Molecular Dynamics and Optimization Studies of Horse Prion Protein Wild Type and Its S167D Mutant
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
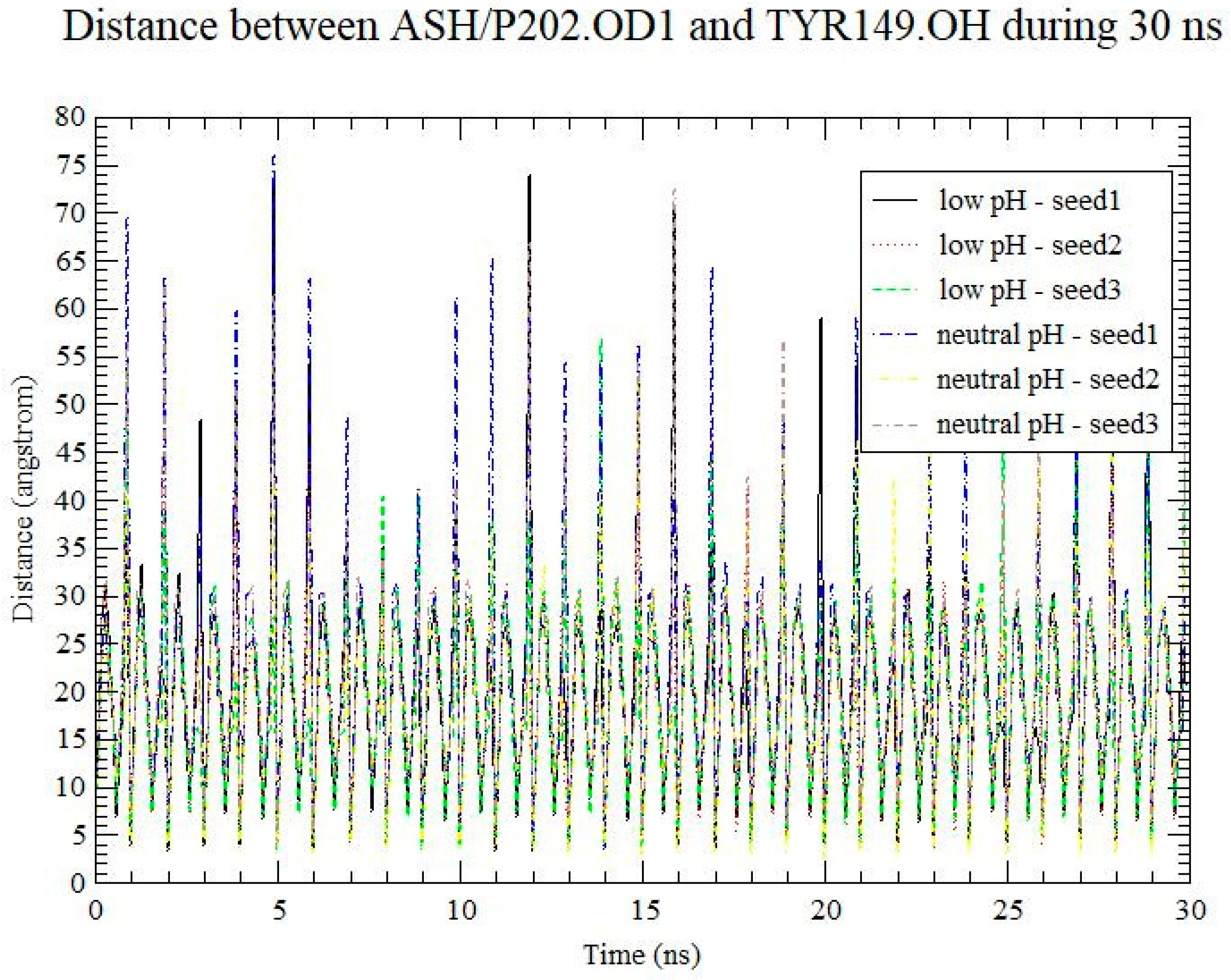
3. Results and Discussion
4. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviation
| SBs | salt bridges |
| HBs | hydrogen bonds |
| HYDs | hydrophobic interactions |
| vdWs | van der Waals |
| aa | amino acid (or residue) |
| S-S | disulfide bond |
References
- Vorberg, I.; Martin, H.G.; Eberhard, P.; Suzette, A.P. Multiple amino acid residues within the rabbit prion protein inhibit formation of its abnormal isoform. J. Virol. 2003, 77, 2003–2009. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Li, J.; Xiong, M.Q.; Peng, Y.; Yao, W.M.; Hong, J.; Lin, D.H. Solution structure and dynamics of the I214V mutant of the rabbit prion protein. PLoS ONE 2010, 5, e13273. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Li, J.; Yao, W.M.; Xiong, M.Q.; Hong, J.; Peng, Y.; Xiao, G.F.; Lin, D.H. Unique structural characteristics of the rabbit prion protein. J. Biol. Chem. 2010, 285, 31682–31693. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.Q.; Sweeting, B.; Mulligan, V.K.; Arslan, P.E.; Cashman, N.R.; Pai, E.F.; Chakrabartty, A. Prion disease susceptibility is affected by β-structure folding propensity and local side-chain interactions in PrP. Proc. Natl. Acad. Sci. USA 2010, 107, 19808–19813. [Google Scholar] [CrossRef] [PubMed]
- Sweeting, B.; Brown, E.; Khan, M.Q.; Chakrabartty, A.; Pai, E.F. N-terminal helix-cap in α-helix 2 modulates β-state misfolding in rabbit and hamster prion proteins. PLoS ONE 2013, 8, e63047. [Google Scholar] [CrossRef] [PubMed]
