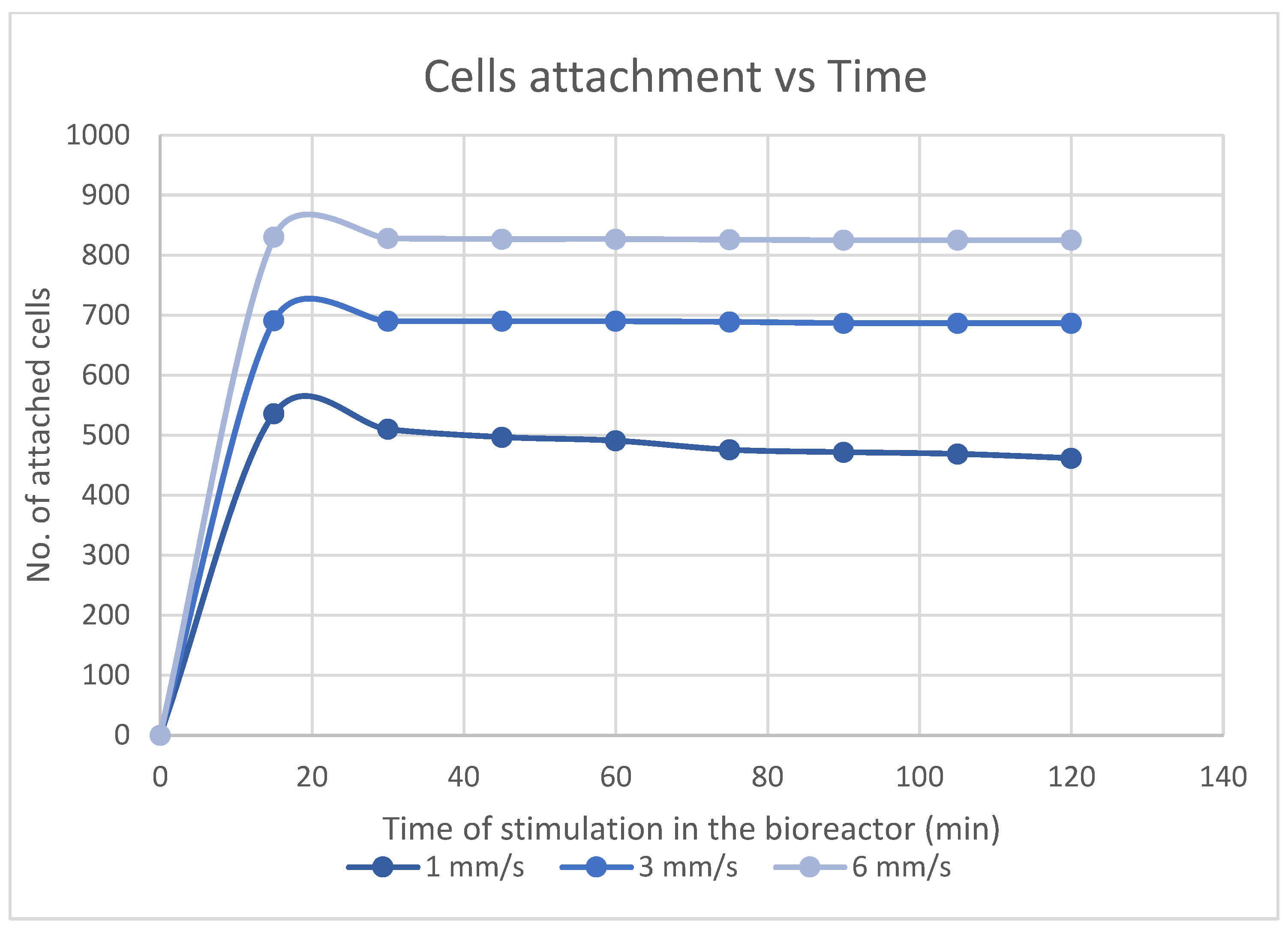
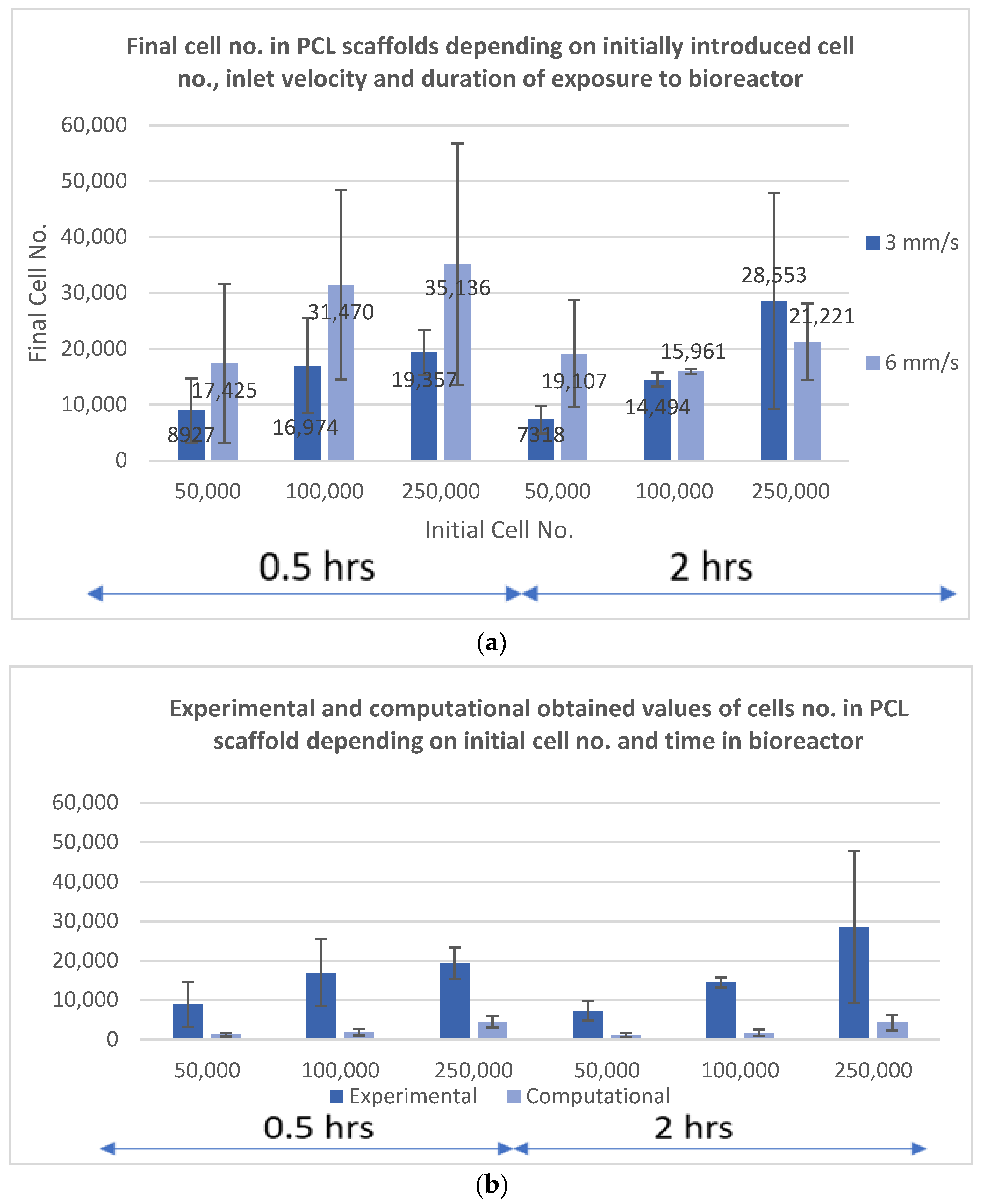
- The physical parameters influencing cells deposition in the scaffolds under dynamic conditions (bioreactor) are decisive within the first 17 to 20 min for cells long-term proliferation and tissue formation. The Comsol software modelling is recommended to predict cellular events in scaffolds, in a bioreactor, within the first half of an hour after contact between cells and substrate. In addition, the Comsol model correctly predicted the events related to cells adherence at lower inlet velocity (3 mm/s).
- According to the Comsol findings, the shear rate increases in the space of the scaffold pores, while the fluid velocity doubles inside the scaffold, which determines some changes in cells distribution, making it less uniform and efficient between layers 2 and 5 of the scaffold structure.
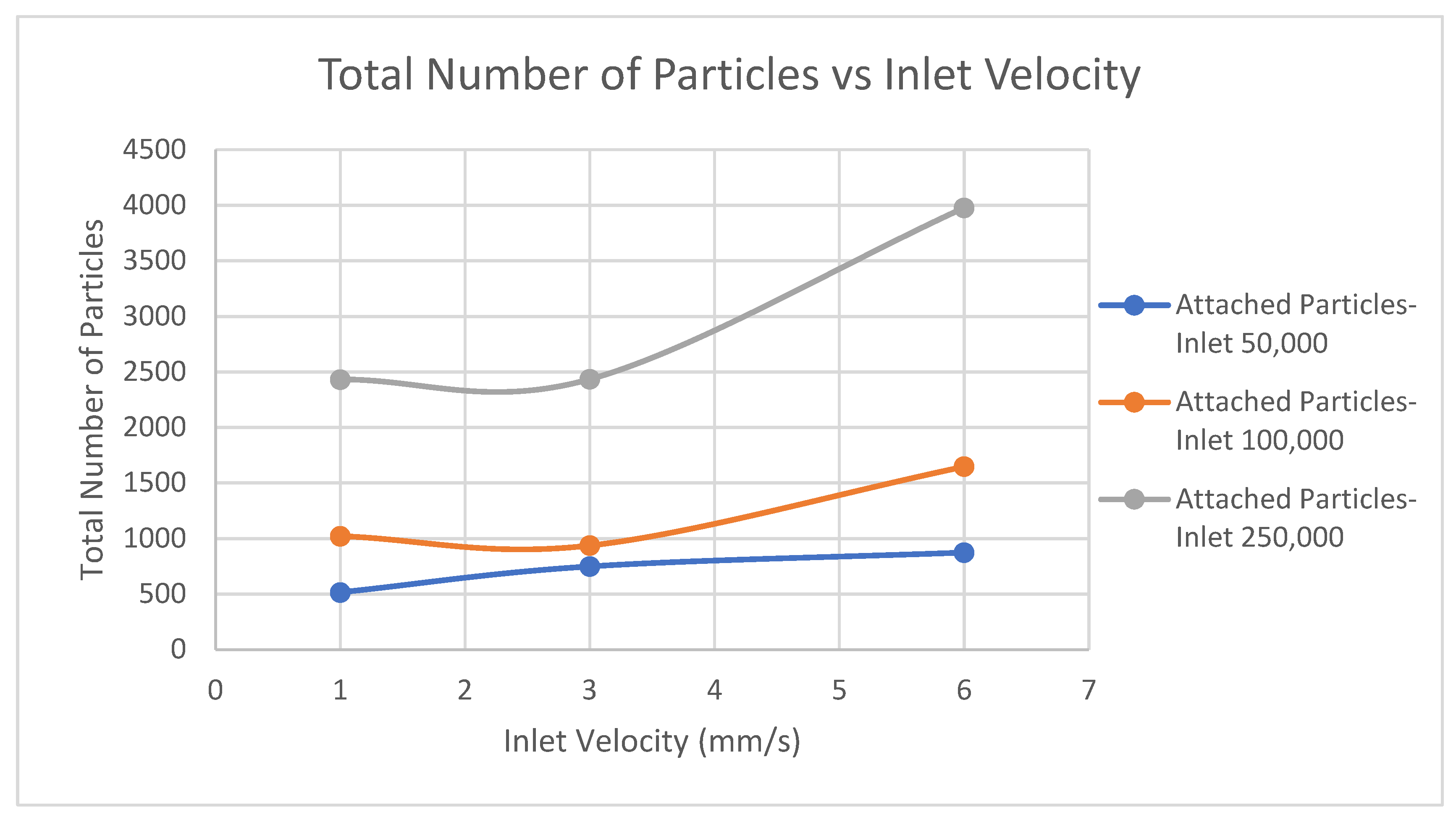
- Computationally, the inlet velocity of 6 mm/s proved to be the optimum in order to better distribute the cells on the scaffolds, but it did not coincide with the optimum experimental inlet velocity (3 mm/s). The reason is that the model does not account for the biological parameters which are crucial for cell attachment in the first minutes after contact with the substrate.
- It was computationally predicted that only 1.5% of the inserted particles attach to the scaffold. Experimentally, a solution to avoid cell loss from the scaffold due to physical events is the seeding of cells on the substrates in static conditions and their subsequent incubation for hours to days, before the exposure to a bioreactor.
- In the computational modelling, cells were assumed to attach to the substrate as soon as they hit the scaffold wall. In reality, the cell–biomaterial interactions are biologically complex and need to be taken into account.
5.2. Experimental Conclusions
