Antimicrobial Resistance and Novel Alternative Approaches to Conventional Antibiotics
Abstract
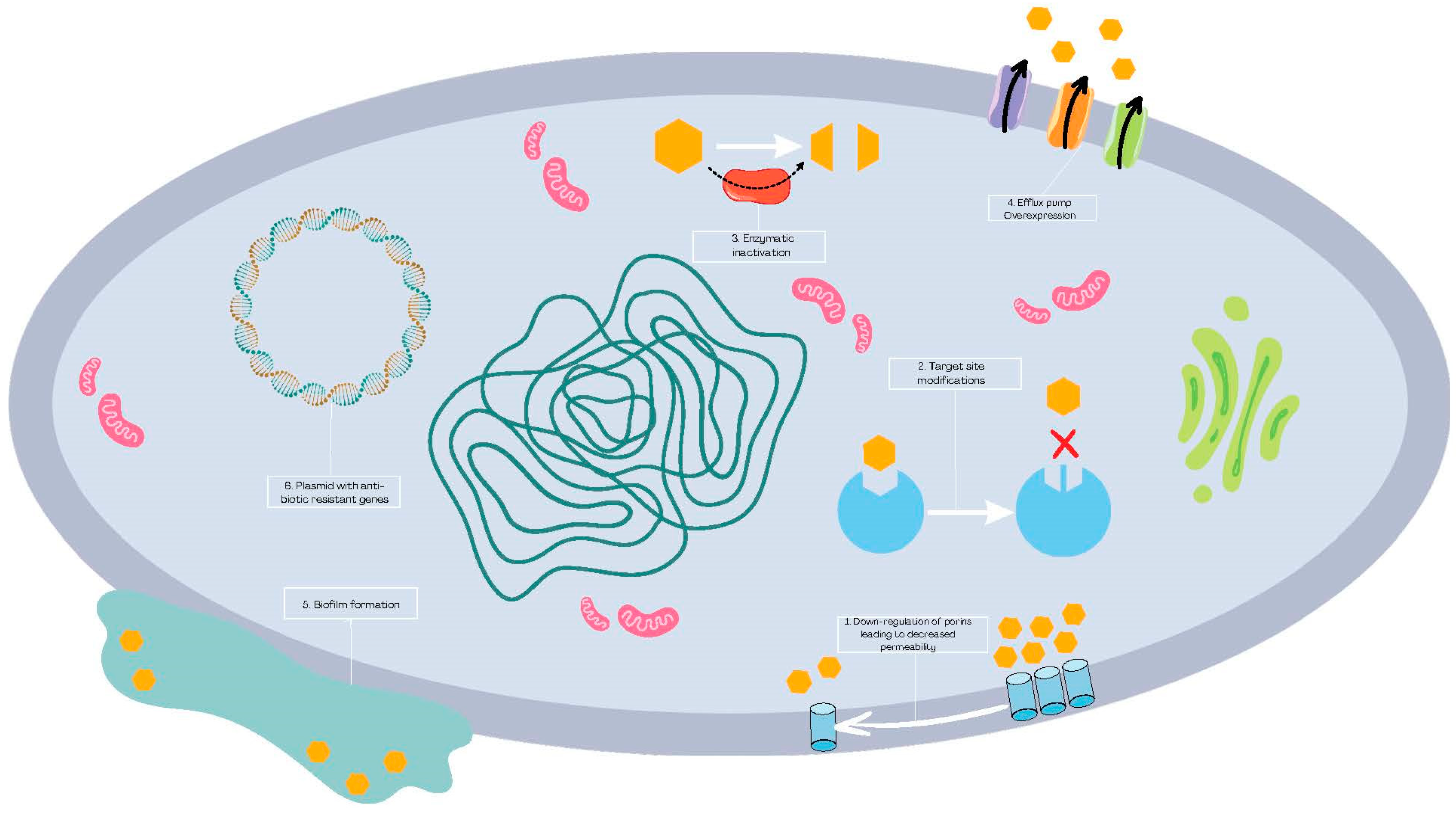
1. Introduction
2. Methods
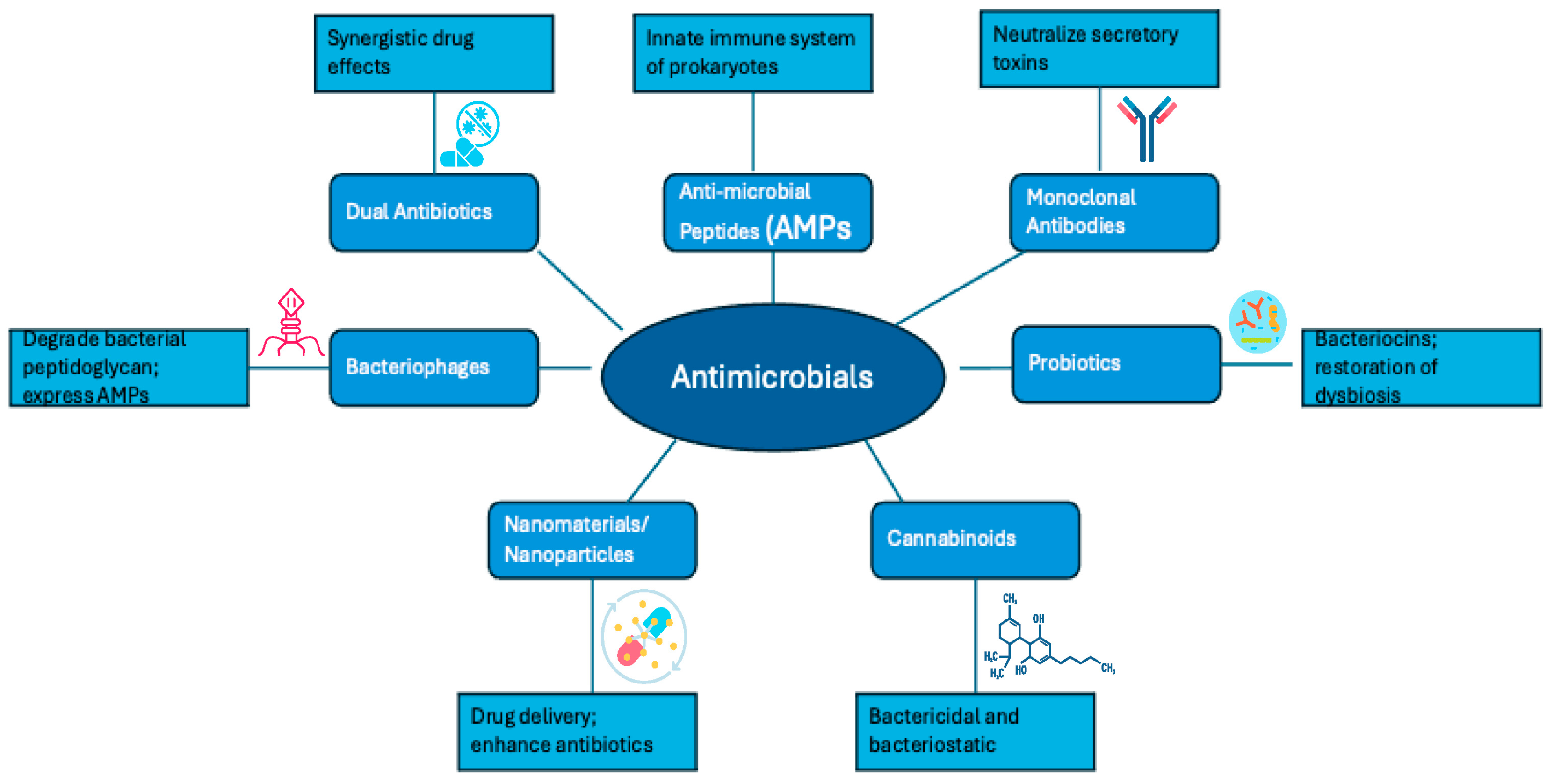
3. Alternatives Methods to Antibiotics in the Face of Antibiotic Resistance
3.1. Dual Antimicrobial Therapy and Drug–Adjuvant Combinations
3.2. Antimicrobial Peptides
3.3. Monoclonal Antibodies
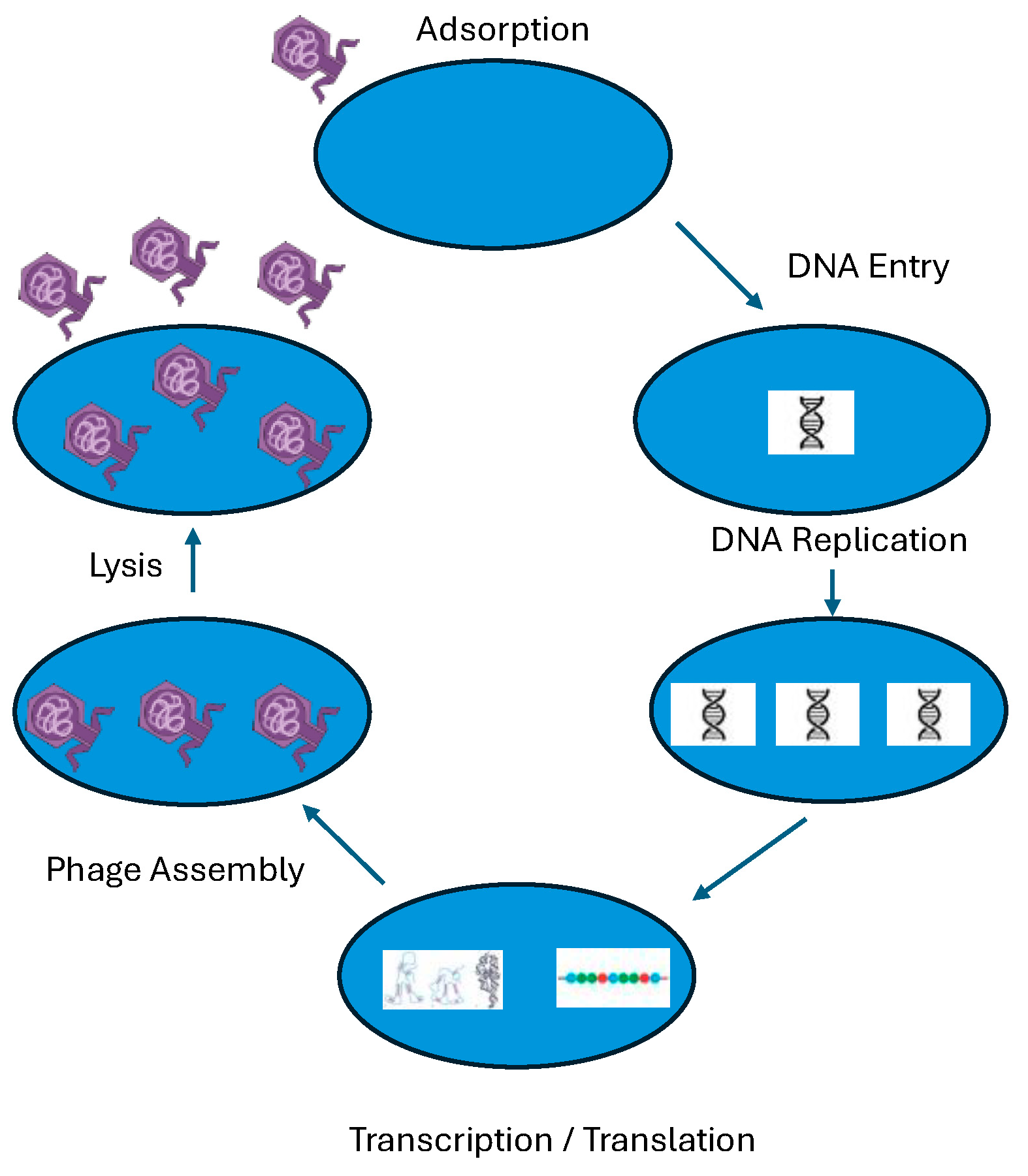
3.4. Bacteriophages
3.5. Probiotics
3.6. Nanomaterials and Nanoparticles
3.7. Cannabinoids
4. Conclusions
Funding
Conflicts of Interest
References
- Breijyeh, Z.; Jubeh, B.; Karaman, R. Resistance of gram-negative bacteria to current antibacterial agents and approaches to resolve it. Molecules 2020, 25, 1340. [Google Scholar] [CrossRef] [PubMed]
- Cappiello, F.; Loffredo, M.R.; Del Plato, C.; Cammarone, S.; Casciaro, B.; Quaglio, D.; Mangoni, M.L.; Botta, B.; Ghirga, F. The Revaluation of Plant-Derived Terpenes to Fight Antibiotic-Resistant Infections. Antibiotics 2020, 9, 325. [Google Scholar] [CrossRef] [PubMed]
- Martinez, J.L. General principles of antibiotic resistance in bacteria. Drug Discov. Today Technol. 2014, 11, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Pulingam, T.; Parumasivam, T.; Gazzali, A.M.; Sulaiman, A.M.; Chee, J.Y.; Lakshmanan, M.; Chin, C.F.; Sudesh, K. Antimicrobial resistance: Prevalence, economic burden, mechanisms of resistance and strategies to overcome. Eur. J. Pharm. Sci. 2022, 170, 106103. [Google Scholar] [CrossRef] [PubMed]
- Baharoglu, Z.; Garriss, G.; Mazel, D. Multiple pathways of genome plasticity leading to development of antibiotic resistance. Antibiotics 2013, 2, 288–315. [Google Scholar] [CrossRef] [PubMed]
- Uddin, T.M.; Chakraborty, A.J.; Khusro, A.; Zidan, B.R.; Mitra, S.; Bin Emran, T.; Dhama, K.; Ripon, K.H.; Gajdács, M.; Sahibzada, M.U.K.; et al. Antibiotic resistance in microbes: History, mechanisms, therapeutic strategies and future prospects. J. Infect. Public Health 2021, 14, 1750–1766. [Google Scholar] [CrossRef]