- Polymenidoua, M.; Trusheimb, H.; Stallmacha, L.; Moosa, R.; Julius, J.A.; Mielea, G.; Lenzbauerb, C.; Aguzzia, A. Canine MDCK cell lines are refractory to infection with human and mouse prions. Vaccine 2008, 26, 2601–2614. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, K.; Mohri, S.; Yokoyama, T. Comparison of the local structural stabilities of mammalian prion protein (PrP) by fragment molecular orbital calculations. Prion 2013, 7, 185–191. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sanchez-Garcia, J.; Jensen, K.; Zhang, Y.; Rincon-Limas, D.E.; Fernandez-Funez, P. A single amino acid (Asp159) from the dog prion protein suppresses the toxicity of the mouse prion protein in Drosophila. Neurobiol. Dis. 2016, 95, 204–209. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Borges, N.; Parra, B.; Vidal, E.; Eraña, H.; Sánchez-Martín, M.A.; de Castro, J.; Elezgarai, S.R.; Pumarola, M.; Mayoral, T.; Castilla, J. Unraveling the key to the resistance of canids to prion diseases. PLoS Pathog. 2017, 13, e1006716. [Google Scholar] [CrossRef] [PubMed]
- Lysek, D.A.; Schorn, C.; Nivon, L.G.; Esteve-Moya, V.; Christen, B.; Calzolai, L.; von Schroetter, C.; Fiorito, F.; Herrmann, T.; Guntert, P. Prion protein NMR structures of cats, dogs, pigs, and sheep. Proc. Natl. Acad. Sci. USA 2005, 102, 640–645. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.P. Molecular dynamics studies of dog prion protein wild-type and its D159N mutant. J. Biomol. Struct. Dyn. 2021, 39, 4234–4242. [Google Scholar] [CrossRef] [PubMed]
- Perez, D.R.; Damberger, F.F.; Wuthrich, K. Horse prion protein NMR structure and comparisons with related variants of the mouse prion protein. J. Mol. Biol. 2010, 400, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.P. The structural stability of wild-type horse prion protein. J. Biomol. Struct. Dyn. 2011, 29, 369–377. [Google Scholar] [CrossRef]
- Fernández-Funez, P. The Prion Protein Zoo: Identification of Protective Residues from Prion-Resistant Animals. Available online: http://www.cajal.csic.es/actividades2017/2017-10-30-PedroFernandezSketch.pdf (accessed on 29 September 2021).