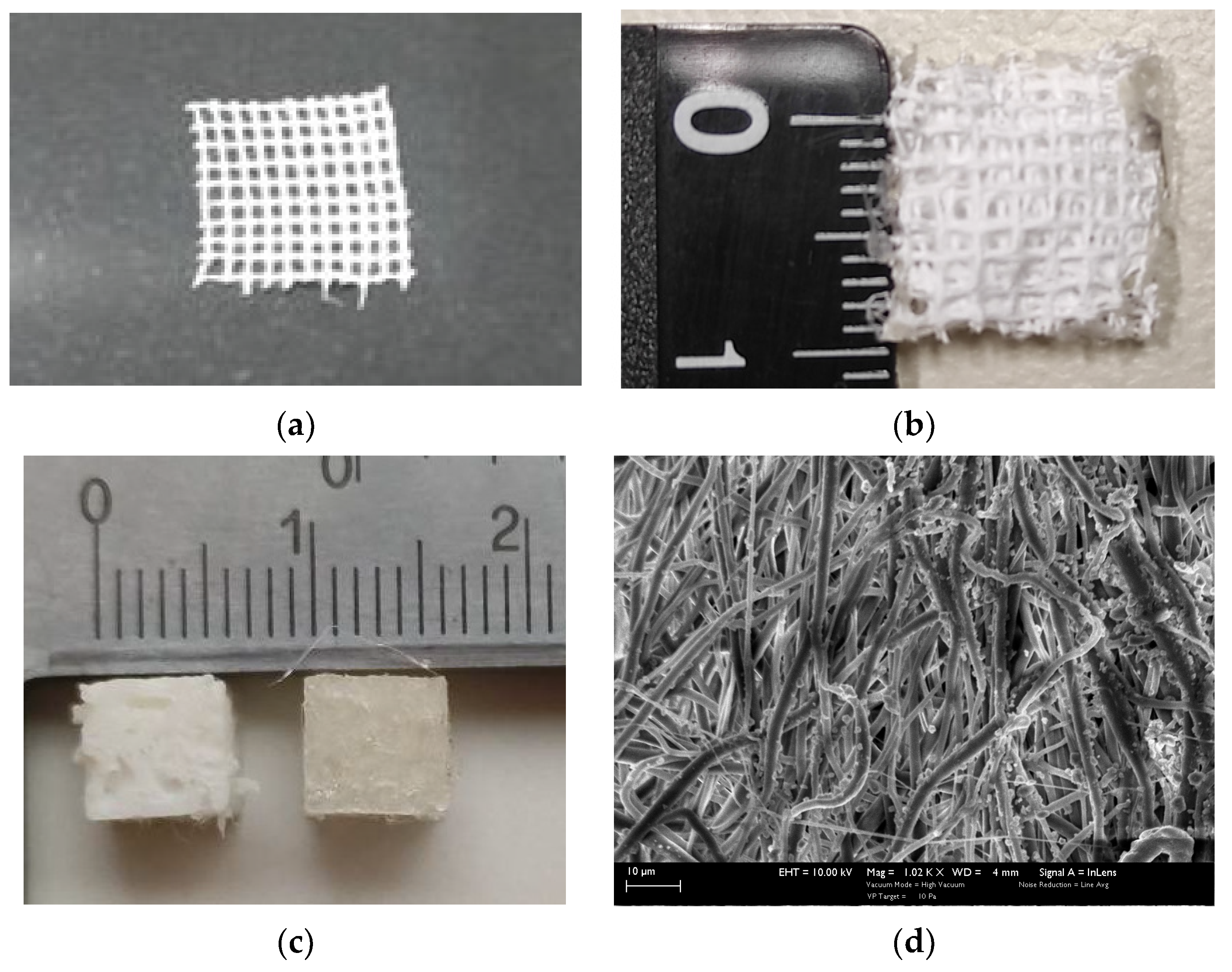
- Three types of 3D scaffolds were fabricated: CNTs-reinforced PCL by electrospinning, as well as PLA and PU by 3D printing. The electrospun scaffolds were multi-layered. All scaffolds had micro-level pores. The pores of the 3D printed scaffolds were irregularly shaped and biomimetic.
- The PCL has weak overall mechanical properties, but it performs well in tensile, having a good elasticity modulus. PLA compression modulus of elasticity is considerably higher than the one of PU. The ultimate strength of PCL and PU is almost similar, but PU exhibits a rubber-like behavior while PLA a plastic one. These properties are extremely important to understand cell feedback to scaffolds fabricated from these two types of thermoplastics.
- The evaluation of the computational study was made with stem cells seeded in PCL electrospun scaffolds; it was shown that the most appropriate inlet velocity was 3 mm/s while the best duration of exposure in a bioreactor was 0.5 h. The procedure consisted in placing the scaffold in the bioreactor and applying a flow of culture medium containing cells, in order to mimic the dynamic micro-environment of the body and how floating cells will end up attaching to the 3D structure. Cells attached in a much higher number than theoretically predicted which indicates the crucial role of biological adhesion in vitro and in vivo.
- The optimum number of deposited cells depends very much on the material of the scaffold and the duration of the experiment. Short experiments (0.5 to 2 h) may involve 2.5 × 105 cells if the surface area of the 3D structure is at least 1 × 1 cm2. For longer incubation times (1 to 3 days), 105 cells in population is the optimum.
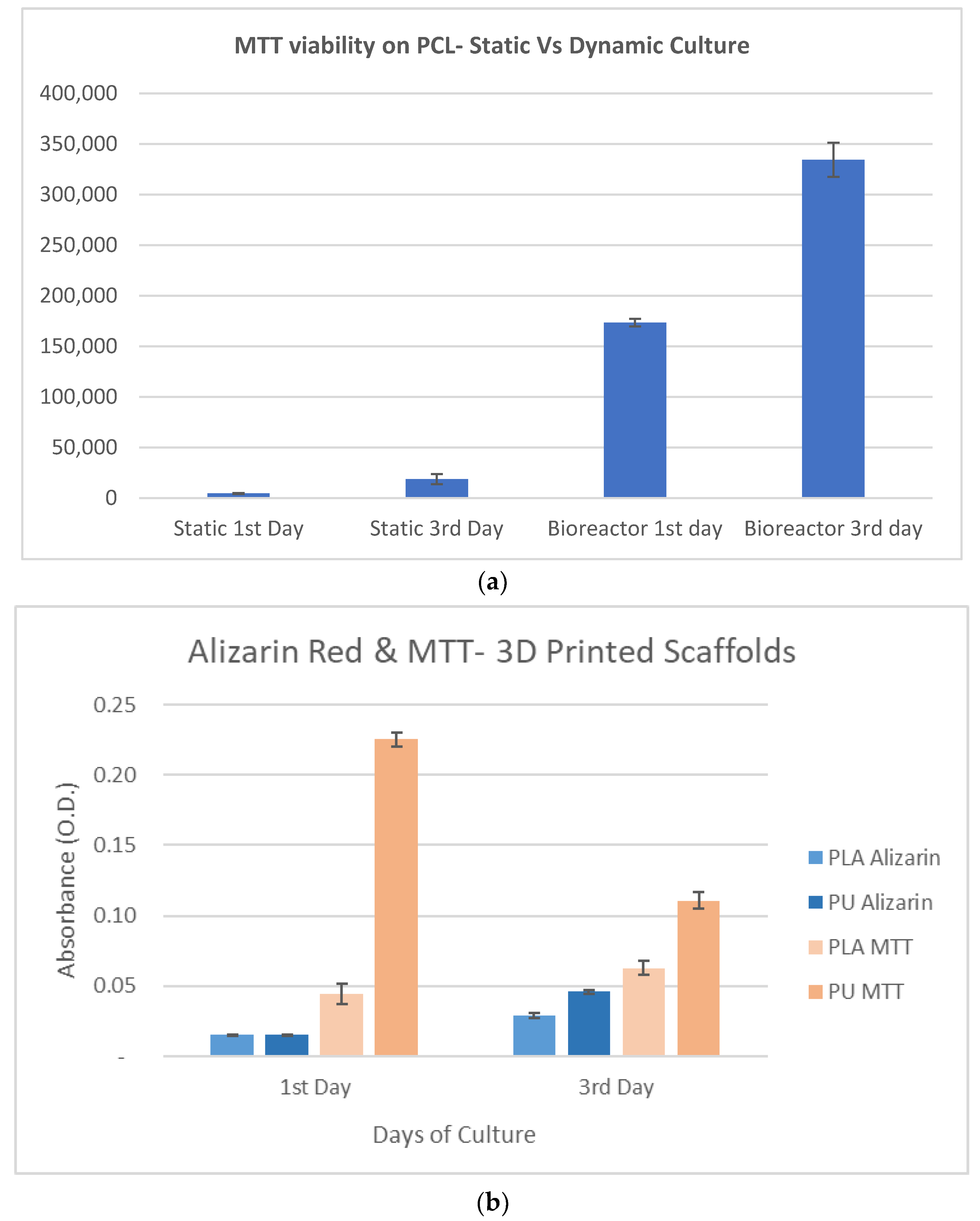