- Darby, E.M.; Trampari, E.; Siasat, P.; Gaya, M.S.; Alav, I.; Webber, M.A.; Blair, J.M.A. Molecular mechanisms of antibiotic resistance revisited. Nat. Rev. Microbiol. 2023, 21, 280–295. [Google Scholar] [CrossRef] [PubMed]
- Mancuso, G.; Midiri, A.; Gerace, E.; Biondo, C. Bacterial antibiotic resistance: The most critical pathogens. Pathogens 2021, 10, 1310. [Google Scholar] [CrossRef]
- Marturano, J.E.; Lowery, T.J. ESKAPE pathogens in bloodstream infections are associated with higher cost and mortality but can be predicted using diagnoses upon admission. Open Forum Infect. Dis. 2019, 6, ofz503. [Google Scholar] [CrossRef]
- Abushaheen, M.A.; Muzaheed; Fatani, A.J.; Alosaimi, M.; Mansy, W.; George, M.; Acharya, S.; Rathod, S.; Divakar, D.D.; Jhugroo, C.; et al. Antimicrobial resistance, mechanisms and its clinical significance. Disease-a-Month 2020, 66, 100971. [Google Scholar] [CrossRef]
- Reygaert, W.C. An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol. 2018, 4, 482–501. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Raudonis, R.; Glick, B.R.; Lin, T.J.; Cheng, Z. Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 2019, 37, 177–192. [Google Scholar] [CrossRef] [PubMed]
- Munita, J.M.; Arias, C.A. Mechanisms of antibiotic resistance. In Virulence Mechanisms of Bacterial Pathogens; American Society for Microbiology: Washington DC, USA, 2016; pp. 481–511. [Google Scholar]
- Urban-Chimel, R.; Marek, A.; Stępień-Pyśniak, D.; Wieczorek, K.; Dec, M.; Nowaczek, A.; Osek, J. Antibiotic resistance in bacteria—A review. Antibiotics 2022, 11, 1079. [Google Scholar]
- Akram, F.; Imtiaz, M.; Haq, I. Emergent crisis of antibiotic resistance: A silent pandemic threat to 21st century. Microb. Pathog. 2023, 174, 105923. [Google Scholar] [CrossRef] [PubMed]
