Isolation and Identification of Autochthonous Lactic Acid Bacteria from Commonly Consumed African Indigenous Leafy Vegetables in Kenya
Abstract
:1. Introduction
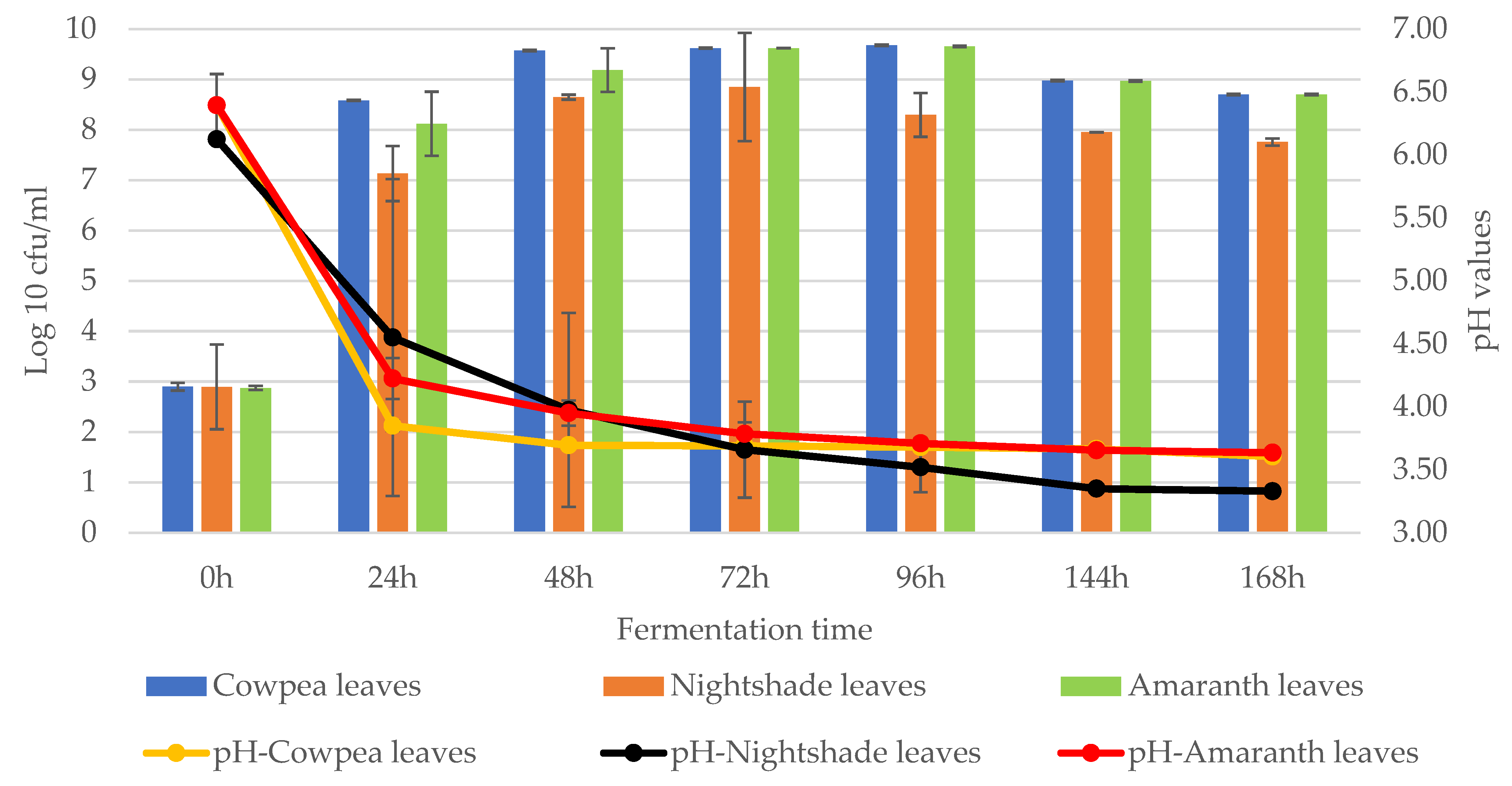
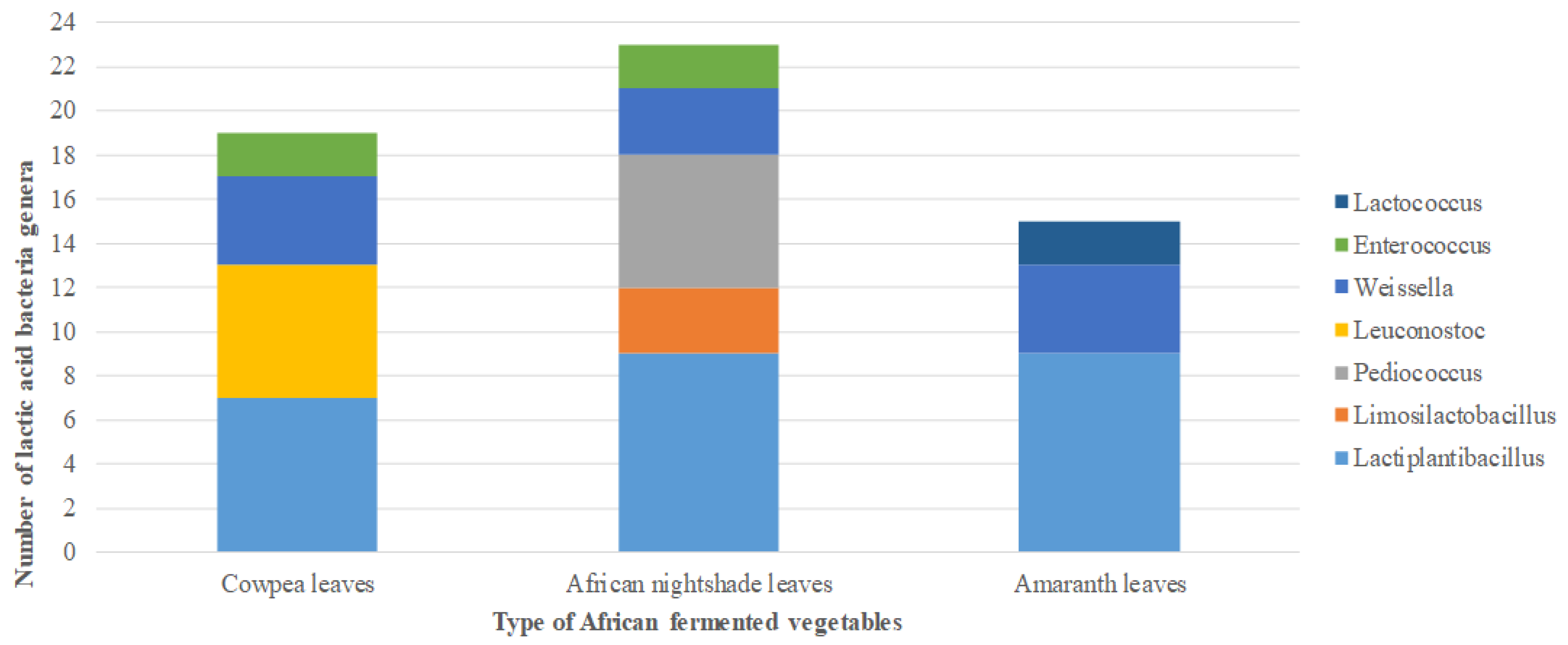
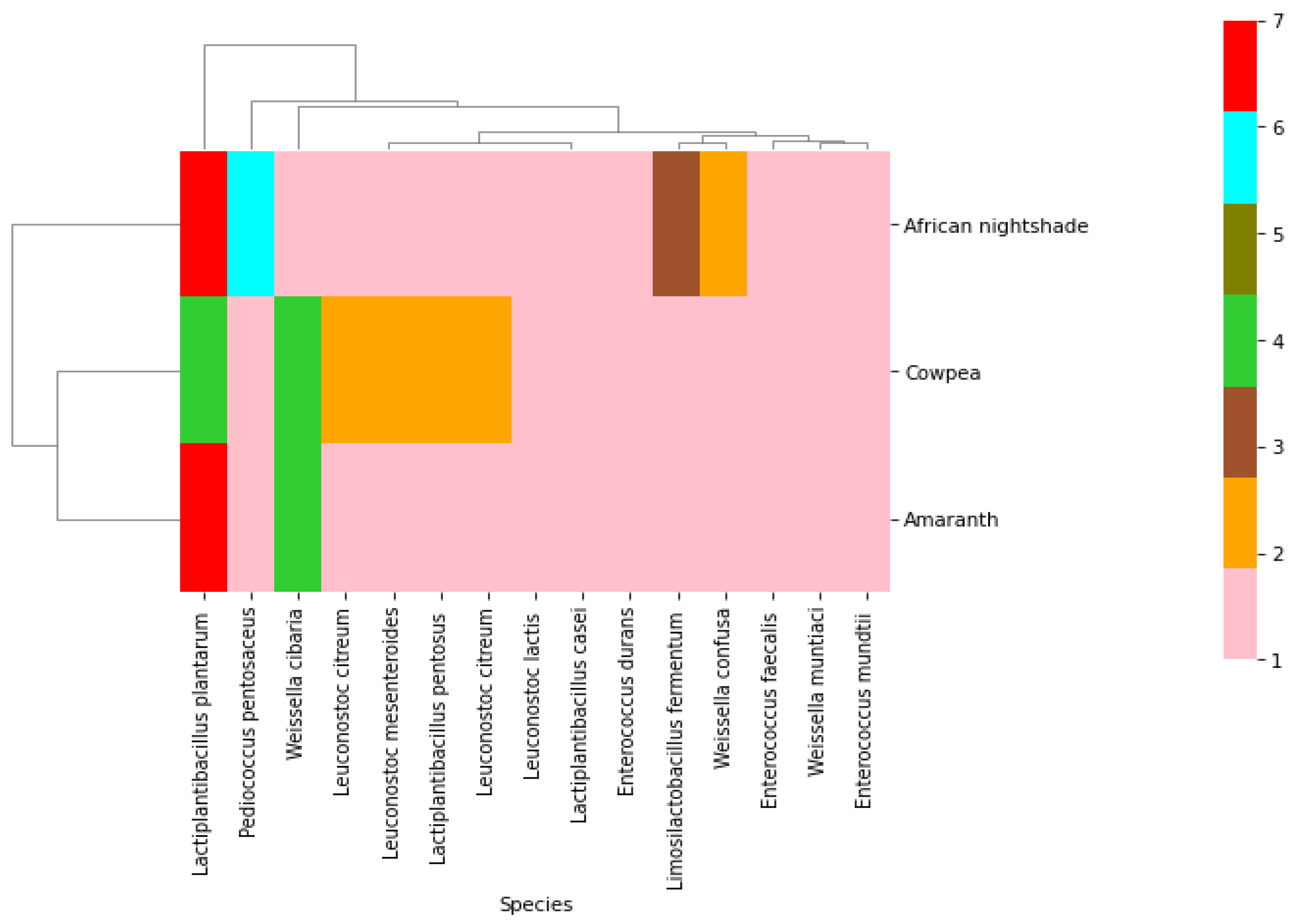
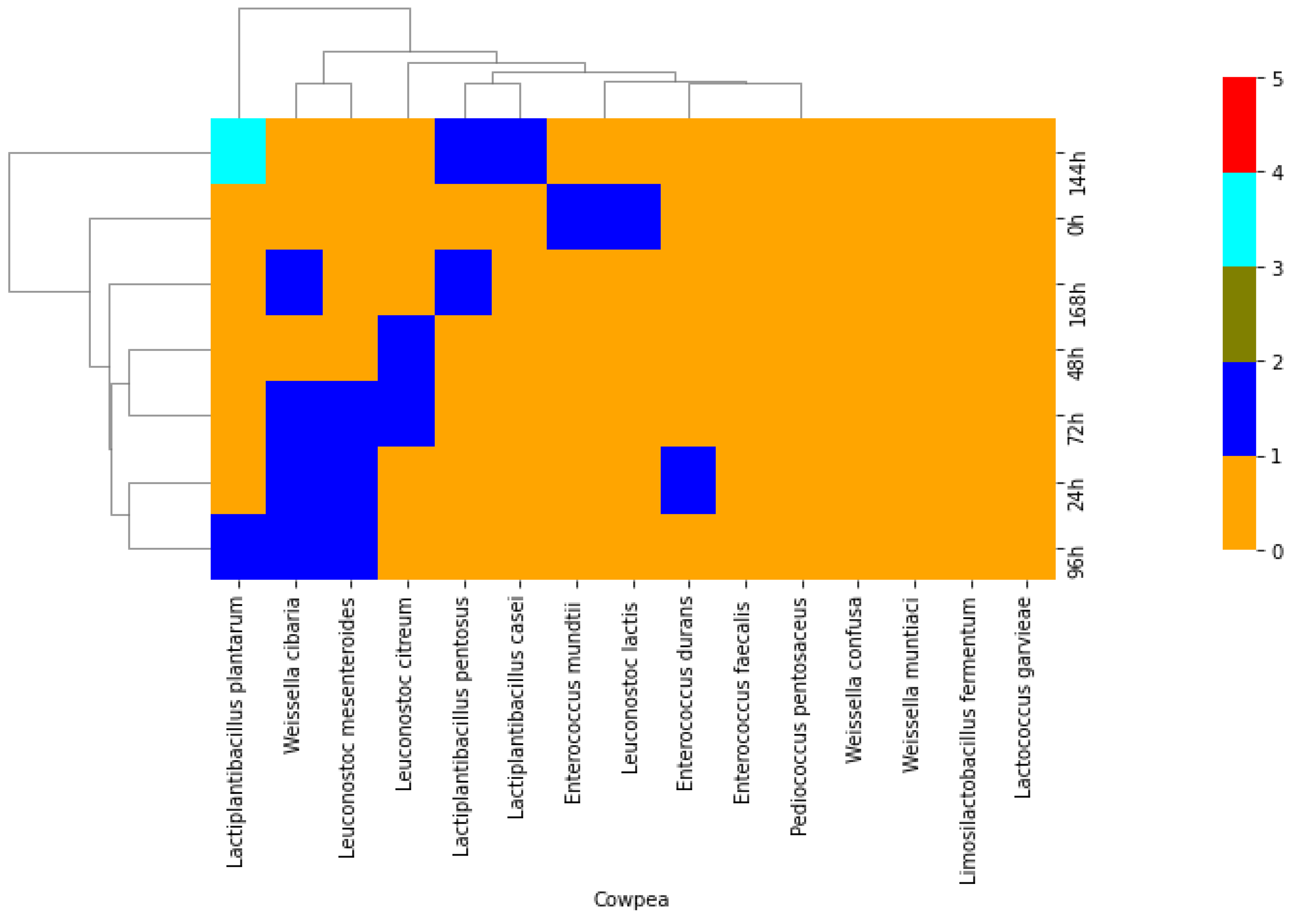
2. Results
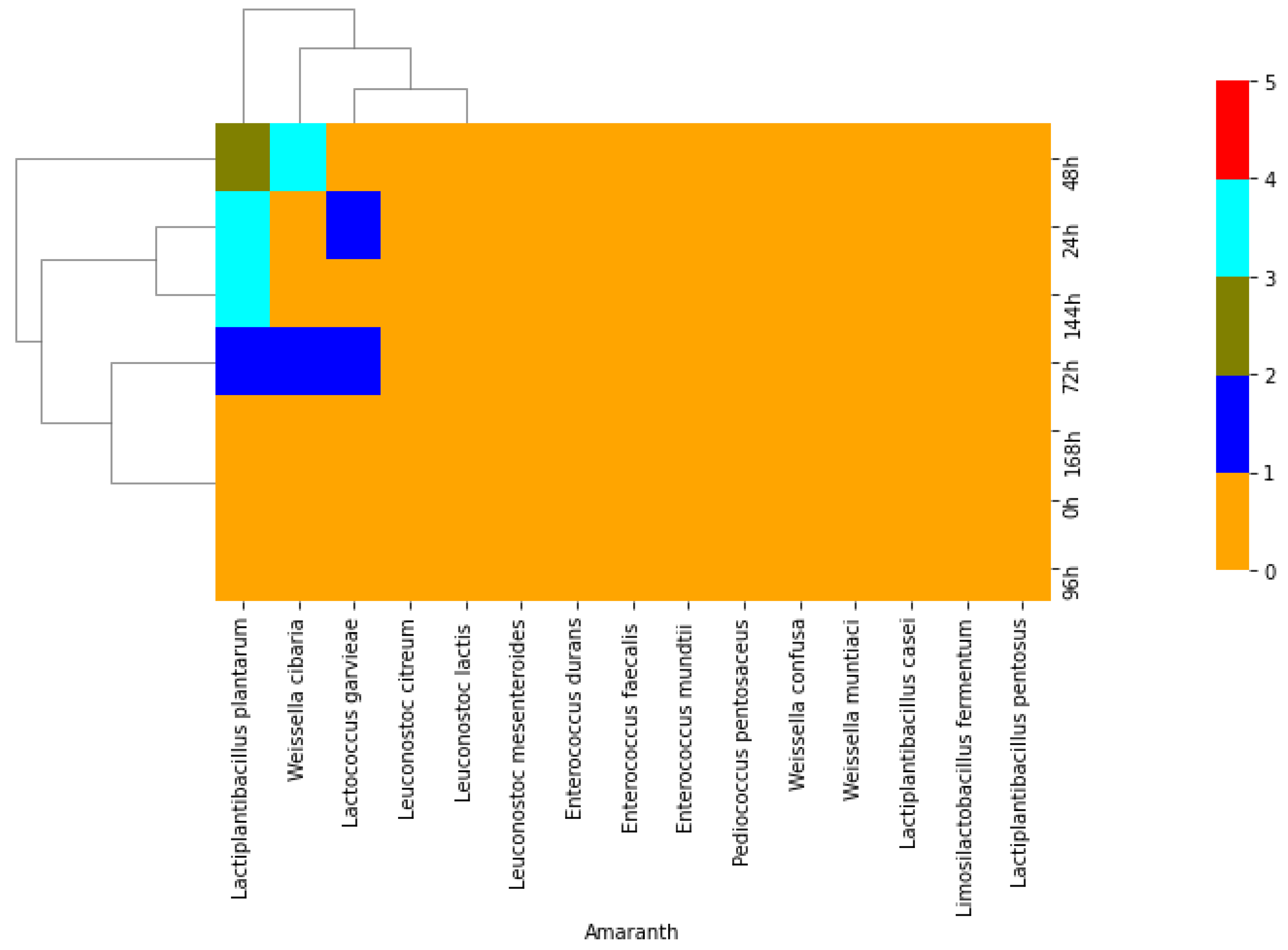
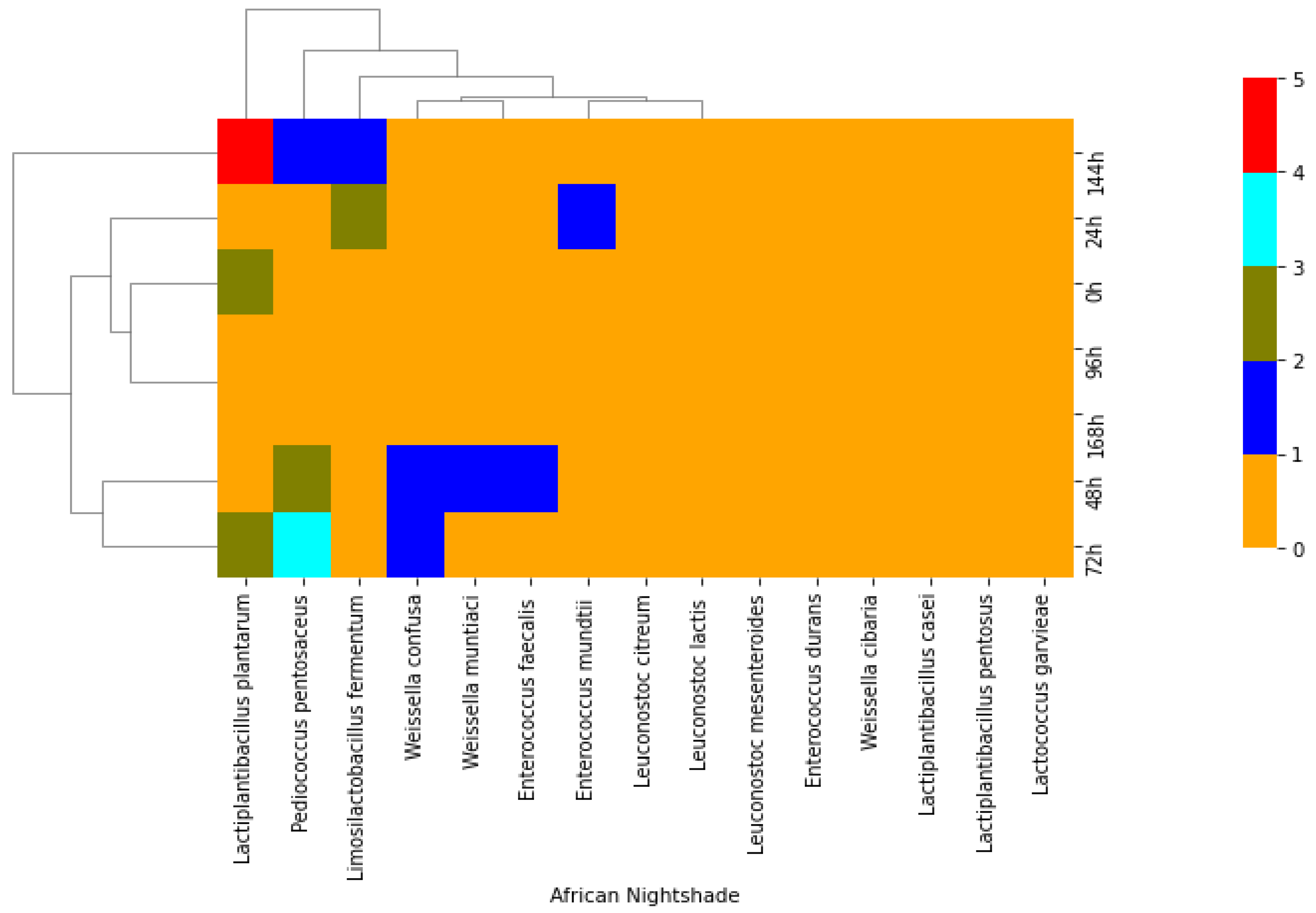
2.1. Isolation of Lactic Acid Bacteria from African Indigenous Vegetables
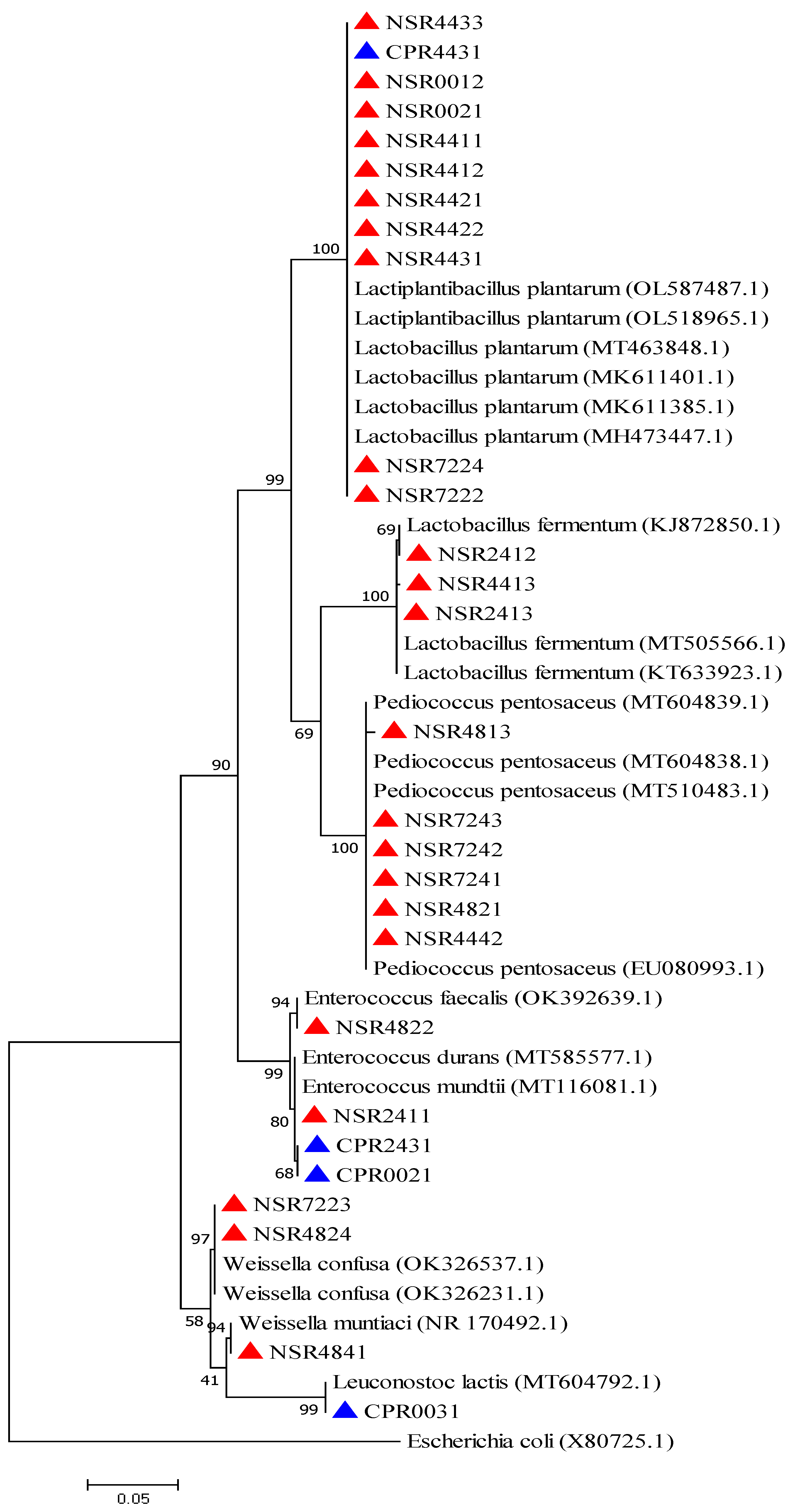
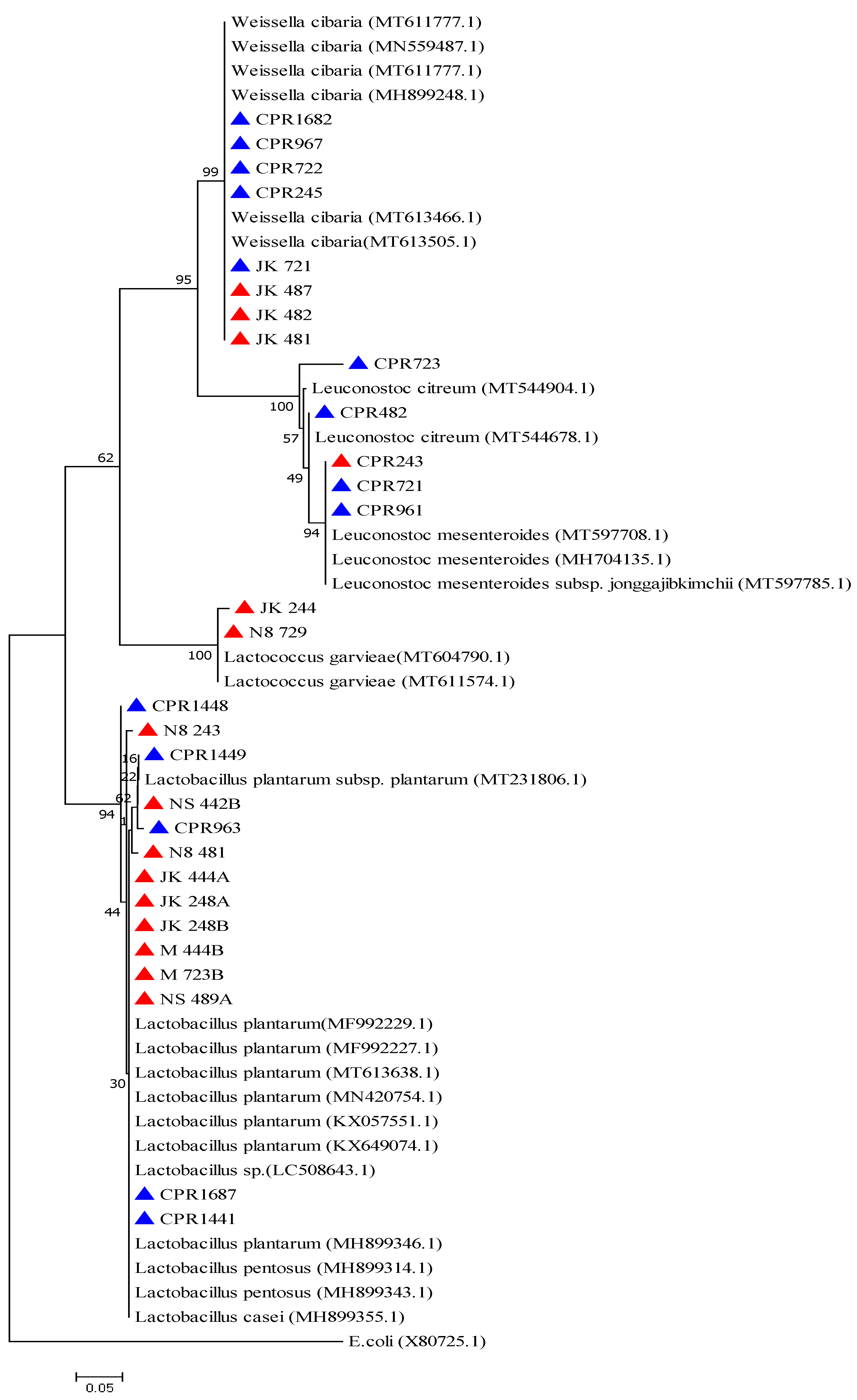
2.2. Phenotypic and Molecular Identification of Lactic Acid Bacteria Strains
3. Discussion
4. Materials and Methods
4.1. Growth and Preparation of Plant Materials
4.2. Fermentation of African Indigenous Leafy Vegetables
4.3. Microbiological Testing
4.4. Phenotypic Characterization
4.5. Genotypic Characterization
4.6. Phylogenetic Analysis
4.7. Data Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abukutsa-Onyango, M. Strategic repositioning of African indigenous vegetables in the horticulture sector. In Proceedings of the Second RUFORUM Biennial Meeting, Entebbe, Uganda, 20–24 September 2010; pp. 1413–1419. [Google Scholar]
- Irakoze, M.L.; Wafula, E.N.; Owaga, E. Potential role of african fermented indigenous vegetables in maternal and child nutrition in sub-saharan Africa. Int. J. Food Sci. 2021, 2021, 3400329. [Google Scholar] [CrossRef] [PubMed]
- Oguntoyinbo, F.A.; Cho, G.-S.; Trierweiler, B.; Kabisch, J.; Rösch, N.; Neve, H.; Bockelmann, W.; Frommherz, L.; Nielsen, D.S.; Krych, L.; et al. Fermentation of African Kale ( Brassica Carinata ) Using L. plantarum BFE 5092 and L. fermentum BFE 6620 starter strains. Int. J. Food Microbiol. 2016, 238, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Mulaw, G.; Sisay Tessema, T.; Muleta, D.; Tesfaye, A. In vitro evaluation of probiotic properties of lactic acid bacteria isolated from some traditionally fermented ethiopian food products. Int. J. Microbiol. 2019, 2019, 7179514. [Google Scholar] [CrossRef] [Green Version]
- Wafula, E.; Franz, C.M.A.P.; Rohn, S.; Huch, M.; Mathara, J.; Trierweiler, B. Fermentation of African indigenous leafy vegetables to lower post-harvest losses, maintain quality and increase product safety. Africa J. Hortic. Sci. 2016, 9, 1–13. [Google Scholar]
