Environmental and Anthropogenic Influence on the Core Beneficial Honeybee Gut Microbiota—A Short Communication from Bulgaria
Abstract
:1. Introduction
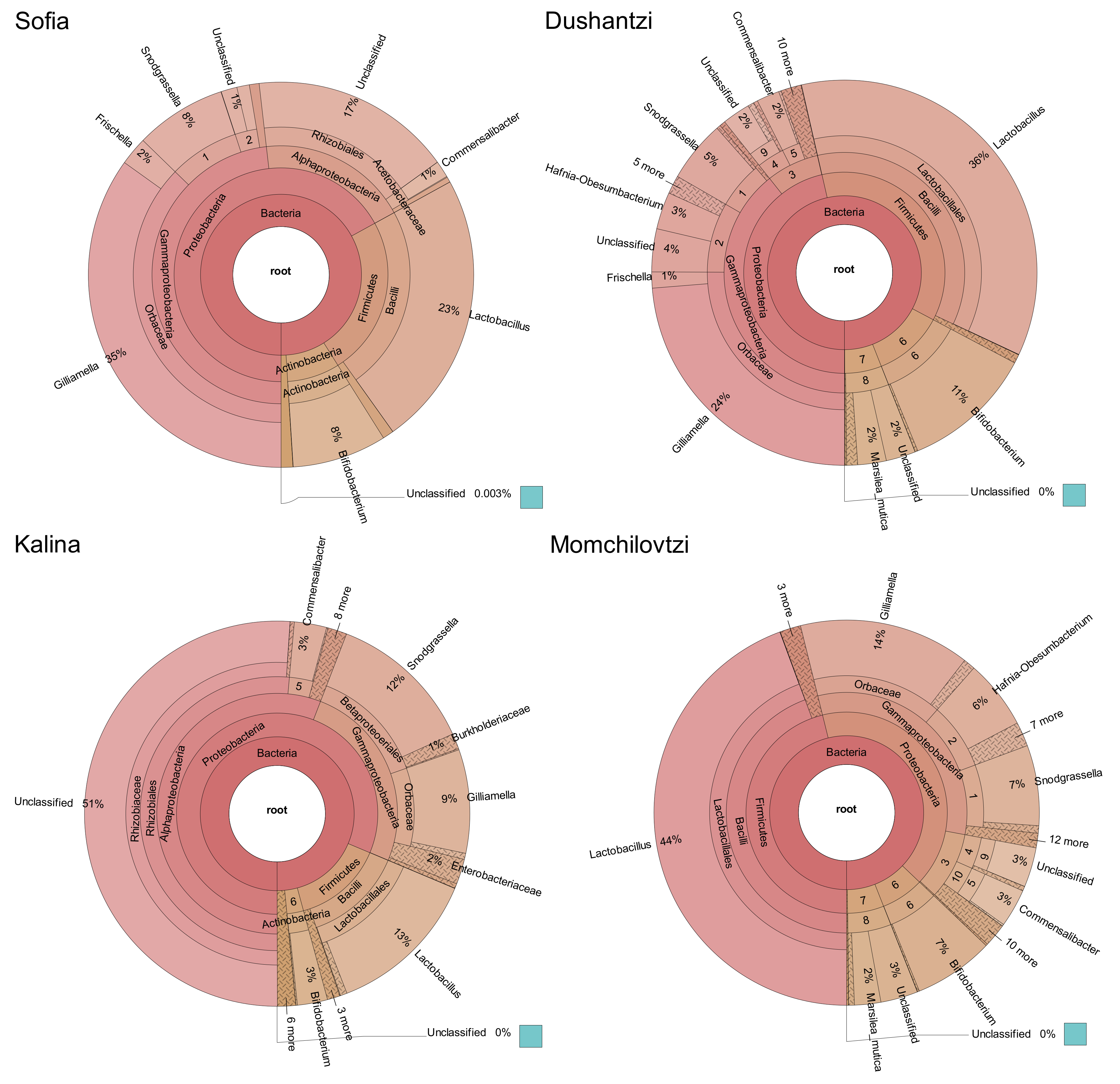
2. Results
3. Discussion
4. Materials and Methods
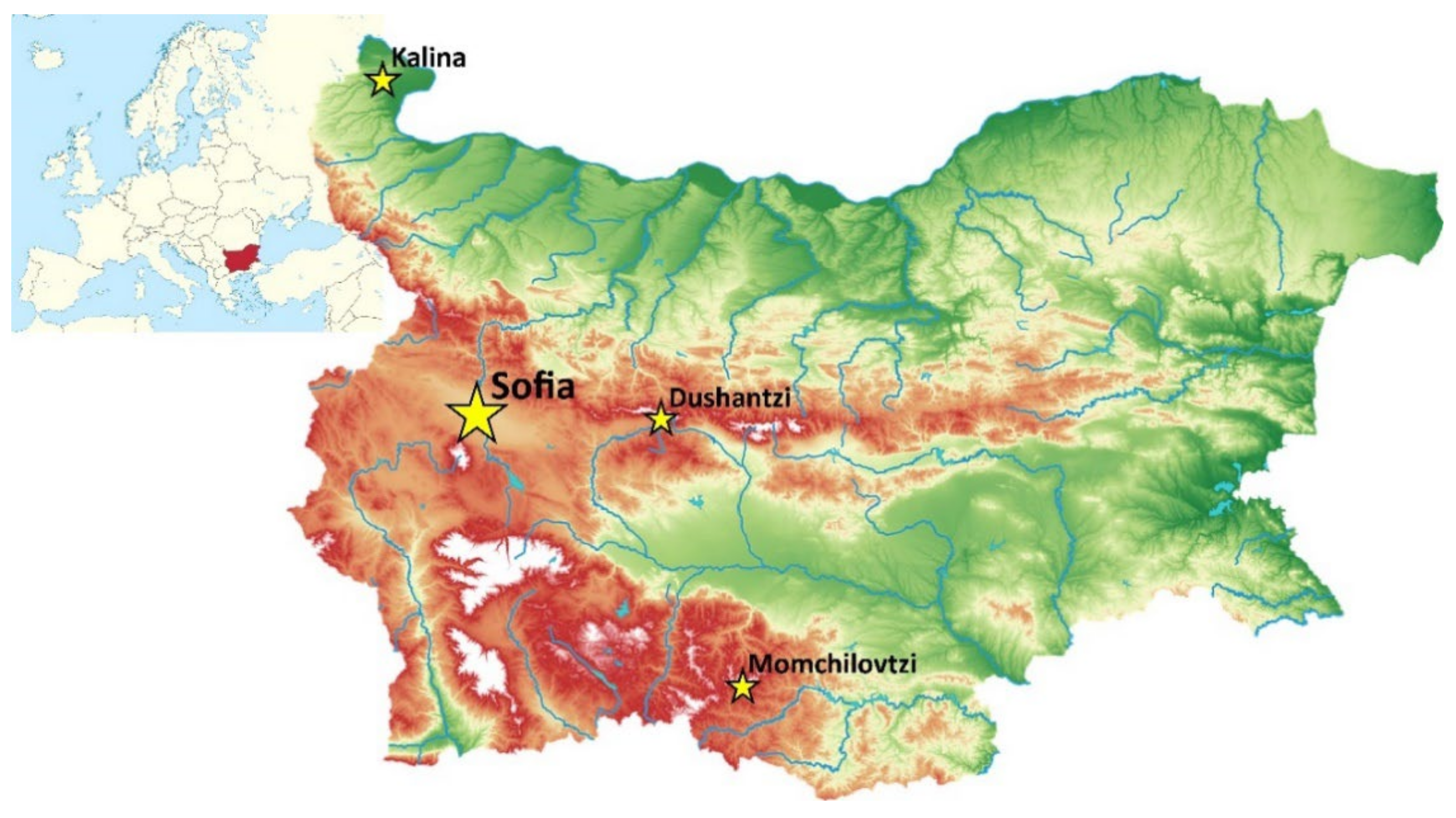
4.1. Sampling Locations and Their Environmental Characteristics
4.1.1. Sofia
4.1.2. Dushantzi
4.1.3. Kalina
4.1.4. Momchilovtzi
4.2. DNA-Based Techniques
4.3. Next-Generation Sequencing
4.4. Bioinformatics Analyses
5. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hein, L. The economic value of the pollination service, a review across scales. Open Ecol. J. 2009, 2, 74–82. [Google Scholar] [CrossRef]
- Cilia, G.; Fratini, F.; Tafi, E.; Mancini, S.; Turchi, B.; Sagona, S.; Cerri, D.; Felicioli, A.; Nanetti, A. Changes of western honey bee apis mellifera ligustica (spinola, 1806) ventriculus microbial profile related to their in-hive tasks. J. Apic. Res. 2021, 60, 198–202. [Google Scholar] [CrossRef]
- Hroncova, Z.; Havlik, J.; Killer, J.; Doskocil, I.; Tyl, J.; Kamler, M.; Titera, D.; Hakl, J.; Mrazek, J.; Bunesova, V.; et al. Variation in honey bee gut microbial diversity affected by ontogenetic stage, age and geographic location. PLoS ONE 2015, 10, e0118707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinson Vincent, G.; Moy, J.; Moran Nancy, A. Establishment of characteristic gut bacteria during development of the honeybee worker. Appl. Environ. Microbiol. 2012, 78, 2830–2840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, K.E.; Sheehan, T.H.; Eckholm, B.J.; Mott, B.M.; DeGrandi-Hoffman, G. An emerging paradigm of colony health: Microbial balance of the honey bee and hive (Apis mellifera). Insectes Sociaux 2011, 58, 431. [Google Scholar] [CrossRef]