- The measurements of MTT and Alizarin red indicated that the PU scaffold with rubber-like behavior enabled enhanced feedback with respect to cells viability, followed by intense mineralization. It worth mentioning that PU comparing to PLA has properties closer to the natural tissue. The viability and the mineralization in cells in PLA scaffold were enabled simultaneously and were moderate in intensity comparing to the PU case.
5.3. Highlights
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hutmacher, D.W.; Singh, H. Computational fluid dynamics for improved bioreactor design and 3D culture. Trends Biotechnol. 2008, 26, 166–172. [Google Scholar] [CrossRef] [PubMed]
- Singh, H.; Hutmacher, D.W. Bioreactor Studies and Computational Fluid Dynamics. In Bioreactor Systems for Tissue Engineering; Kasper, C., van Griensven, M., Pörtner, R., Eds.; Springer: Berlin/Heidelberg, Germany, 2009; pp. 231–249. [Google Scholar]
- Guller, A.; Igrunkova, A. Engineered Microenvironments for 3D Cell Culture and Regenerative Medicine: Challenges, Advances, and Trends. Bioengineering 2023, 10, 17. [Google Scholar] [CrossRef] [PubMed]
- Nikolova, M.P.; Chavali, M.S. Recent advances in biomaterials for 3D scaffolds: A review. Bioact. Mater. 2019, 4, 271–292. [Google Scholar] [CrossRef]
- Sampson, K.; Koo, S.; Gadola, C.; Vasiukhina, A.; Singh, A.; Spartano, A.; Gollapudi, R.; Duley, M.; Mueller, J.; James, P.F.; et al. Cultivation of hierarchical 3D scaffolds inside a perfusion bioreactor: Scaffold design and finite-element analysis of fluid flow. SN Appl. Sci. 2021, 3, 884. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, Y.; Yan, J.; Zhang, K.; Lin, F.; Xiang, L.; Deng, L.; Guan, Z.; Cui, W.; Zhang, H. Pharmaceutical electrospinning and 3D printing scaffold design for bone regeneration. Adv. Drug Deliv. Rev. 2021, 174, 504–534. [Google Scholar] [CrossRef] [PubMed]
- Chia, H.N.; Wu, B.M. Recent advances in 3D printing of biomaterials. J. Biol. Eng. 2015, 9, 4. [Google Scholar] [CrossRef]
