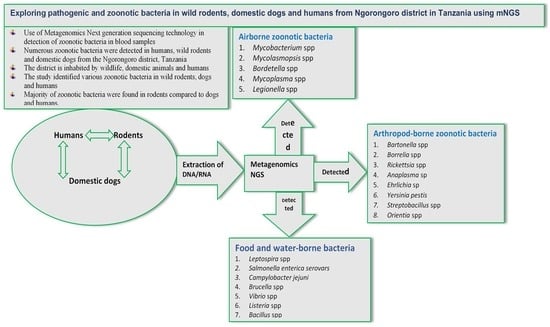
Exploring Pathogenic and Zoonotic Bacteria from Wild Rodents, Dogs, and Humans of the Ngorongoro District in Tanzania Using Metagenomics Next-Generation Sequencing
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
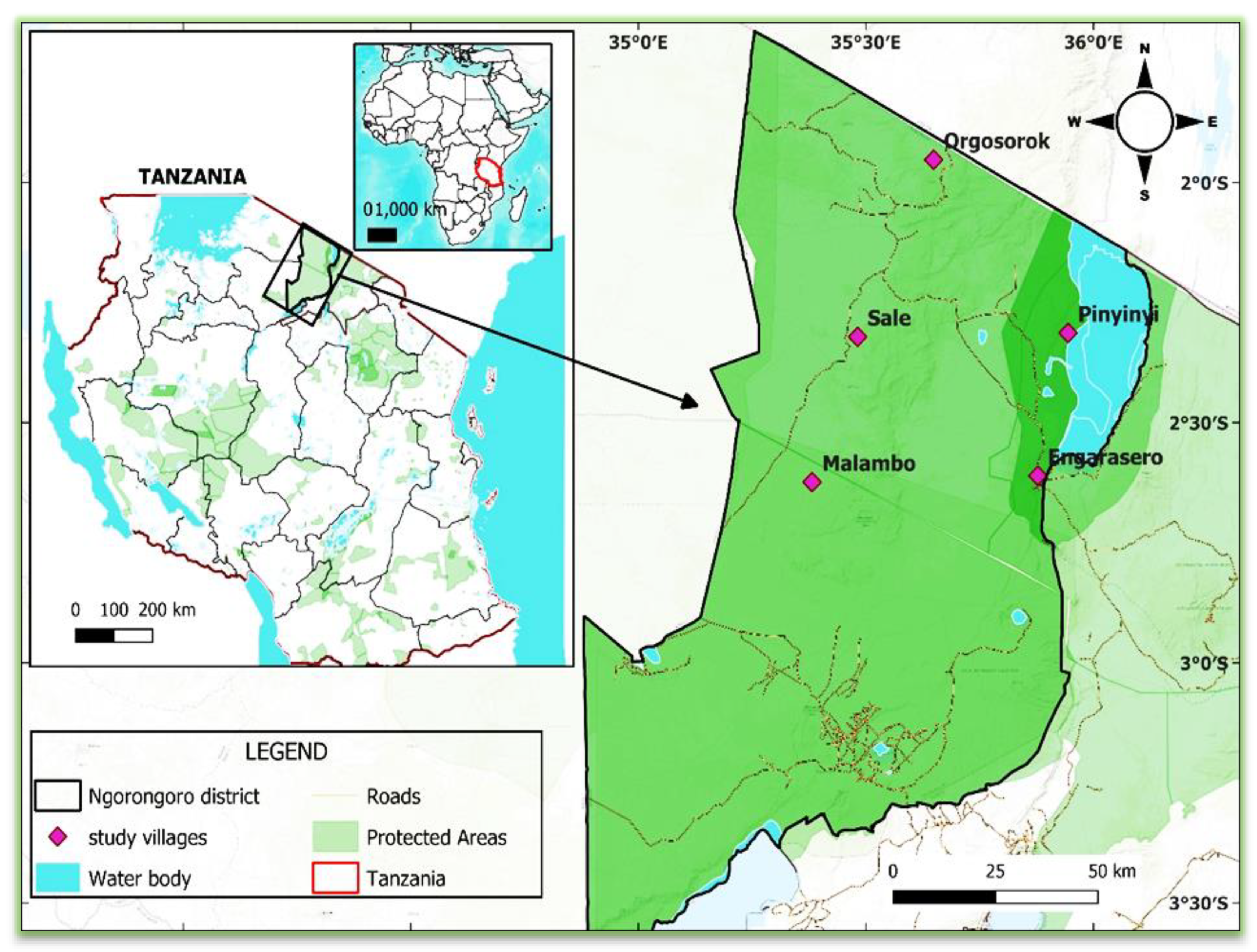
2.1. Description of the Study Area
2.2. Study Design and Sampling Procedures
2.3. Trapping of Rodents
2.4. Collection and Handling of Samples (from Wild Rodents)
2.5. Collection of Blood Samples from Humans and Domestic Dogs
2.6. Preparation of Pools of Blood Samples
2.7. Nucleic Acids Extraction, Libraries Preparation, and Sequencing
2.8. Bioinformatics Analysis
3. Results
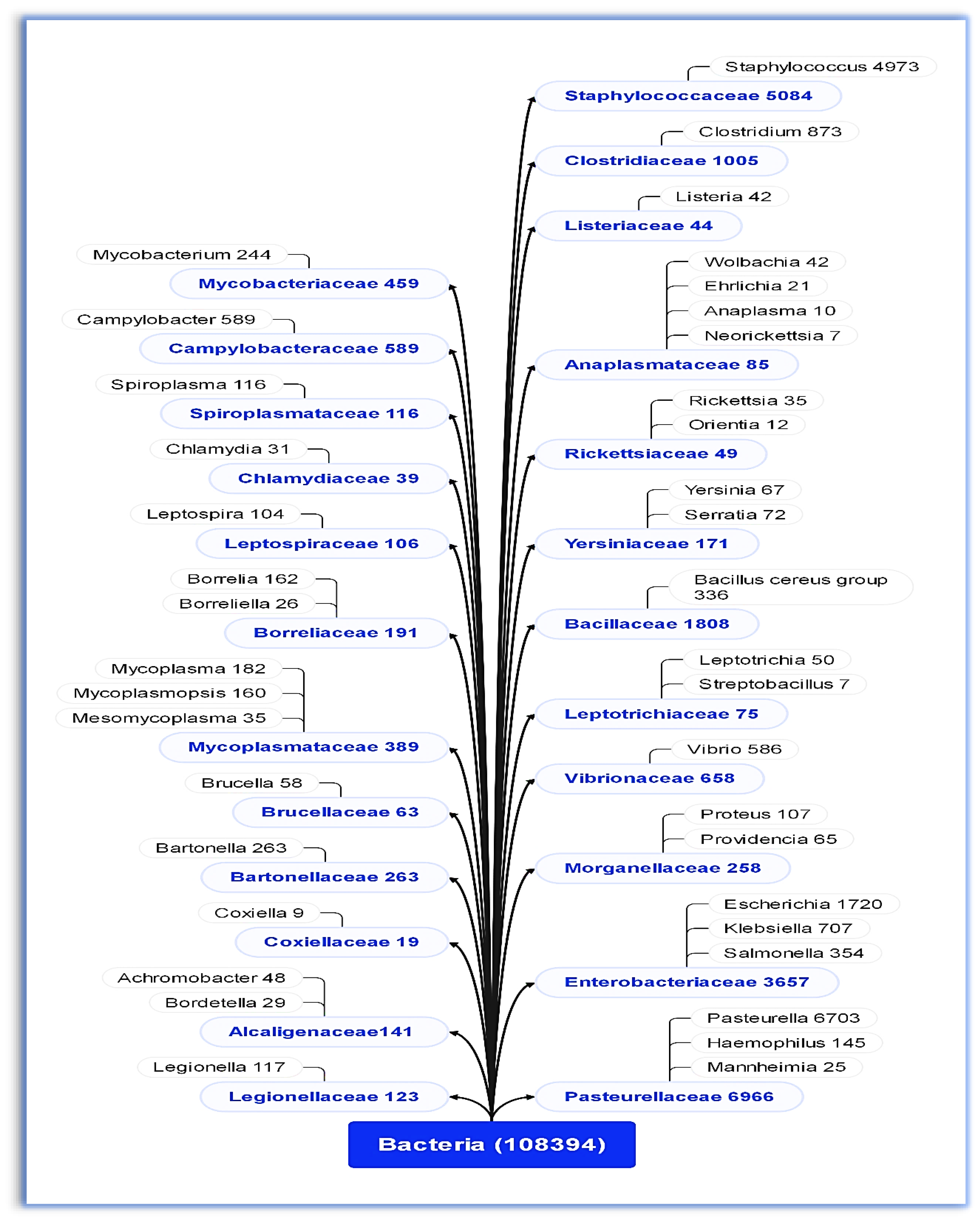
3.1. Bacterial Families and Genera Identified
3.2. Airborne, Contagious, and Arthropod-Borne Zoonotic Bacteria
3.3. Pathogenic and Zoonotic Bacteria Detected in Humans, Wild Rodents, and Domestic Dogs
4. Discussion
Authors’ Reflection Based on the Finding
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- White, R.J.; Razgour, O. Emerging zoonotic diseases originating in mammals: A systematic review of effects of anthropogenic land-use change. Mammal Rev. 2020, 50, 336–352. [Google Scholar] [CrossRef] [PubMed]
- Daszak, P.; Cunningham, A.A.; Hyatt, A.D. Anthropogenic environmental change and the emergence of infectious diseases in wildlife. Acta Trop. 2001, 78, 103–116. [Google Scholar] [CrossRef]
- Rahman, M.T.; Sobur, M.A.; Islam, M.S.; Ievy, S.; Hossain, M.J.; El Zowalaty, M.E.; Rahman, A.T.; Ashour, H.M. Zoonotic diseases: Etiology, impact, and control. Microorganisms 2020, 8, 1405. [Google Scholar] [CrossRef]
- Mgode, G.F.; Katakweba, A.S.; Mhamphi, G.G.; Fwalo, F.; Bahari, M.; Mdangi, M.; Kilonzo, B.S.; Mulungu, L.S. Prevalence of leptospirosis and toxoplasmosis: A study of rodents and shrews in cultivated and fallow land, Morogoro rural district, Tanzania. Tanzan. J. Health Res. 2014, 16, 3. [Google Scholar] [CrossRef]
- Theonest, N.O.; Carter, R.W.; Amani, N.; Doherty, S.L.; Hugho, E.; Keyyu, J.D.; Mable, B.K.; Shirima, G.M.; Tarimo, R.; Thomas, K.M.; et al. Molecular detection and genetic characterization of Bartonella species from rodents and their associated ectoparasites from northern Tanzania. PLoS ONE 2019, 14, e0223667. [Google Scholar] [CrossRef]
- Durnez, L.; Eddyani, M.; Mgode, G.F.; Katakweba, A.; Katholi, C.R.; Machang’u, R.R.; Kazwala, R.R.; Portaels, F.; Leirs, H. First detection of mycobacteria in African rodents and insectivores, using stratified pool screening. Appl. Environ. Microbiol. 2008, 74, 768–773. [Google Scholar] [CrossRef]
- Song, H.; Kim, J.; Guk, J.H.; Kim, W.H.; Nam, H.; Suh, J.G.; Seong, J.K.; Cho, S. Metagenomic analysis of the gut microbiota of wild mice, a newly identified reservoir of Campylobacter. Front. Cell. Infect. Microbiol. 2021, 10, 596149. [Google Scholar] [CrossRef]
- Hieronimo, P.; Kimaro, D.N.; Kihupi, N.I.; Gulinck, H.; Mulungu, L.S.; Msanya, B.M.; Leirs, H.; Deckers, J.A. Land use determinants of small mammals’ abundance and distribution in a plague endemic area of Lushoto District, Tanzania. Tanzan. J. Health Res. 2014, 16, 3. [Google Scholar] [CrossRef][Green Version]