- Myers, R.R.; Sanchez-Garcia, J.; Leving, D.C.; Fernandez-Funez, P. New Drosophila models to uncover the intrinsic and extrinsic factors mediating the toxicity of the human prion protein. Dis. Model. Mech. 2022, 15, dmm049184. [Google Scholar] [CrossRef] [PubMed]
- Myers, R.; Cembran, A.; Fernandez-Funez, P. Insight from animals resistant to prion diseases: Deciphering the genotype—Morphotype–phenotype code for the prion protein. Front. Cell Neurosci. 2020, 14, 254. [Google Scholar] [CrossRef]
- Zhang, J.P. Optimization-Based Molecular Dynamics Studies of SARS-CoV-2 Molecular Structures—Research on COVID-19; Springer: Cham, Switzerland, 2023. [Google Scholar]
- Zhang, J.P. Structural Dynamics Analyses of Prion Protein Structures—The Resistance to Prion Diseases Down Under; Springer: Singapore, 2018; pp. 5–12. [Google Scholar]
- Zhang, J.P. Molecular Structures and Structural Dynamics of Prion Proteins and Prions—Mechanism Underlying the Resistance to Prion Diseases; Springer: Dordrecht, The Netherlands, 2015. [Google Scholar]
- Zazeri, G.; Povinelli, A.P.R.; Pavan, N.M.; Jones, A.M.; Ximenes, V.F. Solvent-induced lag phase during the formation of lysozyme amyloid fibrils triggered by sodium dodecyl sulfate: Biophysical experimental and in silico study of solvent effects. Molecules 2023, 28, 6891. [Google Scholar] [CrossRef]



| HBs Kept | Locations | HBs Lost | Locations |
|---|---|---|---|
| ARG228.N-ALA224.O | in α3 | TYR149.OH-ASP202.OD1 | α1–α3 |
| ASN181.N-HIS177.O | in α2 | ASN153.ND2-TYR149.O | α1-310H1-loop-α1 |
| CYS179.N-PHE175.O | in α2 | ARG156.N-ASN153.O | 310H1-α1-310H1-loop |
| CYS214.N-VAL210.O | in α3 | ARG164.NE-GLU168.OE2 | β2-310H2-loop-310H2 |
| GLN217.N-MET213.O | in α3 | ARG164.NH2-GLU168.OE1 | β2-310H2-loop-310H2 |
| GLU207.N-VAL203.O | in α3 | HIS177.ND1-ASP178.OD1 | in α2 |
| GLU211.N-GLU207.O | in α3 | ASP178.N-ASN174.O | in α2 |
| GLU221.N-GLN217.O | in α3 | HIS187.N-THR183.O | in α2 |
| GLY131.N-VAL161.O | β1–β2 | THR192.N-THR188.O | in α2 |
| HIS177.N-LYS173.O | in α2 | LYS194.N-THR191.O | α2-α3-loop-α2 |
| ILE182.N-ASP178.O | in α2 | GLU196.N-THR191.OG1 | α2-α3-loop-α2 |
| ILE205.N-THR201.O | in α3 | PHE198.N-THR192.OG1 | α2-α3-loop-α2 |
| ILE215.N-GLU211.O | in α3 | GLN212.NE2-GLU211.OE2 | in α3 |
| LYS185.N-ASN181.O | in α2 | GLN217.NE2-TYR163.OH | α3-β2 |
| LYS204.N-GLU200.O | in α3 | GLN219.NE2-ILE215.O | in α3 |
| MET129.N-TYR163.O | β1–β2 | GLN227.NE2-GLU223.O | in α3 |
| MET134.N-ASN159.O | β1-α1-loop-310H1-β2-loop | SER231.OG-GLN227.O | C-terminal-α3 |
| MET206.N-ASP202.O | in α3 | ||
| MET213.N-VAL209.O | in α3 | new HBs | locations |