- Hall, C.W.; Mah, T.F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 2017, 41, 276–301. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. 2021 Antibacterial Agents in Clinical and Preclinical Development: An Overview and Analysis; World Health Organization: Geneva, Switzerland, 2022. [Google Scholar]
- Gupta, R.; Sharma, S. Role of alternatives to antibiotics in mitigating the antimicrobial resistance crisis. Indian J. Med. Res. 2022, 156, 464–477. [Google Scholar] [CrossRef] [PubMed]
- Chin, K.W.; Tiong, H.L.; Luang-In, V.; Ma, N.L. An overview of antibiotic and antibiotic resistance. Environ. Adv. 2023, 11, 100331. [Google Scholar] [CrossRef]
- León-Buitimea, A.; Garza-Cárdenas, C.R.; Garza-Cervantes, J.A.; Lerma-Escalera, J.A.; Morones-Ramírez, J.R. The demand for new antibiotics: Antimicrobial peptides, nanoparticles, and combinatorial therapies as future strategies in antibacterial agent design. Front. Microbiol. 2020, 11, 1669. [Google Scholar] [CrossRef] [PubMed]
- Fischbach, M.A. Combination therapies for combating antimicrobial resistance. Curr. Opin. Microbiol. 2011, 14, 519–523. [Google Scholar] [CrossRef]
- Konwar, A.N.; Hazarika, S.N.; Bharadwaj, P.; Thakur, D. Emerging Non-Traditional Approaches to Combat Antibiotic Resistance. Curr. Microbiol. 2022, 79, 330. [Google Scholar] [CrossRef]
- Lister, P.D.; Wolter, D.J.; Hanson, N.D. Antibacterial-resistant Pseudomonas aeruginosa: Clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin. Microbiol. Rev. 2009, 22, 582–610. [Google Scholar] [CrossRef] [PubMed]
- Mushtaq, S.; Warner, M.; Livermore, D.M. In vitro activity of ceftazidime+ NXL104 against Pseudomonas aeruginosa and other non-fermenters. J. Antimicrob. Chemother. 2010, 65, 2376–2381. [Google Scholar] [CrossRef] [PubMed]