- Gido, E.O.; Ayuya, O.I.; Owuor, G.; Bokelmann, W. Consumption intensity of leafy african indigenous vegetables: Towards enhancing nutritional security in rural and urban dwellers in Kenya. Agric. Food Econ. 2017, 5, 14. [Google Scholar] [CrossRef]
- Uusiku, N.P.; Oelofse, A.; Duodu, K.G.; Bester, M.J.; Faber, M. Nutritional value of leafy vegetables of sub-saharan Africa and their potential contribution to human health: A review. J. Food Compos. Anal. 2010, 23, 499–509. [Google Scholar] [CrossRef]
- Franz, C.M.A.P.; Huch, M.; Mathara, J.M.; Abriouel, H.; Benomar, N.; Reid, G.; Galvez, A.; Holzapfel, W.H. African fermented foods and probiotics. Int. J. Food Microbiol. 2014, 190, 84–96. [Google Scholar] [CrossRef]
- Stoll, D.A.; Wafula, E.N.; Mathara, J.M.; Trierweiler, B.; Kulling, S.E.; Huch, M. Fermentation of African nightshade leaves with lactic acid bacterial starter cultures. Int. J. Food Microbiol. 2021, 342, 109056. [Google Scholar] [CrossRef]
- Liu, S.; Han, Y.; Zhou, Z. Lactic acid bacteria in traditional fermented Chinese foods. Food Res. Int. 2011, 44, 643–651. [Google Scholar] [CrossRef]
- Owade, J.O.; Abong’, G.O.; Okoth, M.W.; Mwang’ombe, A.W.; Jobor, J.O. Comparative profiling of lactic acid bacteria isolates in optimized and spontaneous fermentation of cowpea leaves. Food Sci. Nutr. 2021, 9, 1651–1664. [Google Scholar] [CrossRef]
- Ibinabo, T.I.; Wafula, E.N.; Josiah, K.; Julius, M.M. Phenotypic and genotypic characterization of lactic acid bacteria isolated from spontaneously fermented vegetable amaranth. African J. Food Sci. 2021, 15, 254–261. [Google Scholar] [CrossRef]
- Xiong, T.; Li, X.; Guan, Q.; Peng, F.; Xie, M. Starter culture fermentation of chinese sauerkraut: Growth, acidification and metabolic analyses. Food Control 2014, 41, 122–127. [Google Scholar] [CrossRef]
- Bautista-Gallego, J.; Medina, E.; Sánchez, B.; Benítez-Cabello, A.; Arroyo-López, F.N. Role of lactic acid bacteria in fermented vegetables. Grasas Aceites 2020, 71, 358. [Google Scholar] [CrossRef]
- Jay, J.M.; Lossener, M.J.; Golden, D.A. Mordern Food Microbiology, 7th ed.; Springer Science: New York, NY, USA, 2005; ISBN 0321109317. [Google Scholar]
- Stiles, M.E.; Holzapfel, W.H. Lactic acid bacteria of foods and their current taxonomy. Int. J. Food Microbiol. 1997, 36, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Holzapfel, W.H.; Wood, B.J.B. Lactic Acid Bacteria: Biodiversity and Taxonomy, 1st ed.; Holzapfel, W.H., Wood, B.J.B., Eds.; John Wiley & Sons, Ltd: Chichester, UK, 2014; Volume 9781444333, ISBN 9781118655252. [Google Scholar]