| PHE225.N-GLU221.O | in α3 | TYR150.N-GLU146.O | in α1 |
| THR183.OG1-CYS179.O | in α2 | ARG151.N-ASP147.O | in α1 |
| THR188.N-VAL184.O | in α2 | GLU152.N-ARG148.O | α1-310H1-loop-α1 |
| THR190.N-GLN186.O | in α2 | ASN171.ND2-ASN174.OD1 | 310H2-α2-loop-α2 |
| THR216.N-GLN212.O | in α3 | GLN186.N-ILE182.O | in α2 |
| TYR149.N-TYR145.O | in α1 | ||
| TYR150.OH-PRO137.O | α1-β1-α1-loop | ||
| TYR162.N-THR183.OG1 | β2-α2 | ||
| TYR218.N-CYS214.O | in α3 | ||
| TYR222.N-TYR218.O | in α3 | ||
| VAL176.N-GLN172.O | in α2 | ||
| VAL180.N-VAL176.O | in α2 | ||
| VAL184.N-VAL180.O | in α2 | ||
| VAL189.N-LYS185.O | in α2 | ||
| VAL203.N-THR199.O | α3-α2-α3-loop | ||
| VAL209.N-ILE205.O | in α3 | ||
| VAL210.N-MET206.O | in α3 |
| B β-Bridge | G 310-Helix | I π-Helix | H α-Helix | E β-Sheet | T Turn | S Bend | |||
|---|---|---|---|---|---|---|---|---|---|
| 300 K | Low pH | Seed1 | 0.00% | 4.88% | 0.00% | 50.41% | 3.54% | 7.79% | 7.65% |
| Seed2 | 0.26% | 3.32% | 0.09% | 48.54% | 4.63% | 8.34% | 9.00% | ||
| Seed3 | 0.21% | 4.44% | 0.00% | 46.63% | 4.07% | 8.93% | 10.89% | ||
| Neutral pH | Seed1 | 0.00% | 4.53% | 0.00% | 51.20% | 3.54% | 5.04% | 8.73% | |
| Seed2 | 0.13% | 3.94% | 0.03% | 50.15% | 3.55% | 9.00% | 7.94% | ||
| Seed3 | 0.50% | 2.90% | 0.00% | 51.17% | 3.73% | 8.34% | 8.83% | ||
| 350 K | Low pH | Seed1 | 0.21% | 2.85% | 0.00% | 49.39% | 3.42% | 8.34% | 10.18% |
| Seed2 | 0.34% | 3.55% | 0.03% | 48.73% | 4.11% | 9.05% | 9.11% | ||
| Seed3 | 0.37% | 3.24% | 0.00% | 49.29% | 4.14% | 8.67% | 8.20% | ||
| Neutral pH | Seed1 | 0.47% | 4.90% | 0.00% | 46.79% | 4.96% | 9.62% | 7.92% | |
| Seed2 | 0.18% | 3.60% | 0.03% | 49.18% | 3.49% | 9.04% | 9.60% | ||
| Seed3 | 0.67% | 4.70% | 0.03% | 49.53% | 4.31% | 6.76% | 9.19% | ||
| 450 K | Low pH | Seed1 | 1.09% | 3.47% | 0.09% | 42.18% | 1.17% | 12.46% | 12.70% |
| Seed2 | 0.93% | 4.85% | 0.12% | 41.37% | 3.60% | 14.31% | 12.84% | ||
| Seed3 | 0.76% | 4.25% | 0.06% | 38.34% | 3.14% | 14.80% | 15.43% | ||
| Neutral pH | Seed1 | 0.81% | 4.35% | 0.00% | 38.38% | 0.64% | 16.43% | 14.61% | |
| Seed2 | 0.73% | 4.09% | 0.12% | 42.47% | 3.33% | 11.43% | 14.64% | ||
| Seed3 | 0.28% | 4.07% | 0.09% | 45.13% | 4.05% | 10.64% | 7.31% |
| HBs at 300 K under Neutral pH Environment | HBs at 350 K under Neutral pH Environment | HBs at 450 K under Neutral pH Environment |
|---|---|---|
| ASP202@OD1-TYR157@OH.HH 29.19, 84.64, 44.75 | GLU196@O-ARG+156@NE.HE 10.27, 12.50, 0 | ASP202@OD1-ARG+156@NE.HE 0, 0, 11.57 |
| ASP202@OD2-TYR157@OH.HH 62.79, 0, 49.53 | GLU196@O-ARG+156@NH1.HH12 0, 0, 17.27 | ASP202@OD1-ARG+156@NH1.HH12 23.27, 28.72, 14.80 |