- Levasseur, P.; Girard, A.M.; Claudon, M.; Goossens, H.; Black, M.T.; Coleman, K.; Miossec, C. In Vitro Antibacterial Activity of the Ceftazidime-Avibactam (NXL104) Combination against Pseudomonas aeruginosa Clinical Isolates. Antimicrob. Agents Chemother. 2012, 56, 1606–1608. Available online: https://pubmed.ncbi.nlm.nih.gov/22214778/ (accessed on 20 June 2024). [CrossRef] [PubMed]
- Kirienko, N.V.; Rahme, L.; Cho, Y.H. Beyond antimicrobials: Non-traditional approaches to combating multidrug-resistant bacteria. Front. Cell. Infect. Microbiol. 2019, 9, 343. [Google Scholar] [CrossRef] [PubMed]
- Mazurkiewicz-Pisarek, A.; Baran, J.; Ciach, T. Antimicrobial Peptides: Challenging Journey to the Pharmaceutical, Biomedical, and Cosmeceutical Use. Int. J. Mol. Sci. 2023, 24, 9031. [Google Scholar] [CrossRef] [PubMed]
- Mahlapuu, M.; Björn, C.; Ekblom, J. Antimicrobial peptides as therapeutic agents: Opportunities and challenges. Crit. Rev. Biotechnol. 2020, 40, 978–992. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.H.; Lu, T.K. Development and challenges of antimicrobial peptides for therapeutic applications. Antibiotics 2020, 9, 24. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.M.; Hwang, Y.C.; Liu, I.J.; Lee, C.C.; Tsai, H.-Z.; Li, H.-J.; Wu, H.-C. Development of therapeutic antibodies for the treatment of diseases. J. Biomed. Sci. 2020, 27, 1. [Google Scholar] [CrossRef]
- Streicher, L.M. Exploring the future of infectious disease treatment in a post-antibiotic era: A comparative review of alternative therapeutics. J. Glob. Antimicrob. Resist. 2021, 24, 285–295. [Google Scholar] [CrossRef]
- Ye, J.; Chen, X. Current Promising Strategies against Antibiotic-Resistant Bacterial Infections. Antibiotics 2022, 12, 67. [Google Scholar] [CrossRef]