- Kwong, W.K.; Mancenido, A.L.; Moran, N.A. Immune system stimulation by the native gut microbiota of honey bees. R. Soc. Open Sci. 2017, 4, 170003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, H.; Steele, M.I.; Leonard, S.P.; Motta, E.V.S.; Moran, N.A. Honey bees as models for gut microbiota research. Lab Anim. 2018, 47, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.C.; Fruciano, C.; Hildebrand, F.; Al Toufalilia, H.; Balfour, N.J.; Bork, P.; Engel, P.; Ratnieks, F.L.; Hughes, W.O. Gut microbiota composition is associated with environmental landscape in honey bees. Ecol. Evol. 2018, 8, 441–451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, B.R.; Shin, J.; Guevarra, R.; Lee, J.H.; Kim, D.W.; Seol, K.H.; Lee, J.H.; Kim, H.B.; Isaacson, R. Deciphering diversity indices for a better understanding of microbial communities. J. Microbiol. Biotechnol. 2017, 27, 2089–2093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ondov, B.D.; Bergman, N.H.; Phillippy, A.M. Interactive metagenomic visualization in a web browser. BMC Bioinform. 2011, 12, 385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raymann, K.; Moran, N.A. The role of the gut microbiome in health and disease of adult honey bee workers. Curr. Opin. Insect Sci. 2018, 26, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Papp, M.; Békési, L.; Farkas, R.; Makrai, L.; Maróti, G.; Tőzsér, D.; Solymosi, N. Natural diversity of honey bee (Apis mellifera) gut bacteriome in various climatic and seasonal states. bioRxiv 2021. [Google Scholar] [CrossRef]
- Ribière, C.; Hegarty, C.; Stephenson, H.; Whelan, P.; O’Toole, P.W. Gut and whole-body microbiota of the honey bee separate thriving and non-thriving hives. Microb. Ecol. 2019, 78, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Horak, R.D.; Leonard, S.P.; Moran, N.A. Symbionts shape host innate immunity in honeybees. Proc. R. Soc. B Biol. Sci. 2020, 287, 20201184. [Google Scholar] [CrossRef] [PubMed]