| ASP202@OD1-ARG+156@NH1.HH11 0, 67.38, 0 | GLU196@O-ARG+156@NH2.HH21 21.24, 0, 0 | ASP202@OD1-ARG+156@NH2.HH21 0, 5.00, 14.12 |
| ASP202@OD1-ARG+156@NH1.HH12 60.02, 0, 42.64 | GLU196@O-ARG+156@NH2.HH22 0, 0, 12.43 | ASP202@OD1-ARG+156@NH2.HH22 16.40, 20.87, 16.53 |
| ASP202@OD2-ARG+156@NH1.HH12 28.89, 0, 42.91 | GLU196@OE1-ARG+156@NH2.HH21 0, 17.91, 0 | ASP202@OD2-ARG+156@NE.HE 0, 0, 10.60 |
| ASP202@OD1-ARG+156@NH2.HH22 33.65, 0, 48.65 | GLU196@OE2-ARG+156@NE.HE 0, 10.62, 13.05 | ASP202@OD2-ARG+156@NH1.HH12 17.80, 22.15, 17.12 |
| ASP202@OD2-ARG+156@NH2.HH22 60.77, 0, 43.96 | GLU196@OE2-ARG+156@NH2.HH21 0, 0, 11.42 | ASP202@OD2-ARG+156@NH2.HH21 0, 0, 15.88 |
| ASP202@OD1-TYR149@OH.HH 0, 12.02, 0 | ASP202@OD1-TYR149@OH.HH 0, 19.46, 18.03 | ASP202@OD2-ARG+156@NH2.HH22 21.58, 27.68, 13.57 |
| ASP202@OD2-TYR149@OH.HH 0, 67.42, 0 | ASP202@OD2-TYR149@OH.HH 0, 22.37, 41.93 | ASP202@OD1-TYR157@OH.HH 0, 20.78, 32.18 |
| ASP178@OD2-TYR128@OH.HH 27.04, 0, 0 | ASP202@OD1-ARG+156@NE.HE 0, 0, 10.28 | ASP202@OD2-TYR157@OH.HH 0, 22.68, 29.00 |
| ASP178@OD1-TYR128@OH.HH 25.90, 0, 12.08 | ASP202@OD1-ARG+156@NH1.HH11 0, 0, 45.58 | ASP202@OD1-TYR149@OH.HH 0, 7.25, 10.40 |
| ASP178@OD2-ARG+164@NH2.HH21 0, 82.87, 40.21 | ASP202@OD1-ARG+156@NH1.HH12 49.23, 24.93, 0 | GLU196@OE1-GLY119@N.H1 10.47, 0, 0 |
| ASP178@OD1-ARG+164@NE.HE 0, 61.45, 30.03 | ASP202@OD1-ARG+156@NH2.HH21 0, 0, 10.11 | GLU196@OE1-SER120@OG.HG 12.35, 0, 0 |
| ASP178@OD1-ARG+164@NH2.HH21 0, 22.19, 30.73 | ASP202@OD1-ARG+156@NH2.HH22 31.87, 28.07, 0 | ASP178@OD1-ARG+164@NE.HE 0, 11.82, 8.88 |
| ASP178@OD2-ARG+164@NE.HE 0, 0, 13.35 | ASP202@OD2-ARG+156@NH1.HH11 0, 20.97, 0 | ASP178@OD1-ARG+164@NH1.HH11 0, 0, 18.53 |
| GLU146@OE1-LYS+204@NZ.HZ1 0, 16.53, 0 | ASP202@OD2-ARG+156@NH1.HH12 28.89, 26.21, 0 | ASP178@OD1-ARG+164@NH1.HH12 11.90, 0, 0 |
| GLU146@OE2-LYS+204@NZ.HZ1 0, 16.10, 0 | ASP202@OD2-ARG+156@NH2.HH21 0, 0, 14.52 | ASP178@OD1-ARG+164@NH2.HH21 0, 17.60, 12.73 |
| GLU146@OE2-LYS+204@NZ.HZ2 0, 16.09, 0 | ASP202@OD2-ARG+156@NH2.HH22 45.29, 18.65, 0 | ASP178@OD2-ARG+164@NH1.HH11 0, 0, 16.20 |
| GLU146@OE1-LYS+204@NZ.HZ2 0, 15.53, 0 | ASP202@OD1-TYR157@OH.HH 43.71, 27.18, 66.71 | ASP178@OD2-ARG+164@NH1.HH12 10.07, 0, 0 |
| GLU146@OE1-LYS+204@NZ.HZ3 0, 17.57, 0 | ASP202@OD2-TYR157@OH.HH 30.47, 57.38, 0 | ASP178@OD2-ARG+164@NH2.HH21 7.88, 18.63, 13.17 |
| GLU146@OE2-LYS+204@NZ.HZ3 0, 17.39, 0 | ASP178@OD1-ARG+164@NE.HE 13.48, 23.84, 15.06 | ASP178@OD1-TYR169@OH.HH 0, 27.60, 15.75 |
| GLN172@OE1-GLN219@NE2.HE21 10.89, 0, 0 | ASP178@OD1-ARG+164@NH2.HH21 24.08, 15.08, 23.03 | ASP178@OD2-TYR169@OH.HH 0, 24.72, 17.75 |