- Kostinek, M.; Ban-Koffi, L.; Ottah-Atikpo, M.; Teniola, D.; Schillinger, U.; Holzapfel, W.H.; Franz, C.M.A.P. Diversity of predominant lactic acid bacteria associated with cocoa fermentation in Nigeria. Curr. Microbiol. 2008, 56, 306–314. [Google Scholar] [CrossRef] [PubMed]
- Holzapfel, W. Use of starter cultures in fermentation on a household scale. Food Control 1997, 8, 241–258. [Google Scholar] [CrossRef]
- Kostinek, M.; Specht, I.; Edward, V.A.; Schillinger, U.; Hertel, C.; Holzapfel, W.H.; Franz, C.M.A.P. Diversity and technological properties of predominant lactic acid bacteria from fermented cassava used for the preparation of gari, a traditional African food. Syst. Appl. Microbiol. 2005, 28, 527–540. [Google Scholar] [CrossRef]
- McFeeters, R.F.; Pérez-Díaz, I.; Lee, C.-H.; Breidt, F. Fermented vegetables. Food Microbiol. 2013, 27603, 841–855. [Google Scholar] [CrossRef]
- Kostinek, M.; Specht, I.; Edward, V.A.; Pinto, C.; Egounlety, M.; Sossa, C.; Mbugua, S.; Dortu, C.; Thonart, P.; Taljaard, L.; et al. Characterisation and biochemical properties of predominant lactic acid bacteria from fermenting cassava for selection as starter cultures. Int. J. Food Microbiol. 2007, 114, 342–351. [Google Scholar] [CrossRef]
- Kim, M.; Chun, J. Bacterial community structure in kimchi, a korean fermented vegetable food, as revealed by 16S rRNA gene analysis. Int. J. Food Microbiol. 2005, 103, 91–96. [Google Scholar] [CrossRef]
- Lee, J.-Y.; Kim, C.-J.; Kunz, B. Identification of lactic acid bacteria isolated from kimchi and studies on their suitability for application as starter culture in the production of fermented sausages. Meat Sci. 2006, 72, 437–445. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.R.; Moss, M.O. Food Microbiology, 3rd ed.; RSC Publishing: Cambridge, UK, 2008. [Google Scholar]
- Halász, A.; Baráth, A.; Holzapfel, W. The influence of starter culture selection on sauerkraut fermentation. Z. Fur. Leb. Und-Forsch. 1999, 208, 434–438. [Google Scholar] [CrossRef]
- Jung, J.Y.; Lee, S.H.; Kim, J.M.; Park, M.S.; Bae, J.-W.; Hahn, Y.; Madsen, E.L.; Jeon, C.O. Metagenomic analysis of kimchi, a traditional Korean fermented food. Appl. Environ. Microbiol. 2011, 77, 2264–2274. [Google Scholar] [CrossRef] [Green Version]