- Butler, M.S.; Gigante, V.; Sati, H.; Paulin, S.; Al-Sulaiman, L.; Rex, J.H.; Fernandes, P.; Arias, C.A.; Paul, M.; Thwaites, G.E.; et al. Analysis of the clinical pipeline of treatments for drug-resistant bacterial infections: Despite progress, more action is needed. Antimicrob. Agents Chemother. 2022, 66, e01991-21. [Google Scholar] [CrossRef] [PubMed]
- Voelker, R. FDA approves bacteriophage trial. JAMA 2019, 321, 638. [Google Scholar] [CrossRef]
- Golkar, Z.; Bagasra, O.; Pace, D.G. Bacteriophage therapy: A potential solution for the antibiotic resistance crisis. J. Infect. Dev. Ctries. 2014, 8, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Hu, K.; Xie, Y.; Liu, Y.; Mu, D.; Guo, H.; Zhang, Z.; Zhang, Y.; Chang, D.; Shi, Y. A Novel Phage PD-6A3, and Its Endolysin Ply6A3, with Extended Lytic Activity Against Acinetobacter baumannii. Front. Microbiol. 2019, 9, 3302, Erratum in Front. Microbiol. 2019, 10, 196. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jun, S.Y.; Jang, I.J.; Yoon, S.; Jang, K.; Yu, K.-S.; Cho, J.Y.; Seong, M.-W.; Jung, G.M.; Yoon, S.J.; Kang, S.H. Pharmacokinetics and tolerance of the phage endolysin-based candidate drug SAL200 after a single intravenous administration among healthy volunteers. Antimicrob. Agents Chemother. 2017, 61, e02629-16. [Google Scholar] [CrossRef]
- Fowler, V.G.; Das, A.F.; Lipka-Diamond, J.; Schuch, R.; Pomerantz, R.; Jáuregui-Peredo, L.; Bressler, A.; Evans, D.C.; Moran, G.J.; Rupp, M.E.; et al. Exebacase for patients with Staphylococcus aureus bloodstream infection and endocarditis. J. Clin. Investig. 2020, 130, 3750–3760. [Google Scholar] [CrossRef]
- Piewngam, P.; Zheng, Y.; Nguyen, T.H.; Dickey, S.W.; Joo, H.-S.; Villaruz, A.E.; Glose, K.A.; Fisher, E.L.; Hunt, R.L.; Li, B.; et al. Pathogen elimination by probiotic Bacillus via signaling interference. Nature 2018, 562, 532–537. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.H.; Kim, B.S.; Kang, S.S. Bacteriocin of Pediococcus acidilactici HW01 inhibits biofilm formation and virulence factor production by Pseudomonas aeruginosa. Probiot. Antimicrob. Proteins 2020, 12, 73–81. [Google Scholar] [CrossRef]