| ASP178OD1-TYR169@HH 35.55, 0, 0 | ASP178@OD2-ARG+164@NE.HE 36.29, 0, 0 | ASP178@OD1-TYR128@OH.HH 0, 12.25, 6.50 |
| ASP178OD2-TYR169@HH 40.47, 0, 0 | ASP178@OD2-ARG+164@NH2.HH21 30.03, 23.94, 20.27 | ASP178@OD2-TYR128@OH.HH 0, 12.47, 5.90 |
| ASP178@OD1-TYR169@OH.HH 32.27, 25.65, 57.75 | GLU221@OE1-SER167@OG.HG 11.70, 0, 0 | |
| ASP178@OD2-TYR169@OH.HH 0, 11.96, 30.44 | GLU221@OE2-SER167@OG.HG 10.38, 0, 0 | |
| GLU221@OE1-TYR163@OH.HH 23.40, 0, 26.15 | LEU138@O-TYR150@OH.HH 0, 10.75, 0 | |
| GLU221@OE2-TYR163@OH.HH 24.55, 14.65, 25.87 | GLY126@O-ARG+164@NH2.HH21 0, 0, 15.48 | |
| PRO158@O-ARG+136@NE.HE 0, 10.81, 0 | GLY127@O-ARG+164@NE.HE 0, 0, 14.48 | |
| ASP178@OD1-TYR128@OH.HH 6.19, 35.51, 50.43 | GLY131@O-GLN160@NE2.HE21 0, 0, 13.70 | |
| ASP178@OD2-TYR128@OH.HH 5.35, 22.53, 38.97 | GLU146@OE1-ARG+208@NE.HE 0, 0, 11.57 | |
| GLU146@OE1-ARG+208@NH2.HH21 5.88, 5.28, 11.72 | ||
| GLU146@OE2-ARG+208@NH2.HH21 5.18, 0, 13.53 |
| SBs at 300 K under Neutral pH Environment | SBs at 350 K under Neutral pH Environment | SBs at 450 K under Neutral pH Environment |
|---|---|---|
| ASP147@CG-ARG+148@CA.CZ 100, 100, 100 | ASP147@CG-ARG+148@CA.CZ 100, 100, 100 | ASP147@CG-ARG+148@CA.CZ 100, 100, 100 |
| GLU211@CD-ARG+208@CA.CZ 99.99, 98.53, 99.88 | GLU211@CD-ARG+208@CA.CZ 99.75, 99.49, 99.19 | GLU211@CD-ARG+208@CA.CZ 98.50, 99.32, 97.43 |
| GLU207@CD-LYS+204@CA.NZ 99.70, 92.81, 99.90 | GLU207@CD-LYS+204@CA.NZ 99.58, 95.18, 94.64 | GLU207@CD-LYS+204@CA.NZ 81.43, 95.43, 95.37 |
| GLU221@CD-LYS+220@CA.NZ 99.18, 95.05, 99.53 | GLU221@CD-LYS+220@CA.NZ 95.19, 75.51, 95.72 | GLU152@CD-ARG+148@CA.CZ 79.35, 37.07, 30.50 |
| GLU207@CD-ARG+208@CA.CZ 97.93, 89.95, 90.66 | GLU223@CD-LYS+220@CA.NZ 84.79, 88.04, 89.61 | GLU223@CD-LYS+220@CA.NZ 52.90, 84.60, 85.40 |
| GLU223@CD-LYS+220@CA.NZ 83.49, 86.61, 84.31 | HID177@CG-LYS+173@CA.NZ 89.28, 68.55, 83.69 | GLU207@CD-ARG+208@CA.CZ 78.65, 77.78, 71.73 |
| HID177@CG-LYS+173@CA.NZ 76.03, 54.27, 59.23 | HID177@NE2-LYS+173@CA.NZ 84.88, 59.56, 79.02 | GLU152@CD-ARG+151@CA.CZ 64.87, 41.73, 48.42 |
| ASP178@CG-ARG+164@CA.CZ 8.54, 80.97, 72.61 | GLU207@CD-ARG+208@CA.CZ 82.31, 75.41, 70.49 | GLU221@CD-LYS+220@CA.NZ 59.77, 67.45, 71.52 |
| ASP144@CG-ARG+148@CA.CZ 56.94, 63.26, 81.40 | ASP144@CG-ARG+148@CA.CZ 69.22, 66.07, 69.63 | HID177@CG-LYS+173@CA.NZ 35.13, 61.08, 61.32 |
| HID187@NE2-ARG+156@CA.CZ 75.33, 79.47, 78.88 | GLU196@CD-LYS+194@CA.NZ 80.79, 46.75, 13.83 | HID177@NE2-LYS+173@CA.NZ 24.10, 50.62, 48.95 |
| HID177@NE2-LYS+173@CA.NZ 73.13, 44.92, 48.37 | GLU152@CD-ARG+148@CA.CZ 19.88, 51.91, 50.99 | ASP147@CG-HID+140@ND1.HD1 57.98, 56.63, 71.32 |