- Emery, O.; Schmidt, K.; Engel, P. Immune system stimulation by the gut symbiont frischella perrara in the honey bee (Apis mellifera). Mol. Ecol. 2017, 26, 2576–2590. [Google Scholar] [CrossRef] [PubMed]
- Dimov, S.G.; Zagorchev, L.; Iliev, M.; Dekova, T.; Ilieva, R.; Kitanova, M.; Georgieva-Miteva, D.; Dimitrov, M.; Peykov, S. A snapshot picture of the fungal composition of bee bread in four locations in bulgaria, differing in anthropogenic influence. J. Fungi 2021, 7, 845. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The silva ribosomal rna gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive bayesian classifier for rapid assignment of rrna sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Kalina | Sofia | Momchilovtzi | Dushantzi | ||
|---|---|---|---|---|---|
| Quality control statistics of the NGS data | Number of raw paired-end reads | 139,384 | 138,342 | 106,761 | 130,041 |
| Number of raw tags | 107,840 | 114,233 | 88,950 | 96,505 | |
| Number of clean tags | 105,840 | 112,012 | 86,962 | 95,339 | |
| Number of effective tags | 92,726 | 100,564 | 70,750 | 92,140 | |
| Total number of bases of effective tags | 38,396,863 | 42,476,920 | 29,967,266 | 37,496,294 | |
| Average length of effective tags | 414 | 422 | 424 | 407 | |
| Q20 1 | 98.17 | 98.16 | 98.07 | 98.14 | |
| Q30 1 | 94.20 | 94.15 | 93.93 | 94.07 | |
| Percentage of GC content | 52.78 | 51.85 | 51.54 | 54.99 | |
| Percentage of effective tags in raw paired-end reads | 66.53 | 72.69 | 66.27 | 70.85 | |
| Tags data and tags annotation data | Total tags | 92,726 | 100,564 | 70,750 | 184,280 |