- Alam, F.; Shukla, V.R.; Varadarajan, K.; Kumar, S. Microarchitected 3D printed polylactic acid (PLA) nanocomposite scaffolds for biomedical applications. J. Mech. Behav. Biomed. Mater. 2020, 103, 103576. [Google Scholar] [CrossRef]
- Bruyas, A.; Lou, F.; Stahl, A.M.; Gardner, M.; Maloney, W.; Goodman, S.; Yang, Y.P. Systematic characterization of 3D-printed PCL/β-TCP scaffolds for biomedical devices and bone tissue engineering: Influence of composition and porosity. J. Mater. Res. 2018, 33, 1948–1959. [Google Scholar] [CrossRef]
- Hu, X.; Zhao, W.; Zhang, Z.; Xie, J.; He, J.; Cao, J.; Li, Q.; Yan, Y.; Xiong, C.; Li, K. Novel 3D printed shape-memory PLLA-TMC/GA-TMC scaffolds for bone tissue engineering with the improved mechanical properties and degradability. Chin. Chem. Lett. 2023, 34, 107451. [Google Scholar] [CrossRef]
- Nair, A.; Tang, L. Influence of scaffold design on host immune and stem cell responses. Semin. Immunol. 2017, 29, 62–71. [Google Scholar] [CrossRef]
- Florczyk, S.J.; Hotaling, N.A.; Simon, M.; Chalfoun, J.; Horenberg, A.L.; Schaub, N.J.; Wang, D.; Szczypiński, P.M.; DeFelice, V.L.; Bajcsy, P.; et al. Measuring dimensionality of cell-scaffold contacts of primary human bone marrow stromal cells cultured on electrospun fiber scaffolds. J. Biomed. Mater. Res. Part A 2023, 111, 106–117. [Google Scholar] [CrossRef]
- Thomas, V.; Jose, M.V.; Chowdhury, S.; Sullivan, J.F.; Dean, D.R.; Vohra, Y.K. Mechano-morphological studies of aligned nanofibrous scaffolds of polycaprolactone fabricated by electrospinning. J. Biomater. Sci. Polym. Ed. 2006, 17, 969–984. [Google Scholar] [CrossRef] [PubMed]
- Suwantong, O. Biomedical applications of electrospun polycaprolactone fiber mats. Polym. Adv. Technol. 2016, 27, 1264–1273. [Google Scholar] [CrossRef]