- McCauley, D.J.; Salkeld, D.J.; Young, H.S.; Makundi, R.; Dirzo, R.; Eckerlin, R.P.; Lambin, E.F.; Gaffikin, L.; Barry, M.; Helgen, K.M. Effects of land use on plague (Yersinia pestis) activity in rodents in Tanzania. Am. J. Trop. Med. Hyg. 2015, 92, 776. [Google Scholar] [CrossRef]
- Katakweba, A.A.; Mulungu, L.S.; Eiseb, S.J.; Mahlaba, T.A.; Makundi, R.H.; Massawe, A.W.; Borremans, B.; Belmain, S.R. Prevalence of haemoparasites, leptospires and coccobacilli with potential for human infection in the blood of rodents and shrews from selected localities in Tanzania, Namibia and Swaziland. Afr. Zool. 2012, 47, 119–127. [Google Scholar]
- Chipwaza, B.; Sumaye, R.D.; Weisser, M.; Gingo, W.; Yeo, N.K.; Amrun, S.N.; Okumu, F.O.; Ng, L.F. Occurrence of 4 dengue virus serotypes and chikungunya virus in Kilombero Valley, Tanzania, during the dengue outbreak in 2018. Open Forum Infect. Dis. 2021, 8, 626. [Google Scholar] [CrossRef]
- Issae, A.M. Community Knowledge, Attitudes and Practices on Dog Management and Epidemiology of Parasitic Infestations in Dogs of Mvomero District and Morogoro Municipality, Tanzania. Ph.D. Thesis, Sokoine University of Agriculture, Morogoro, Tanzania, 2018. Available online: https://www.suaire.sua.ac.tz/handle/123456789/2742 (accessed on 12 June 2023).
- De Vries, S.G.; Visser, B.J.; Nagel, I.M.; Goris, M.G.; Hartskeerl, R.A.; Grobusch, M.P. Leptospirosis in Sub-Saharan Africa: A systematic review. Int. J. Infect. Dis. 2014, 28, 47–64. [Google Scholar] [CrossRef]
- Yaovi, A.B.; Sessou, P.; Tonouhewa, A.B.; Hounmanou, G.Y.; Thomson, D.; Pelle, R.; Farougou, S.; Mitra, A. Prevalence of antibiotic-resistant bacteria amongst dogs in Africa: A meta-analysis review. Onderstepoort J. Vet. Res. 2022, 89, 1–12. [Google Scholar] [CrossRef]
- Mwakapeje, E.R.; Høgset, S.; Fyumagwa, R. Anthrax outbreaks in the humans—Livestock and wildlife interface areas of Northern Tanzania: A retrospective record review 2006–2016. BMC Public Health 2018, 18, 106. [Google Scholar] [CrossRef]
- Katale, B.Z.; Mbugi, E.V.; Karimuribo, E.D.; Keyyu, J.D.; Kendall, S.; Kibiki, G.S.; Godfrey-Faussett, P.; Michel, A.L.; Kazwala, R.R.; van Helden, P.; et al. Prevalence and risk factors for infection of bovine tuberculosis in indigenous cattle in the Serengeti ecosystem, Tanzania. BMC Vet Res. 2013, 9, 267. [Google Scholar] [CrossRef]
- Motto, S.K.; Shirima, G.M.; de Clare Bronsvoort, B.M.; Cook, E.A.J. Epidemiology of leptospirosis in Tanzania: A review of the current status, serogroup diversity and reservoirs. PLoS Negl. Trop. Dis. 2021, 15, e0009918. [Google Scholar] [CrossRef]
- Mellau, L.S.; Kuya, S.L.; Wambura, P.N. Seroprevalence of brucellosis in domestic ruminants in livestock-wildlife interface: A case study of Ngorongoro Conservation Area, Arusha, Tanzania. Tanzan. Vet. J. 2009, 26, 44–50. [Google Scholar] [CrossRef]
- Makala, R.; Majigo, M.V.; Bwire, G.M.; Kibwana, U.; Mirambo, M.M.; Joachim, A. Seroprevalence of Brucella infection and associated factors among pregnant women receiving antenatal care around human, wildlife and livestock interface in Ngorongoro ecosystem, Northern Tanzania. A cross-sectional study. BMC Infect. Dis. 2020, 20, 152. [Google Scholar] [CrossRef]