| Taxon tags | 91,561 | 99,301 | 69,413 | 183,708 | |
| Unclassified tags | 0 | 2 | 0 | 8 | |
| Unique tags | 1165 | 1261 | 1337 | 564 | |
| Observed species | 162 | 84 | 231 | 322 | |
| Alpha diversity indexes | Shannon | 3.042 | 3.165 | 4.012 | 2.585 |
| Simpson | 0.712 | 0.819 | 0.875 | 0.728 | |
| Chao1 | 163.500 | 85.000 | 231.000 | 322.000 | |
| ACE | 163.921 | 85.427 | 231.000 | 322.000 | |
| Good’s coverage | 1.000 | 1.000 | 1.000 | 1.000 | |
| Lactobacillus sp. | Bifidobacterium sp. | Snodgrassiella alvi | Gilliamella apicola | Frischella perrara | Commensalibacter sp. | Total % | |
|---|---|---|---|---|---|---|---|
| Sofia | 23 | 8 | 8 | 35 | 2 | 1 | 77 |
| Dushantzi | 36 | 11 | 5 | 24 | 1 | 2 | 79 |
| Momchilovtzi | 44 | 7 | 7 | 14 | 1 | 3 | 76 |
| Kalina | 13 | 3 | 12 | 9 | 1 | 3 | 41 |
| Average % | 29 | 7 | 8 | 21 | 1 | 2 | 68 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dimov, S.G. Environmental and Anthropogenic Influence on the Core Beneficial Honeybee Gut Microbiota—A Short Communication from Bulgaria. Bacteria 2022, 1, 88-95. https://doi.org/10.3390/bacteria1020008
Dimov SG. Environmental and Anthropogenic Influence on the Core Beneficial Honeybee Gut Microbiota—A Short Communication from Bulgaria. Bacteria. 2022; 1(2):88-95. https://doi.org/10.3390/bacteria1020008
Chicago/Turabian StyleDimov, Svetoslav G. 2022. "Environmental and Anthropogenic Influence on the Core Beneficial Honeybee Gut Microbiota—A Short Communication from Bulgaria" Bacteria 1, no. 2: 88-95. https://doi.org/10.3390/bacteria1020008
APA StyleDimov, S. G. (2022). Environmental and Anthropogenic Influence on the Core Beneficial Honeybee Gut Microbiota—A Short Communication from Bulgaria. Bacteria, 1(2), 88-95. https://doi.org/10.3390/bacteria1020008