- Cho, J.; Lee, D.; Yang, C.; Jeon, J.; Kim, J.; Han, H. Microbial population dynamics of kimchi, a fermented cabbage product. FEMS Microbiol. Lett. 2006, 257, 262–267. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.-S.; Heo, G.-Y.; Lee, J.W.; Oh, Y.-J.; Park, J.A.; Park, Y.-H.; Pyun, Y.-R.; Ahn, J.S. Analysis of kimchi microflora using denaturing gradient gel electrophoresis. Int. J. Food Microbiol. 2005, 102, 143–150. [Google Scholar] [CrossRef]
- Sutic, M.; Banina, A. Influence of aflatoxin B1 on gas production by lactic acid bacteria. J Env. Pathol Toxicol Oncol 1990, 10, 149–153. [Google Scholar]
- Fusco, V.; Quero, G.M.; Cho, G.-S.; Kabisch, J.; Meske, D.; Neve, H.; Bockelmann, W.; Franz, C.M.A.P. The genus weissella: Taxonomy, ecology and biotechnological potential. Front. Microbiol. 2015, 6, 155. [Google Scholar] [CrossRef] [Green Version]
- Moumene, M.; Drissi, F.; Croce, O.; Djebbari, B.; Robert, C.; Angelakis, E.; Benouareth, D.E.; Raoult, D.; Merhej, V. Complete genome sequence and description of Lactococcus Garvieae M14 isolated from algerian fermented milk. New Microbes New Infect. 2016, 10, 122–131. [Google Scholar] [CrossRef] [Green Version]
- Mathara, J.M.; Schillinger, U.; Kutima, P.M.; Mbugua, S.K.; Holzapfel, W.H. Isolation, identification and characterisation of the dominant microorganisms of kule naoto: The maasai traditional fermented milk in Kenya. Int. J. Food Microbiol. 2004, 94, 269–278. [Google Scholar] [CrossRef]
- Oguntoyinbo, F.A.; Tourlomousis, P.; Gasson, M.J.; Narbad, A. Analysis of bacterial communities of traditional fermented west African cereal foods using culture independent methods. Int. J. Food Microbiol. 2011, 145, 205–210. [Google Scholar] [CrossRef]
- Cho, G.-S.; Huch, M.; Hanak, A.; Holzapfel, W.H.; Franz, C.M.A.P. Genetic analysis of the plantaricin EFI locus of Lactobacillus plantarum PCS20 reveals an unusual plantaricin E gene sequence as a result of mutation. Int. J. Food Microbiol. 2010, 141, S117–S124. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Barba, J.L.; Jiménez-Díaz, R. A novel Lactobacillus pentosus-paired starter culture for spanish-style green olive fermentation. Food Microbiol. 2012, 30, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Lavermicocca, P.; Dekker, M.; Russo, F.; Valerio, F.; Di Venere, D.; Sisto, A. Lactobacillus paracasei-enriched vegetables containing health promoting molecules. In Probiotics, Prebiotics, and Synbiotics; Elsevier: Amsterdam, The Netherlands, 2016; pp. 361–370. ISBN 9780128023716. [Google Scholar]