- Mbugi, E.V.; Katale, B.Z.; Siame, K.K.; Keyyu, J.D.; Kendall, S.L.; Dockrell, H.M.; Streicher, E.M.; Michel, A.L.; Rweyemamu, M.M.; Warren, R.M.; et al. Genetic diversity of Mycobacterium tuberculosis isolated from tuberculosis patients in the Serengeti ecosystem in Tanzania. Tuberculosis 2015, 95, 170–178. [Google Scholar] [CrossRef][Green Version]
- Swai, E.S.; Mkumbukwa, A.J.; Chaula, S.L.; Leba, B.G. Epidemiological Investigation of Bovine Brucellosis in Indigenous Cattle Herds in Kasulu District of Tanzania. Yale J. Biol. Med. 2021, 94, 285–296. [Google Scholar]
- Niboye, E.P. Vegetation cover changes in Ngorongoro Conservation Area from 1975 to 2000: The importance of remote sensing images. Open Geogr. J. 2010, 3, 15–27. [Google Scholar] [CrossRef]
- Nielsen, H.B.; Almeida, M.; Juncker, A.S.; Rasmussen, S.; Li, J.; Sunagawa, S.; Plichta, D.R.; Gautier, L.; Pedersen, A.G.; Le Chatelier, E. Identification and Assembly of Genomes and Genetic Elements in Complex Metagenomic Samples without Using Reference Genomes. Nat. Biotechnol. 2014, 32, 822–828. [Google Scholar] [CrossRef] [PubMed]
- Amrane, S.; Lagier, J.C. Metagenomic and clinical microbiology. Hum. Microbiome J. 2018, 9, 1–6. [Google Scholar] [CrossRef]
- Nayfach, S.; Roux, S.; Seshadri, R.; Udwary, D.; Varghese, N.; Schulz, F.; Wu, D.; Paez-Espino, D.; Chen, I.M.; Huntemann, M.; et al. A genomic catalog of Earth’s microbiomes. Nat. Biotechnol. 2021, 39, 499–509. [Google Scholar] [CrossRef]
- Mweya, C.N.; Holst, N.; Mboera, L.E.; Kimera, S.I. Simulation modelling of population dynamics of mosquito vectors for Rift Valley fever virus in a disease epidemic setting. PLoS ONE 2014, 9, e108430. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tanzania Ministry of Finance. 2012 Population and Housing Census General Report; National Bureau of Statistics: Dar es Salaam, Tanzania, 2023; 264p. [Google Scholar]
- World Health Organization. WHO Guidelines on Drawing Blood: Best Practices in Phlebotomy; World Health Organization: Rome, Italy, 2010.
- Lima-Oliveira, G.; Lippi, G.; Salvagno, G.L.; Picheth, G.; Guidi, G.C. Laboratory Diagnostics and Quality of Blood Collection. J. Med. Biochem. 2015, 34, 288–294. [Google Scholar] [CrossRef]
- McKernan, K.; Spangler, J.; Helbert, Y.; Lynch, R.C.; Devitt-Lee, A.; Zhang, L.; Orphe, W.; Warner, J.; Foss, T.; Hudalla, C.J.; et al. Metagenomic analysis of medicinal Cannabis samples; pathogenic bacteria, toxigenic fungi, and beneficial microbes grow in culture-based yeast and mold tests. F1000Research 2016, 5, 2471. [Google Scholar] [CrossRef]
- Weber, M.N.; Cibulski, S.P.; Olegario, J.C.; Da Silva, M.S.; Puhl, D.E.; Mosena, A.C.; Alves, C.D.; Paim, W.P.; Baumbach, L.F.; Mayer, F.Q.; et al. Characterization of dog serum virome from Northeastern Brazil. Virology 2018, 525, 192–199. [Google Scholar] [CrossRef]
- Wingett, S.W.; Andrews, S. FastQ Screen: A tool for multi-genome mapping and quality control. F1000Research 2018, 7, 1338. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef] [PubMed]