- Vieira, A.T.; Rocha, V.M.; Tavares, L.; Garcia, C.C.; Teixeira, M.M.; Oliveira, S.C.; Cassali, G.D.; Gamba, C.; Martins, F.S.; Nicoli, J.R. Control of Klebsiella pneumoniae pulmonary infection and immunomodulation by oral treatment with the commensal probiotic Bifidobacterium longum 51A. Microbes Infect. 2016, 18, 180–189. [Google Scholar] [CrossRef]
- Sikorska, H.; Smoragiewicz, W. Role of probiotics in the prevention and treatment of meticillin-resistant Staphylococcus aureus infections. Int. J. Antimicrob. Agents 2013, 42, 475–481. [Google Scholar] [CrossRef]
- Rabetafika, H.N.; Razafindralambo, A.; Ebenso, B.; Razafindralambo, H.L. Probiotics as Antibiotic Alternatives for Human and Animal Applications. Encyclopedia 2023, 3, 561–581. [Google Scholar] [CrossRef]
- Gao, W.; Zhang, L. Nanomaterials arising amid antibiotic resistance. Nat. Rev. Microbiol. 2021, 19, 5–6. [Google Scholar] [CrossRef] [PubMed]
- Jelinkova, P.; Mazumdar, A.; Sur, V.P.; Kociova, S.; Dolezelikova, K.; Jimenez, A.M.J.; Koudelkova, Z.; Mishra, P.K.; Smerkova, K.; Heger, Z.; et al. Nanoparticle-drug conjugates treating bacterial infections. J. Control. Release 2019, 307, 166–185. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, V.; Mishra, N.; Gadani, K.; Solanki, P.S.; Shah, N.A.; Tiwari, M. Mechanism of anti-bacterial activity of zinc oxide nanoparticle against carbapenem-resistant Acinetobacter baumannii. Front. Microbiol. 2018, 9, 1218. [Google Scholar] [CrossRef] [PubMed]
- Fordjour, E.; Manful, C.F.; Sey, A.A.; Javed, R.; Pham, T.H.; Thomas, R.; Cheema, M. Cannabis: A multifaceted plant with endless potentials. Front. Pharmacol. 2023, 14, 1200269. [Google Scholar] [CrossRef] [PubMed]
- Klahn, P. Cannabinoids-promising antimicrobial drugs or intoxicants with benefits? Antibiotics 2020, 9, 297. [Google Scholar] [CrossRef] [PubMed]