- Wang, P.; Pyo, S.-H.; Zhu, W.; Hwang, H.; Chen, S. 3D Printing of Polyurethanes for Biomedical Applications, in Emerging Technologies in Biophysical Sciences: A World Scientific Reference. In Emerging Technologies in Biophysical Sciences: A World Scientific Reference; World Scientific: Singapore, 2020; pp. 389–406. [Google Scholar] [CrossRef]
- Rusu, L.-C.; Ardelean, L.C.; Jitariu, A.-A.; Miu, C.A.; Streian, C.G. An Insight into the Structural Diversity and Clinical Applicability of Polyurethanes in Biomedicine. Polymers 2020, 12, 1197. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Chen, G.; Wang, G.; Zhu, P.; Gao, C. Recent Progress on 3D-Printed Polylactic Acid and Its Applications in Bone Repair. Adv. Eng. Mater. 2020, 22, 1901065. [Google Scholar] [CrossRef]
- Elamparithi, A.; Punnoose, A.M.; Kuruvilla, S.; Ravi, M.; Rao, S.; Paul, S.F.D. Electrospun polycaprolactone matrices with tensile properties suitable for soft tissue engineering. Artif. Cells Nanomed. Biotechnol. 2016, 44, 878–884. [Google Scholar] [CrossRef]
- Hou, Y.; Wang, W.; Bartolo, P. Investigation of polycaprolactone for bone tissue engineering scaffolds: In vitro degradation and biological studies. Mater. Des. 2022, 216, 110582. [Google Scholar] [CrossRef]
- Kanyanta, V.; Ivankovic, A. Mechanical characterisation of polyurethane elastomer for biomedical applications. J. Mech. Behav. Biomed. Mater. 2010, 3, 51–62. [Google Scholar] [CrossRef]
- Kucinska-Lipka, J.; Gubanska, I.; Sienkiewicz, M. Thermal and mechanical properties of polyurethanes modified with L-ascorbic acid. J. Therm. Anal. Calorim. 2017, 127, 1631–1638. [Google Scholar] [CrossRef]
- Wendels, S.; Avérous, L. Biobased polyurethanes for biomedical applications. Bioact. Mater. 2021, 6, 1083–1106. [Google Scholar] [CrossRef]
- Pinto, V.C.; Ramos, T.; Alves, S.; Xavier, J.; Tavares, P.; Moreira, P.; Guedes, R.M. Comparative Failure Analysis of PLA, PLA/GNP and PLA/CNT-COOH Biodegradable Nanocomposites thin Films. Procedia Eng. 2015, 114, 635–642. [Google Scholar] [CrossRef]