- Katale, B.Z.; Mbugi, E.V.; Keyyu, J.D.; Fyumagwa, R.D.; Rweyemamu, M.M.; Van Helden, P.D.; Dockrell, H.M.; Matee, M.I. One Health approach in the prevention and control of mycobacterial infections in Tanzania: Lessons learnt and future perspectives. One Health Outlook 2019, 1, 2. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Goyal, P.; Mattana, J. Bordetella bronchiseptica pneumonia a thread in the diagnosis of human immunodeficiency virus infection. ID Cases 2019, 15, e00509. [Google Scholar] [CrossRef] [PubMed]
- Trainor, E.A.; Nicholson, T.L.; Merkel, T.J. Bordetella pertussis transmission. Pathog. Dis. 2015, 73, ftv068. [Google Scholar] [CrossRef]
- Huchzermeyer, F.W. Diseases of farmed crocodiles and ostriches. Rev. Sci. Tech. Off. Int. Épizooties 2002, 21, 265–276. [Google Scholar] [CrossRef]
- Simionatto, S.; Marchioro, S.B.; Maes, D.; Dellagostin, O.A. Mycoplasma hyopneumoniae: From disease to vaccine development. Vet. Microbiol. 2013, 165, 234–242. [Google Scholar] [CrossRef]
- Hoelzle, L.E.; Zeder, M.; Felder, K.M.; Hoelzle, K. Pathobiology of Mycoplasma suis. Vet. J. 2014, 202, 20–25. [Google Scholar] [CrossRef]
- Acosta, D.B.; Ruiz, M.; Sanchez, J.P. First molecular detection of Mycoplasma suis in the pig louse Haematopinus suis (Phthiraptera: Anoplura) from Argentina. Acta Trop. 2019, 194, 165–168. [Google Scholar] [CrossRef]
- Hernandez, L.; Lopez, J.; St-Jacques, M.; Ontiveros, L.; Acosta, J.; Handel, K. Mycoplasma mycoides subsp. capri associated with goat respiratory disease and high flock mortality. Can. Vet. J. 2006, 47, 366. [Google Scholar]
- Cunha, B.A.; Burillo, A.; Bouza, E. Legionnaires’ disease. Lancet 2016, 387, 376–385. [Google Scholar] [CrossRef]
- Murphy, D.S.; Lee, X.; Larson, S.R.; Johnson, D.K.; Loo, T.; Paskewitz, S.M. Prevalence and distribution of human and tick infections with the Ehrlichia muris-like agent and Anaplasma phagocytophilum in Wisconsin, 2009–2015. Vector-Borne Zoonotic Dis. 2017, 17, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Talagrand-Reboul, E.; Boyer, P.H.; Bergström, S.; Vial, L.; Boulanger, N. Relapsing fevers: Neglected tick-borne diseases. Front. Cell. Infect. Microbiol. 2018, 8, 98. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, R.S.; Santodomingo, A.M.; Muñoz-Leal, S.; Silva-de la Fuente, M.C.; Llanos-Soto, S.; Salas, L.M.; González-Acuña, D. Rodents as potential reservoirs for Borrelia spp. in northern Chile. Rev. Bras. Parasitol. Vet. 2020, 29. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, R.; Signoli, M.; Chevé, D.; Costedoat, C.; Tzortzis, S.; Aboudharam, G.; Raoult, D.; Drancourt, M. Yersinia pestis: The natural history of plague. Clin. Microbiol. Rev. 2020, 34, e00044-19. [Google Scholar] [CrossRef] [PubMed]
- Kolo, A.O.; Collins, N.E.; Brayton, K.A.; Chaisi, M.; Blumberg, L.; Frean, J.; Gall, C.A.; Wentzel, J.M.; Wills-Berriman, S.; de Boni, L.; et al. Anaplasma phagocytophilum and other anaplasma spp. in various hosts in the Mnisi community, Mpumalanga Province, South Africa. Microorganisms 2020, 8, 1812. [Google Scholar] [CrossRef]
- Kamani, J.; Morick, D.; Mumcuoglu, K.Y.; Harrus, S. Prevalence and diversity of Bartonella species in commensal rodents and ectoparasites from Nigeria, West Africa. PLoS Negl. Trop. Dis. 2013, 7, e2246. [Google Scholar] [CrossRef]