- Dellaglio, F.; Torriani, S.; Felis, G. Reclassification of Lactobacillus cellobiosus rogosa et al. 1953 as a later synonym of Lactobacillus fermentum beijerinck 1901. Int. J. Syst. Evol. Microbiol. 2004, 54, 809–812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jimenez, E.; Langa, S.; Martin, V.; Arroyo, R.; Martin, R.; Fernandez, L.; Rodriguez, J.M. Complete genome sequence of lactobacillus fermentum CECT 5716, a probiotic strain isolated from human Milk. J. Bacteriol. 2010, 192, 4800. [Google Scholar] [CrossRef] [Green Version]
- López-Huertas, E. Safety and efficacy of human breast milk Lactobacillus fermentum CECT 5716. A mini-review of studies with infant formulae. Benef. Microbes 2015, 6, 219–224. [Google Scholar] [CrossRef]
- Collins, M.D.; Samelis, J.; Metaxopoulos, J.; Wallbanks, S. Taxonomic studies on some Leuconostoc-like organisms from fermented sausages: Description of a new genus Weissella for the Leuconostoc paramesenteroides group of species. J. Appl. Bacteriol. 1993, 75, 595–603. [Google Scholar] [CrossRef]
- Arendt, E.K.; Lucid, A.; Lucey, B.; Sleator, R.D.; Coffey, A.; Lynch, K.M. Genomics of weissella cibaria with an examination of its metabolic traits. Microbiology 2015, 161, 914–930. [Google Scholar] [CrossRef]
- Kot, W.; Neve, H.; Heller, K.J.; Vogensen, F.K. Bacteriophages of Leuconostoc, Oenococcus, and Weissella. Front. Microbiol. 2014, 5, 186. [Google Scholar] [CrossRef] [Green Version]
- Saravanan, C.; Shetty, P.K.H. Isolation and characterization of exopolysaccharide from Leuconostoc lactis KC117496 isolated from idli batter. Int. J. Biol. Macromol. 2016, 90, 100–106. [Google Scholar] [CrossRef]
- Franz, C.M.A.P.; Endo, A.; Abriouel, H.; Van Reenen, C.; Galvez, A.; Dicks, L.M. The genus Pediococcus. In Lactic Acid Bacteria: Biodiversity and Taxonomy; Holzapfel, W., Wood, B.J., Eds.; Wiley Blackwell: Chichester, UK, 2014; pp. 361–376. [Google Scholar]
- Knorr, D. Technology aspects related to microorganisms in functional foods. Trends Food Sci. Technol. 1998, 9, 295–306. [Google Scholar] [CrossRef]
- Todorov, S.D.; Dicks, L.M.T. Bacteriocin production by Pediococcus pentosaceus isolated from marula (Scerocarya Birrea). Int. J. Food Microbiol. 2009, 132, 117–126. [Google Scholar] [CrossRef]
- Yirga, H. The use of probiotics in animal nutrition. J. Probiotics Health 2015, 3, 1–10. [Google Scholar] [CrossRef]
- Wafula, E.N.; Murunga, S.I. Isolation and Identification of phosphate solubilizing and nitrogen-fixing bacteria from lake ol’ bolossat sediments, Kenya. Mod. Appl. Sci. 2020, 14, 37–51. [Google Scholar] [CrossRef]