- Mei, H.; Yin, X.; Zhang, J.; Zhao, W. Compressive Properties of 3D Printed Polylactic Acid Matrix Composites Reinforced by Short Fibers and SiC Nanowires. Adv. Eng. Mater. 2019, 21, 1800539. [Google Scholar] [CrossRef]
- Stefan, S.; Schirmer, C.; Maschke, R.W.; Rossi, L.; Eibl, R.; Eibl, D. Computational Fluid Dynamics for Advanced Characterisation of Bioreactors Used in the Biopharmaceutical Industry: Part I: Literature Review. In Computational Fluid Dynamics—Recent Advances, New Perspectives and Applications; Guozhao, J., Jingliang, D., Eds.; IntechOpen: Rijeka, Croatia, 2023; Chapter 2. [Google Scholar]
- Meneses, J.; Silva, J.C.; Fernandes, S.R.; Datta, A.; Ferreira, F.C.; Moura, C.; Amado, S.; Alves, N.; Pascoal-Faria, P. A Multimodal Stimulation Cell Culture Bioreactor for Tissue Engineering: A Numerical Modelling Approach. Polymers 2020, 12, 940. [Google Scholar] [CrossRef] [PubMed]
- Choe, R.; Devoy, E.; Kuzemchak, B.; Sherry, M.; Jabari, E.; Packer, J.D.; Fisher, J.P. Computational investigation of interface printing patterns within 3D printed multilayered scaffolds for osteochondral tissue engineering. Biofabrication 2022, 14, 025015. [Google Scholar] [CrossRef] [PubMed]
- Kozaniti, F.K.; Deligianni, D.D.; Georgiou, M.D.; Portan, D.V. The Role of Substrate Topography and Stiffness on MSC Cells Functions: Key Material Properties for Biomimetic Bone Tissue Engineering. Biomimetics 2022, 7, 7. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Zhu, G.; Wang, L.; An, G.; Shi, X.; Wang, Y. Assembling of electrospun meshes into three-dimensional porous scaffolds for bone repair. Biofabrication 2017, 9, 015018. [Google Scholar] [CrossRef]
- ASTM F2150-02; Standard Guide for Characterization and Testing of Biomaterial Scaffolds Used in Regenerative Medicine and Tissue-Engineered Medical Products. ASTM International: West Conshohocken, PA, USA, 2002.