- Kosoy, M.; Bai, Y.; Sheff, K.; Morway, C.; Baggett, H.; Maloney, S.A.; Boonmar, S.; Bhengsri, S.; Dowell, S.F.; Sitdhirasdr, A.; et al. Identification of Bartonella infections in febrile human patients from Thailand and their potential animal reservoirs. Am. J. Trop. Med. Hyg. 2010, 82, 1140. [Google Scholar] [CrossRef]
- Chung, M.H.; Lee, J.S.; Baek, J.H.; Kim, M.; Kang, J.S. Persistence of Orientia tsutsugamushi in humans. J. Korean Med. Sci. 2012, 27, 231–235. [Google Scholar] [CrossRef]
- Madhubashini, M.; George, S.; Chandrasekaran, S. Streptobacillus moniliformis endocarditis: Case report and review of literature. Indian Heart J. 2013, 65, 442–446. [Google Scholar] [CrossRef]
- Stewart, A.G.; Stewart, A.G. An update on the laboratory diagnosis of Rickettsia spp. infection. Pathogens 2021, 10, 1319. [Google Scholar] [CrossRef]
- Pinter, A.; Horta, M.C.; Pacheco, R.C.; Moraes-Filho, J.; Labruna, M.B. Serosurvey of Rickettsia spp. in dogs and humans from an endemic area for Brazilian spotted fever in the State of São Paulo, Brazil. Cad. Saude Publica 2008, 24, 247–252. [Google Scholar] [CrossRef]
- Blanda, V.; Torina, A.; La Russa, F.; D’Agostino, R.; Randazzo, K.; Scimeca, S.; Giudice, E.; Caracappa, S.; Cascio, A.; de la Fuente, J. A retrospective study of the characterization of Rickettsia species in ticks collected from humans. Ticks Tick-Borne Dis. 2017, 8, 610–614. [Google Scholar] [CrossRef]
- Moraga Fernández, A.; Ortiz, J.A.; Jabbar, A.; Ghafar, A.; Cabezas-Cruz, A.; de la Fuente, G.; de la Fuente, J.; Fernández de Mera, I.G. Fatal cases of bovine anaplasmosis in a herd infected with different Anaplasma marginale genotypes in southern Spain. Ticks Tick-Borne Dis. 2022, 13, 101864. [Google Scholar] [CrossRef] [PubMed]
- Arraga-Alvarado, C.M.; Qurollo, B.A.; Parra, O.C.; Berrueta, M.A.; Hegarty, B.C.; Breitschwerdt, E.B. Case report: Molecular evidence of Anaplasma platys infection in two women from Venezuela. Am. J. Trop. Med. Hyg. 2014, 91, 1161. [Google Scholar] [CrossRef]
- Nzalawahe, J.S.; Komba, E.V.; Lupindu, A.M.; Materu, A.E.; Katakweba, A.S.; Mnyone, L.L. Canine Ehrlichiosis in Africa: Epidemiology, Diagnosis, and Control. In Combating and Controlling Nagana and Tick-Borne Diseases in Livestock; IGI Global: Hershey, PA, USA, 2020; pp. 288–310. [Google Scholar]
- Topazio, J.; Tonin, A.A.; Machado, G.; Noll, J.C.; Ribeiro, A.; Moura, A.B.; Carmo, G.M.; Grosskopf, H.M.; Martins, J.L.; Badke, M.R.; et al. Antibodies to Leptospira interrogans in goats and risk factors of the disease in Santa Catarina (West side), Brazil. Res. Vet. Sci. 2015, 99, 53–57. [Google Scholar] [CrossRef]
- Lee, S.A.; Sang, M.K.; Song, J.; Kwon, S.W.; Weon, H.Y. Complete genome sequence of Brucella anthropic strain T16R-87 isolated from tomato (Solanum lycopersicum L.) rhizosphere. Microbiol. Soc. Korea 2020, 56, 430–432. [Google Scholar]
- Lama, M.; Chanakya, P.P.; Khamari, B.; Peketi, A.S.; Kumar, P.; Muddu, G.K.; Nagaraja, V.; Bulagonda, E.P. Genomic analysis of a multidrug-resistant Brucella anthropi strain isolated from a 4-day-old neonatal sepsis patient. J. Glob. Antimicrob. Resist. 2021, 26, 227–229. [Google Scholar] [CrossRef] [PubMed]