- Cappuccino, J.G.; Sherman, N. Microbiology: A Laboratory Manual, 10th ed.; Pearson: New York, NY, USA, 2014; ISBN 0321840224. [Google Scholar]
- Gregersen, T. Rapid method for distinction of gram-negative from gram-positive bacteria. Eur. J. Appl. Microbiol. Biotechnol. 1978, 5, 123–127. [Google Scholar] [CrossRef]
- Brosius, J.; Palmer, M.L.; Kennedy, P.J.; Noller, H.F. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli (recombinant plasmids/DNA sequence analysis/RrnB cistron). Biochemistry 1978, 75, 4801–4805. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783. [Google Scholar] [CrossRef]

| Phenotypic Characterization | Molecular Identification | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sample ID | Cell Shape | Gram Status | Catalase | CO2 | 6.5% NaCl | Growth at 10 °C | Growth at 45 °C | Closest Relatives | % Identity | Accession No. |
| CPR243 | Cocci | + | - | + | + | - | - | Leuconostoc mesenteroides | 99.90 | MT597785.1 |
| CPR245 | Rods | + | - | + | + | + | + | Weissella cibaria | 100.00 | MN559487.1 |
| CPR482 | Cocci | + | - | + | + | - | + | Leuconostoc citreum | 99.54 | MT544678.1 |
| CPR721 | Cocci | + | - | + | + | - | - | Leuconostoc mesenteroides | 99.21 | MH704135.1 |
| CPR722 | Rods | + | - | + | + | - | + | Weissella cibaria | 100.00 | MT611777.1 |
| CPR723 | Cocci | + | - | + | + | + | + | Leuconostoc citreum | 91.35 | MT544904.1 |
| CPR961 | Cocci | + | - | + | - | - | - | Leuconostoc mesenteroides | 99.61 | MT597708.1 |
| CPR963 | Rods | + | - | - | + | - | + | Lactobacillus plantarum | 98.86 | MT231806.1 |
| CPR967 | Rods | + | - | + | + | + | + | Weissella cibaria | 99.72 | MT611777.1 |
| CPR1448 | Rods | + | - | - | + | - | - | Lactobacillus casei | 97.15 | MH899355.1 |
| CPR1449 | Rods | + | - | - | + | - | + | Lactobacillus pentosus | 97.44 | MH899343.1 |
| CPR1682 | Rods | + | - | + | + | - | + | Weissella cibaria | 98.47 | MH899248.1 |
| CPR1687 | Rods | + | - | - | + | + | + | Lactobacillus pentosus | 97.01 | MH899314.1 |
| CPR14410 | Rods | + | - | - | + | + | + | Lactobacillus plantarum | 97.57 | MH899346.1 |
| CPR0021 | Cocci | + | - | - | + | - | + | Enterococcus mundtii | 99.71 | MT116081.1 |
| CPR0031 | Cocci | + | - | + | - | - | - | Leuconostoc lactis | 99.26 | MT604792.1 |
| CPR2431 | Cocci | + | - | - | - | - | + | Enterococcus durans | 99.37 | MT585577.1 |