| ASP147@CG-HID+140@ND1.HD1 32.85, 59.40, 5.03 | HID187@NE2-ARG+156@CA.CZ 8.05, 50.02, 68.03 | ASP147@CG-ARG+151@CA.CZ 53.13, 57.55, 52.87 |
| GLU152@CD-ARG+148@CA.CZ 41.30, 57.81, 22.23 | ASP147@CG-HID+140@ND1.HD1 66.73, 14.95, 50.97 | ASP144@CG-ARG+148@CA.CZ 46.73, 40.42, 42.32 |
| GLU152@CD-ARG+151@CA.CZ 40.62, 24.43, 46.85 | ASP147@CG-ARG+151@CA.CZ 40.29, 51.06, 19.25 | GLU168@CD-ARG+164@CA.CZ 18.28, 42.77, 9.27 |
| ASP147@CG-ARG+151@CA.CZ 24.14, 6.27, 36.33 | GLU168@CD-ARG+164@CA.CZ 38.75, 33.56, 57.47 | GLU146@CD-ARG+208@CA.CZ 5.67, 19.18, 13.50 |
| ASP178@CG-HID+177@ND1.HD1 5.60, 14.60, 21.83 | GLU152@CD-ARG+151@CA.CZ 65.20, 42.93, 42.81 | ASP178@CG-HID+177@ND1.HD1 16.07, 17.85, 19.48 |
| GLU211@CD-HID+177@ND1.HD1 20.18, 8.37, 12.57 | GLU196@CD-ARG+156@CA.CZ 17.17, 41.71, 43.47 | GLU211@CD-HID+177@ND1.HD1 16.42, 16.27, 12.23 |
| GLU196@CD-LYS+194@CA.NZ 0, 63.33, 0 | GLU211@CD-HID+177@ND1.HD1 27.48, 15.45, 17.77 | HID187@CG-LYS+185@CA.NZ 38.85, 5.92, 0 |
| GLU168@CD-ARG+164@CA.CZ 52.00, 0, 8.60 | ASP178@CG-HID+177@ND1.HD1 10.42, 14.12, 8.83 | ASP178@CG-ARG+164@CA.CZ 0, 22.00, 38.58 |
| GLU196@CD-ARG+156@CA.CZ 0, 9.63, 0 | ASP178@CG-ARG+164@CA.CZ 10.45, 33.61, 0 | GLU152@CD-ARG+156@CA.CZ 34.87, 0, 0 |
| ASP202@CG-ARG+156@CA.CZ 0, 6.53, 0 | GLU223@CD-ARG+228@CA.CZ 0, 0, 10.11 | HID187@NE2-LYS+185@CA.NZ 25.88, 0, 0 |
| HID187@CG-ARG+156@CA.CZ 0, 0, 7.57 | GLU196@CD-LYS+194@CA.NZ 0, 24.88, 14.97 | |
| ASP202@CG-ARG+156@CA.CZ 0, 0, 6.69 | HID187@NE2-ARG+156@CA.CZ 0, 15.08, 24.72 | |
| ASP144@CG-HID+140@ND1.HD1 7.40, 0, 0 | GLU146@CD-HID+140@ND1.HD1 24.42, 0, 16.37 | |
| ASP144@CG-HID+140@ND1.HD1 0, 5.23, 19.77 | ||
| GLU196@CD-ARG+156@CA.CZ 0, 16.48, 16.42 | ||
| HID140@NE2-ARG+136@CA.CZ 14.30, 0, 0 | ||
| ASP202@CG-LYS+194@CA.NZ 0, 12.78, 14.15 | ||
| GLU168@CD-ARG+228@CA.CZ 9.57, 0, 0 | ||
| ASP202@CG-ARG+156@CA.CZ 9.35, 0, 0 | ||
| GLU200@CD-HID+187@ND1.HD1 8.30, 0, 0 | ||
| GLU146@CD-LYS+204@CA.NZ 0, 5.88, 5.47 | ||
| HID140@NE2-ARG+208@CA.CZ 0, 0, 11.40 | ||
| HID140@CG-ARG+136@CA.CZ 7.42, 0, 0 | ||
| GLU200@CD-LYS+194@CA.NZ 0, 0, 10.70 | ||
| GLU221@CD-ARG+228@CA.CZ 7.33, 0, 0 | ||
| GLU207@CD-HID+177@ND1.HD1 6.42, 5.13, 0 |
| Under Low-pH Environment | Under Neutral pH Environment | Position in the PrP Structure |
|---|---|---|
| PHE225@CB-ALA224@CA.C | PHE225@CB-ALA224@CA.C | Within α3 |
| ALA224@CB-PHE225@CA.C | ALA224@CB-PHE225@CA.C | Within α3 |
| VAL210@CB-VAL209@CA.C | VAL210@CB-VAL209@CA.C | Within α3 |
| VAL209@CB-VAL210@CA.C | VAL209@CB-VAL210@CA.C | Within α3 |