- Mor, S.M.; Wiethoelter, A.K.; Lee, A.; Moloney, B.; James, D.R.; Malik, R. Emergence of Brucella suis in dogs in New South Wales, Australia: Clinical findings and implications for zoonotic transmission. BMC Vet. Res. 2016, 12, 199. [Google Scholar] [CrossRef]
- Young, K.T.; Davis, L.M.; DiRita, V.J. Campylobacter jejuni: Molecular biology and pathogenesis. Nat. Rev. Microbiol. 2007, 5, 665–679. [Google Scholar] [CrossRef] [PubMed]
- Jordan, K.; McAuliffe, O. Listeria monocytogenes in foods. Adv. Food Nutr. Res. 2018, 86, 181–213. [Google Scholar] [PubMed]
- Dodds, K.L. Clostridium botulinum in foods. In Clostridium botulinum; CRC Press: Boca Raton, FL, USA, 2018; pp. 52–68. [Google Scholar]
- Cabral, J.P. Water microbiology. Bacterial pathogens and water. Int. J. Environ. Res. Public Health 2010, 7, 3657–3703. [Google Scholar] [CrossRef] [PubMed]
| Sample Type | Sex | Number of Samples Studied | Number of Pools | Number of Samples (s) per Pool | Pooling Volume (μL) per Sample | Total Volume (mL) per Pool |
|---|---|---|---|---|---|---|
| Human | Female | 130 | 13 | 10 | 100 | 1 |
| Male | 70 | 9 | 7–8 | 100 | 0.7–0.8 | |
| Total | 200 | 22 | ||||
| Wild Rodents | Total | 230 | 16 | 14–15 | 80 | 1.12–1.2 |
| Domestic Dogs | Female | 57 | 5 | 11–12 | 100 | 1.1–1.2 |
| Male | 43 | 5 | 8–10 | 100 | 0.8–1 | |
| Total | 100 | 10 |
| Hosts | Positive Pools for Airborne and Contagious Bacteria Species | Positive Pools for Arthropod-Borne Bacteria Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mycobacterium sp. | Mycoplasma sp. | Mycoplasmopsis sp. | Bordetella sp. | Legionella sp. | Borrelia sp. | Borreliella sp. | Bartonella sp. | Yersinia pestis | Orientia sp. | Streptobacillus sp. | Rickettsia sp. | Anaplasma sp. | Ehrlichia sp. | |
| Humans | 11 pools | 2 pools | 0 | 4 pools | 2 pools | 1 pool | 0 | 1 pool | 0 | 0 | 0 | 2 pools | 0 | 0 |
| n = 22 pools | (50%) | (9.09%) | (0.0%) | (18.2%) | (9.09%) | (4.54%) | (0.0%) | (4.54%) | (0.0%) | (0.0%) | (0.0%) | (9.09%) | (0.0%) | (0.0%) |
| Rodents | 9 pools | 7 pools | 8 pools | 3 pools | 7 pools | 6 pools | 5 pools | 11 pools | 1 pool | 2 pools | 1 pool | 3 pools | 1 pool | 0 |
| n = 16 pools | (56.25%) | (43.75%) | (50.0%) | (18.75%) | (43.75%) | (37.5%) | (31.25%) | (68.75%) | (6.25%) | (12.5%) | (6.25%) | (18.75%) | (6.25%) | (0.0%) |
| Dogs | 3 pools | 4 pools | 3 pools | 4 pools | 3 pools | 3 pools | 0 | 5 pools | 0 | 1 pool | 0 | 2 pools | 0 | 2 pools |
| n = 10 pools | (30.0%) | (40.0%) | (30.0%) | (40.0%) | (30%) | (30.0%) | (0.0%) | (50.0%) | (0.0%) | (10.0%) | (0.0%) | (20.0%) | (0.0%) | (20.0%) |
| Na | Genus | Isolated Species | Host |
|---|---|---|---|
| 1 | Mycobacterium avium complex | M. avium subsp. Paratuberculosis, M. intracellulare subsp. chimaera | Rodents, humans, and dogs |
| M. avium subsp. Hominissuis, M. kansasii, M. koreense, M. diernhoferi, M. paragordonae, and M. mantenii | Rodents | ||
| Mycobacterium tuberculosis complex | M. canettii and M. tuberculosis | Rodents, dogs, and humans | |
| M. grossiae, M. colombiense, M. mantenii, M. virginiense, M. basiliense, M. paragordonae, M. diernhoferi, M. marseillense, and M. senriense | Rodents | ||