| CPR4431 | Rods | + | - | - | + | - | + | Lactobacillus plantarum | 100.00 | OL587487.1 |
| CPR4433 | Rods | + | - | - | + | - | + | Lactobacillus plantarum | 99.52 | MK611385.1 |
| JK244 | Cocci | + | - | - | + | - | - | Lactococcus garvieae | 98.22 | MT611574.1 |
| JK248A | Rods | + | - | - | + | + | + | Lactobacillus plantarum | 98.94 | MN640561.1 |
| JK248B | Rods | + | - | - | + | + | + | Lactobacillus plantarum | 98.13 | MF992227.1 |
| JK444A | Rods | + | - | - | + | - | + | Lactobacillus plantarum | 100.00 | KX649074.1 |
| JK481 | Rods | + | - | + | + | - | - | Weissella cibaria | 99.63 | MT613505.1 |
| JK482 | Rods | + | - | + | + | + | + | Weissella cibaria | 100 | MT613466.1 |
| JK487 | Rods | + | - | + | + | - | + | Weissella cibaria | 100 | MT613505.1 |
| JK721 | Rods | + | - | + | + | - | + | Weissella cibaria | 99.82 | MT613505.1 |
| M444B | Rods | + | - | - | + | + | + | Lactobacillus plantarum | 99.27 | KX057551.1 |
| M723B | Rods | + | - | - | + | - | + | Lactobacillus plantarum | 99.06 | MF992227.1 |
| N8243 | Rods | + | - | - | + | + | + | Lactobacillus plantarum | 99.72 | MT613638.1 |
| N8481 | Rods | + | - | - | + | - | + | Lactobacillus plantarum | 96.75 | MN420754.1 |
| N8729 | Cocci | + | - | - | + | - | + | Lactococcus garvieae | 100 | MT604790.1 |
| NS442B | Rods | + | - | - | + | + | + | Lactobacillus plantarum | 98.67 | MF992227.1 |
| NS489A | Rods | + | - | - | + | + | + | Lactobacillus plantarum | 99.76 | MF992229.1 |
| Phenotypic Characterization | Molecular Identification | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sample ID | Cell Shape | Gram Status | Catalase | CO2 | 6.5% NaCl | Growth at 10 °C | Growth at 45 °C | Closest Relatives | % Identity | Accession No. |
| NSR0012 | Rods | + | - | - | + | + | + | Lactiplantibacillus plantarum | 99.50 | OL587487.1 |
| NSR0021 | Rods | + | - | - | + | + | + | Lactobacillus plantarum | 99.14 | MT463848.1 |
| NSR2411 | Cocci | + | - | - | + | - | + | Enterococcus mundtii | 99.49 | AP019810.1 |
| NSR2412 | Rods | + | - | + | + | - | + | Lactobacillus fermentum | 99.90 | KJ872850.1 |
| NSR2413 | Rods | + | - | + | - | - | + | Lactobacillus fermentum | 99.07 | MT505566.1 |
| NSR4411 | Rods | + | - | - | + | + | + | Lactobacillus plantarum | 99.61 | MK611401.1 |
| NSR4412 | Rods | + | - | - | + | + | + | Lactobacillus plantarum | 100.00 | OL587487.1 |
| NSR4413 | Rods | + | - | + | + | - | + | Lactobacillus fermentum | 99.65 | KT633923.1 |