| VAL209@CB-MET206@CA.C | VAL209@CB-MET206@CA.C | Within α3 |
| MET206@CB-ILE205@CA.C | MET206@CB-ILE205@CA.C | Within α3 |
| ILE205@CB-MET206@CA.C | ILE205@CB-MET206@CA.C | Within α3 |
| VAL176@CB-PHE175@CA.C | VAL176@CB-PHE175@CA.C | Within α2 |
| PHE175@CB-VAL176@CA.C | PHE175@CB-VAL176@CA.C | Within α2 |
| VAL166@CB-PRO165@CA.C | VAL166@CB-PRO165@CA.C | Within β2-310H2-loop |
| PRO165@CB-VAL166@CA.C | PRO165@CB-VAL166@CA.C | Within β2-310H2-loop |
| ILE139@CB-LEU138@CA.C | ILE139@CB-LEU138@CA.C | Within β1-α1-loop |
| LEU138@CB-ILE139@CA.C | LEU138@CB-ILE139@CA.C | Within β1-α1-loop |
| LEU138@CB-PRO137@CA.C | LEU138@CB-PRO137@CA.C | Within β1-α1-loop |
| PRO137@CB-LEU138@CA.C | PRO137@CB-LEU138@CA.C | Within β1-α1-loop |
| MET134@CB-ALA133@CA.C | MET134@CB-ALA133@CA.C | Linking β1 and β1-α1-loop |
| ALA133@CB-MET134@CA.C | ALA133@CB-MET134@CA.C | Linking β1 and β1-α1-loop |
| LEU130@CB-MET129@CA.C | LEU130@CB-MET129@CA.C | Within β1 |
| MET129@CB-LEU130@CA.C | MET129@CB-LEU130@CA.C | Within β1 |
| VAL122@CB-VAL121@CA.C | VAL122@CB-VAL121@CA.C | Within N-terminal |
| VAL121@CB-VAL122@CA.C | VAL121@CB-VAL122@CA.C | Within N-terminal |
| MET213@CB-VAL210@CA.C 99.98, 100, 99.98 | MET213@CB-VAL210@CA.C 99.97, 100, 100 | Within α3 |
| MET206@CB-VAL203@CA.C 95.20, 100, 99.93 | MET206@CB-VAL203@CA.C 100, 99.90, 100 | Within α3 |
| 300 K—Under Neutral pH Environment | 350 K—Under Neutral pH Environment | Position in the PrP Structure | |
|---|---|---|---|
| VAL210@CB-VAL180@CA.C | VAL210@CB-VAL180@CA.C | 100% exist at 300 K—low pH | Linking α3 and α2 |
| MET213@CB-VAL161@CA.C | MET213@CB-VAL161@CA.C | Linking α3 and β2 | |
| VAL161@CB-MET213@CA.C | VAL161@CB-MET213@CA.C | Linking β2 and α3 | |
| VAL176@CB-ILE215@CA.C | Linking α2 and α3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J. Molecular Dynamics and Optimization Studies of Horse Prion Protein Wild Type and Its S167D Mutant. Zoonotic Dis. 2024, 4, 187-200. https://doi.org/10.3390/zoonoticdis4030017
Zhang J. Molecular Dynamics and Optimization Studies of Horse Prion Protein Wild Type and Its S167D Mutant. Zoonotic Diseases. 2024; 4(3):187-200. https://doi.org/10.3390/zoonoticdis4030017
Chicago/Turabian StyleZhang, Jiapu. 2024. "Molecular Dynamics and Optimization Studies of Horse Prion Protein Wild Type and Its S167D Mutant" Zoonotic Diseases 4, no. 3: 187-200. https://doi.org/10.3390/zoonoticdis4030017
APA StyleZhang, J. (2024). Molecular Dynamics and Optimization Studies of Horse Prion Protein Wild Type and Its S167D Mutant. Zoonotic Diseases, 4(3), 187-200. https://doi.org/10.3390/zoonoticdis4030017