| Mycobacterium simiae complex | M. simiae, and M. rufum | Rodents and humans | |
| M. kubicae, M. lentiflavum, and M. saskatchewanense | Rodents | ||
| Mycobacterium ulcerans group | M. ulcerans subsp. Shinshuense. M. spongiae M. paraseoulense, M. dioxanotrophicus, M. shinjukuense, M. ostraviense, M. kansasii, M. holsaticum, M. leprae, and M. goodii | Rodents | |
| M. seoulense, M. lacus, and M. cookii | Rodents and humans | ||
| M. xenopi | Rodents, dogs, and humans | ||
| M. virginiense and M. heidelbergense | Humans | ||
| 2 | Mycoplasma | M. miroungigenitalium, M. fastidiosum, M. hyopneumoniae M. putrefaciens, M. haemofelis, M. wenyonii, M. parvum, and M. iguanae | Rodents |
| M. crocodyli, M. pneumoniae, M. suis, and M. tauri | Rodents and humans | ||
| M. mycoides subsp. Capri and M. haemocanis | Dogs | ||
| 3 | Mycoplasmopsis | M. arginini | Rodents and dogs |
| M. bovirhinis, M. gallopavonis, M. agalactiae M. synoviae, M. felis, M. equigenitalium, and M. meleagridis | Rodents | ||
| M. glycophila, M. canis, M. bovis, and M. gallinacea | Dogs | ||
| 4 | Bordetella | B. bronchiseptica | Rodents, dogs, and humans |
| B. bronchialis, B. parapertussis, B. avium, and B. pseudohinzii | Dogs | ||
| B. genomosp. 6, B. flabilis, and B. trematum | Dogs and humans | ||
| B. hinzii | Humans | ||
| 5 | Legionella | L. pneumophila and L. sainthelensi | Rodent and humans |
| L. antarctica and L. lytica | Dogs |
| Na | Genera | Species | Hosts |
|---|---|---|---|
| 1 | Bartonella (21 species) | B. krasnovii and B. tribocorum | Rodents and dogs |
| B. taylorii | Rodents and humans | ||
| B. quintana, B. ancashensis, B. henselae, B. machadoae, B. clarridgeiae, B. vinsonii, B. bovis, B. birtlesii, B. elizabethae, B. taylorii, B. alsatica, B. bacilliformis, B. harrusi, B. grahamii, B. australis, B. schoenbuchensis, B. kosoyi, and B. apihabitans | Rodents | ||
| 2 | Borrelia (6 species) | Borrelia miyamotoi, | Rodents, humans, and dogs |
| B. turcica, B. parkeri, B. anserina B. coriaceae, and B. crocidurae | Rodents | ||
| 3 | Borreliella (5 species) | B. burgdorferi, B. afzelii, B. bissettiae B. valaisiana, and B. mayonii | Rodents |
| 4 | Streptobacillus | S. moniliformis | Rodents and dogs |
| 5 | Rickettsia | R. rhipicephali | Rodents and humans |
| R. typhi and R. prowazekii | Rodents and dogs | ||
| R. tillamookensis, R. asiatica, R. slovaca R. australis, and R. bellii | Rodents | ||
| 6 | Spiroplasma | S. corruscae | Rodents, dogs, and humans |
| S. cantharicola | Humans | ||
| 7 | Mycoplasma | M. suis | Humans |
| 8 | Anaplasma | A. platys, A. phagocytophilum, and A. marginale | Rodents |
| 9 | Ehrlichia | E. canis and E. muris | Dogs |
| 10 | Yersinia | Y. pestis subsp. Pestis | Rodents |
| 11 | Orientia | O. tsutsugamushi | Rodents |
| Hosts | Bacteria Genera | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Leptospira | Brucella | Bacillus | Vibrio | Listeria | Campylobacter | Salmonella | Clostridium | Pasteurella | Chlamydia | |
| Humans | 4 pools | 2 pools | 7 pools | 6 pools | 1 pool | 3 pools | 5 pools | 6 pools | 9 pools | 2 pools |