| NSR4421 | Rods | + | - | - | + | + | + | Lactobacillus plantarum | 99.50 | MH473447.1 |
| NSR4422 | Rods | + | - | - | + | + | + | Lactiplantibacillus plantarum | 100.00 | OL518965.1 |
| NSR4431 | Rods | + | - | - | + | - | + | Lactobacillus plantarum | 99.25 | MK611401.1 |
| NSR4442 | Cocci | + | - | - | + | + | + | Pediococcus pentosaceus | 99.90 | MT510483.1 |
| NSR4813 | Cocci | + | - | - | + | + | + | Pediococcus pentosaceus | 98.67 | EU080993.1 |
| NSR4821 | Cocci | + | - | - | + | + | + | Pediococcus pentosaceus | 99.90 | MT604839.1 |
| NSR4822 | Cocci | + | - | - | + | - | + | Enterococcus faecalis | 99.88 | OK392639.1 |
| NSR4824 | Rods | + | - | + | + | + | + | Weissella confusa | 100.00 | OK326537.1 |
| NSR4841 | Rods | + | - | + | + | + | - | Weissella muntiaci | 99.71 | NR_170492.1 |
| NSR7222 | Rods | + | - | - | + | + | + | Lactobacillus plantarum | 99.70 | OL587487.1 |
| NSR7223 | Rods | + | - | + | + | - | - | Weissella confusa | 100.00 | OK326231.1 |
| NSR7224 | Rods | + | - | - | + | + | + | Lactiplantibacillus plantarum | 100.00 | OL587487.1 |
| NSR7241 | Cocci | + | - | - | + | + | + | Pediococcus pentosaceus | 100.00 | MT604838.1 |
| NSR7242 | Cocci | + | - | - | + | + | + | Pediococcus pentosaceus | 99.90 | MT604839.1 |
| NSR7243 | Cocci | + | - | - | + | + | + | Pediococcus pentosaceus | 100.00 | MT604839.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wafula, E.N.; Kuja, J.O.; Wekesa, T.B.; Wanjala, P.M. Isolation and Identification of Autochthonous Lactic Acid Bacteria from Commonly Consumed African Indigenous Leafy Vegetables in Kenya. Bacteria 2023, 2, 1-20. https://doi.org/10.3390/bacteria2010001
Wafula EN, Kuja JO, Wekesa TB, Wanjala PM. Isolation and Identification of Autochthonous Lactic Acid Bacteria from Commonly Consumed African Indigenous Leafy Vegetables in Kenya. Bacteria. 2023; 2(1):1-20. https://doi.org/10.3390/bacteria2010001
Chicago/Turabian StyleWafula, Eliud N., Josiah O. Kuja, Tofick B. Wekesa, and Paul M. Wanjala. 2023. "Isolation and Identification of Autochthonous Lactic Acid Bacteria from Commonly Consumed African Indigenous Leafy Vegetables in Kenya" Bacteria 2, no. 1: 1-20. https://doi.org/10.3390/bacteria2010001
APA StyleWafula, E. N., Kuja, J. O., Wekesa, T. B., & Wanjala, P. M. (2023). Isolation and Identification of Autochthonous Lactic Acid Bacteria from Commonly Consumed African Indigenous Leafy Vegetables in Kenya. Bacteria, 2(1), 1-20. https://doi.org/10.3390/bacteria2010001