- ASTM D882-18; Standard Test Method for Tensile Properties of Thin Plastic Sheeting. ASTM International: West Conshohocken, PA, USA, 2002.
- Bušs, A.; Suleiko, A.; Rugele, K.; Vanags, J. CFD Analysis of a Stirred Vessel Bioreactor with Double Pitch Blade and Rushton Type Impellers. In Proceedings of the 2017 COMSOL Conference, Rotterdam, the Netherlands, 18 October 2017. [Google Scholar]
- Chatzistamatiou, T.K.; Papassavas, A.C.; Michalopoulos, E.; Gamaloutsos, C.; Mallis, P.; Gontika, I.; Panagouli, E.; Koussoulakos, S.L.; Stavropoulos-Giokas, C. Optimizing isolation culture and freezing methods to preserve Wharton’s jelly’s mesenchymal stem cell (MSC) properties: An MSC banking protocol validation for the Hellenic Cord Blood Bank. Transfusion 2014, 54, 3108–3120. [Google Scholar] [CrossRef]
- Gregory, C.A.; Gunn, W.G.; Peister, A.; Prockop, D.J. An Alizarin red-based assay of mineralization by adherent cells in culture: Comparison with cetylpyridinium chloride extraction. Anal. Biochem. 2004, 329, 77–84. [Google Scholar] [CrossRef]
- Wilson, C.; Clegg, R.E.; Leavesley, D.I.; Pearcy, M.J. Mediation of Biomaterial–Cell Interactions by Adsorbed Proteins: A Review. Tissue Eng. 2005, 11, 1–18. [Google Scholar] [CrossRef]
- Sikavitsas, V.; Bancroft, G.N.; Holtorf, H.L.; Jansen, J.A.; Mikos, A.G. Mineralized matrix deposition by marrow stromal osteoblasts in 3D perfusion culture increases with increasing fluid shear forces. Proc. Natl. Acad. Sci. USA 2004, 100, 14683–14688. [Google Scholar] [CrossRef]
- Pawluchin, A.; Galic, M. Moving through a changing world: Single cell migration in 2D vs. 3D. Front. Cell Dev. Biol. 2022, 10, 1080995. [Google Scholar] [CrossRef] [PubMed]
- Crétel, E.; Pierres, A.; Benoliel, A.-M.; Bongrand, P. How Cells Feel Their Environment: A Focus on Early Dynamic Events. Cell. Mol. Bioeng. 2008, 1, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, A.; Agarwal, M.; Mukherjee, R.; Sen, P.; Sinha, D.K. 3D micro-environment regulates NF-κβ dependent adhesion to induce monocyte differentiation. Cell Death Dis. 2018, 9, 914. [Google Scholar] [CrossRef]
- Cámara-Torres, M.; Sinha, R.; Mota, C.; Moroni, L. Improving cell distribution on 3D additive manufactured scaffolds through engineered seeding media density and viscosity. Acta Biomater. 2020, 101, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Feier, A.; Manu, D.R.; Strnad, G.; Dobreanu, M.; Russu, O.M.; Portan, D.; Bataga, T. A Step Forward Standardization of Biocompatibility Testing on Tissue Culture Polystyrene. Mater. Plast. 2018, 55, 303–307. [Google Scholar] [CrossRef]
- Portan, D.V.; Kroustalli, A.A.; Deligianni, D.D.; Papanicolaou, G.C. On the biocompatibility between TiO2 nanotubes layer and human osteoblasts. J. Biomed. Mater. Res. Part A 2012, 100, 2546–2553. [Google Scholar] [CrossRef]
- Hendriks, M.; Ramasamy, S.K. Blood Vessels and Vascular Niches in Bone Development and Physiological Remodeling. Front. Cell Dev. Biol. 2020, 8, 602278. [Google Scholar] [CrossRef]
- Fritton, S.P.; Weinbaum, S. Fluid and Solute Transport in Bone: Flow-Induced Mechanotransduction. Annu. Rev. Fluid Mech. 2008, 41, 347–374. [Google Scholar] [CrossRef]
- Lee, S.J.; Oh, S.H.; Liu, J.; Soker, S.; Atala, A.; Yoo, J.J. The use of thermal treatments to enhance the mechanical properties of electrospun poly(ɛ-caprolactone) scaffolds. Biomaterials 2008, 29, 1422–1430. [Google Scholar] [CrossRef]
- Lu, L.; Zhang, Q.; Wootton, D.M.; Chiou, R.; Li, D.; Lu, B.; Lelkes, P.I.; Zhou, J. Mechanical Study of Polycaprolactone-hydroxyapatite Porous Scaffolds Created by Porogen-based Solid Freeform Fabrication Method. J. Appl. Biomater. Funct. Mater. 2014, 12, 145–154. [Google Scholar] [CrossRef]