| n = 22 pools | (18.18%) | (9.09%) | (31.81%) | (27.27%) | (4.54%) | (13.63%) | (22.72%) | (27.27%) | (40.9%) | (9.09%) |
| Wild rodents | 6 pools | 7 pools | 3 pools | 2 pools | 8 pools | 4 pools | 7 pools | 9 pools | 11 pools | 0 |
| n = 16 pools | (37.5%) | (43.75%) | (18.75%) | (12.5%) | (50%) | (25%) | (43.75%) | (56.25%) | (68.75%) | (0.0%) |
| Domestic dogs | 3 pools | 2 pools | 6 pools | 5 pools | 4 pools | 4 pools | 3 pools | 4 pools | 4 pools | 3 pools |
| n = 10 pools | (30%) | (20%) | (60%) | (50%) | (40%) | (40%) | (30%) | (40%) | (40%) | (30%) |
| Bacterial Communities | |||
|---|---|---|---|
| Na | Genus | Species | Host |
| 1 | Leptospira | L. santarosai, L. kmetyi, and L. weilii | Rodents and dogs |
| L. interrogans | Rodents, dogs, and humans | ||
| L. kobayashii | Rodents and humans | ||
| L. kirschneri, L. mayottensis, L. borgpeterseni, L. tipperaryensis, and L. noguchii | Rodents | ||
| 2 | Brucella | B. anthropi | Rodents and humans |
| B. suis | Rodents and dogs | ||
| B. pseudogrignonensis | Dogs | ||
| 3 | Bacillus | B. cereus | Humans, rodents, and dogs |
| B. cytotoxicus and | Rodents and dogs | ||
| 4 | Vibrio | V. anguillarum | Humans, rodents, and dogs |
| V. vulnificus | Rodents | ||
| 5 | Listeria | L. monocytogenes | Rodents and humans |
| 6 | Campylobacter | C. jejuni | Rodents and dogs |
| 7 | Salmonella | S. enterica subsp. Enterica | Humans, rodents, and dogs |
| 8 | Clostridium | C. botulinum | Rodents, dogs, and humans |
| 9 | Pasteurella | P. multocida subsp. multocida | Humans, rodents, and dogs |
| 10 | Chlamydia | C. crocodile and C. abortus | Humans |
| C. gallinacean, C. trachomatis, C. pecorum, and C. felis. C. avium | Dogs | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Issae, A.R.; Katakweba, A.S.; Kicheleri, R.P.; Chengula, A.A.; van Zwetselaar, M.; Kasanga, C.J. Exploring Pathogenic and Zoonotic Bacteria from Wild Rodents, Dogs, and Humans of the Ngorongoro District in Tanzania Using Metagenomics Next-Generation Sequencing. Zoonotic Dis. 2023, 3, 226-242. https://doi.org/10.3390/zoonoticdis3030019
Issae AR, Katakweba AS, Kicheleri RP, Chengula AA, van Zwetselaar M, Kasanga CJ. Exploring Pathogenic and Zoonotic Bacteria from Wild Rodents, Dogs, and Humans of the Ngorongoro District in Tanzania Using Metagenomics Next-Generation Sequencing. Zoonotic Diseases. 2023; 3(3):226-242. https://doi.org/10.3390/zoonoticdis3030019
Chicago/Turabian StyleIssae, Amina Ramadhani, Abdul Selemani Katakweba, Rose Peter Kicheleri, Augustino Alfred Chengula, Marco van Zwetselaar, and Christopher Jacob Kasanga. 2023. "Exploring Pathogenic and Zoonotic Bacteria from Wild Rodents, Dogs, and Humans of the Ngorongoro District in Tanzania Using Metagenomics Next-Generation Sequencing" Zoonotic Diseases 3, no. 3: 226-242. https://doi.org/10.3390/zoonoticdis3030019
APA StyleIssae, A. R., Katakweba, A. S., Kicheleri, R. P., Chengula, A. A., van Zwetselaar, M., & Kasanga, C. J. (2023). Exploring Pathogenic and Zoonotic Bacteria from Wild Rodents, Dogs, and Humans of the Ngorongoro District in Tanzania Using Metagenomics Next-Generation Sequencing. Zoonotic Diseases, 3(3), 226-242. https://doi.org/10.3390/zoonoticdis3030019