- Barak, T.; Sharon, E.; Steinberg, D.; Feldman, M.; Sionov, R.V.; Shalish, M. Anti-bacterial effect of cannabidiol against the cariogenic Streptococcus mutans bacterium: An In Vitro Study. Int. J. Mol. Sci. 2022, 23, 15878. [Google Scholar] [CrossRef] [PubMed]
- Saleemi, M.A.; Yahaya, N.; Zain, N.N.; Raoov, M.; Yong, Y.K.; Noor, N.S.; Lim, V. Antimicrobial and cytotoxic effects of Cannabinoids: An updated review with future perspectives and current challenges. Pharmaceuticals 2022, 15, 1228. [Google Scholar] [CrossRef]
- Śledziński, P.; Nowak-Terpiłowska, A.; Zeyland, J. Cannabinoids in medicine: Cancer, immunity, and microbial diseases. Int. J. Mol. Sci. 2021, 22, 263. [Google Scholar] [CrossRef]
- Luz-Veiga, M.; Amorim, M.; Pinto-Ribeiro, I.; Oliveira, A.L.; Silva, S.; Pimentel, L.L.; Rodríguez-Alcalá, L.M.; Madureira, R.; Pintado, M.; Azevedo-Silva, J.; et al. Cannabidiol and cannabigerol exert antimicrobial activity without compromising skin microbiota. Int. J. Mol. Sci. 2023, 24, 2389. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berger, I.; Loewy, Z.G. Antimicrobial Resistance and Novel Alternative Approaches to Conventional Antibiotics. Bacteria 2024, 3, 171-182. https://doi.org/10.3390/bacteria3030012
Berger I, Loewy ZG. Antimicrobial Resistance and Novel Alternative Approaches to Conventional Antibiotics. Bacteria. 2024; 3(3):171-182. https://doi.org/10.3390/bacteria3030012
Chicago/Turabian StyleBerger, Irene, and Zvi G. Loewy. 2024. "Antimicrobial Resistance and Novel Alternative Approaches to Conventional Antibiotics" Bacteria 3, no. 3: 171-182. https://doi.org/10.3390/bacteria3030012
APA StyleBerger, I., & Loewy, Z. G. (2024). Antimicrobial Resistance and Novel Alternative Approaches to Conventional Antibiotics. Bacteria, 3(3), 171-182. https://doi.org/10.3390/bacteria3030012