- Marolt, D.; Campos, I.M.; Bhumiratana, S.; Koren, A.; Petridis, P.; Zhang, G.; Spitalnik, P.F.; Grayson, W.L.; Vunjak-Novakovic, G. Engineering bone tissue from human embryonic stem cells. Proc. Natl. Acad. Sci. USA 2012, 109, 8705–8709. [Google Scholar] [CrossRef] [PubMed]







| Inlet Velocity (mm/s) | |||||||
|---|---|---|---|---|---|---|---|
| 1 | 3 | 6 | |||||
| Particles no. | 5 × 104 | 30 | 120 | 30 | 120 | 30 | 120 |
| 105 | 30 | 120 | 30 | 120 | 30 | 120 | |
| 2.5 × 105 | 30 | 120 | 30 | 120 | 30 | 120 | |
| Stimulation duration (min) | |||||||
| Scaffold Type | Total Protein | Osteopontin | Osteocalcin |
|---|---|---|---|
| PU Control 1 | 0.87 | 0.106 | 0.158 |
| PLA Control | 0.93 | 0.105 | 0.205 |
| PU Bioreactor 2 | 0.909 | 0.116 | 0.172 |
| PLA Bioreactor | 0.93 | 0.105 | 0.114 |
| Cells Feedback in a Static Experiment | ||
| Biomarker/Material | PU (polyurethane) | PLA (polylactic acid) |
| Total Protein (depends on substrate stiffness) | Moderate due to weak adhesion. | Pronounced due to strong adhesion. |
| Osteopontin (depends of cells no. and quality of adhesion) | Regulated by development of extracellular matrix due to increased proliferation of cells. | Regulated by development of extracellular matrix due to strong adhesion of cells. |
| Osteocalcin (depends on differentiation) | Moderate due to weak differentiation. | Pronounced due to strong differentiation. |
| Cells Feedback in a Dynamic Experiment with Bioreactor | ||
| Biomarker/Material | PU (polyurethane) | PLA (polylactic acid) |
| MTT Viability | High cell no. because of biomimetic material properties. | Low cell no. because of high stiffness |
| Alizarin red | Mineralization is induced due to confluency of cells in scaffold. | Mineralization has been induced in static conditions due to the strong adhesion. |
| Total Protein (level has been adjusted during static conditions) | Stays steady comparing to static conditions. | Stays steady comparing to static conditions. |
| Osteopontin | Considerably increased level due to introduction of stresses through fluid flow. | No significant change comparing to static experiment. |
| Osteocalcin | Mineralization is induced due to confluency of cells in scaffold. | Mineralization has been induced in static conditions due to the strong adhesion. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kozaniti, F.K.; Manara, A.E.; Kostopoulos, V.; Mallis, P.; Michalopoulos, E.; Polyzos, D.; Deligianni, D.D.; Portan, D.V. Computational and Experimental Investigation of the Combined Effect of Various 3D Scaffolds and Bioreactor Stimulation on Human Cells’ Feedback. Appl. Biosci. 2023, 2, 249-277. https://doi.org/10.3390/applbiosci2020018
Kozaniti FK, Manara AE, Kostopoulos V, Mallis P, Michalopoulos E, Polyzos D, Deligianni DD, Portan DV. Computational and Experimental Investigation of the Combined Effect of Various 3D Scaffolds and Bioreactor Stimulation on Human Cells’ Feedback. Applied Biosciences. 2023; 2(2):249-277. https://doi.org/10.3390/applbiosci2020018
Chicago/Turabian StyleKozaniti, Foteini K., Aikaterini E. Manara, Vassilis Kostopoulos, Panagiotis Mallis, Efstathios Michalopoulos, Demosthenes Polyzos, Despina D. Deligianni, and Diana V. Portan. 2023. "Computational and Experimental Investigation of the Combined Effect of Various 3D Scaffolds and Bioreactor Stimulation on Human Cells’ Feedback" Applied Biosciences 2, no. 2: 249-277. https://doi.org/10.3390/applbiosci2020018
APA StyleKozaniti, F. K., Manara, A. E., Kostopoulos, V., Mallis, P., Michalopoulos, E., Polyzos, D., Deligianni, D. D., & Portan, D. V. (2023). Computational and Experimental Investigation of the Combined Effect of Various 3D Scaffolds and Bioreactor Stimulation on Human Cells’ Feedback. Applied Biosciences, 2(2), 249-277. https://doi.org/10.3390/applbiosci2020018










