Bayesian Morphometric Analysis for Archaeological Seed Identification: Phoenix (Arecaceae) Palms from the Canary Islands (Spain)
Abstract
1. Introduction
1.1. Taxonomic Overview of Phoenix Palms
1.2. Morphological and Genetic Characteristics of P. canariensis
1.3. Ethnobotanical Significance and Cultural Integration
1.4. Taxonomic Complexity and Hybridization
1.5. Theoretical Framework: Domestication as Coevolution
1.6. Geographical Context
1.7. Research Objectives and Methodological Approach
2. Materials and Methods
2.1. Archaeological Context
2.2. Phoenix Seed Morphology
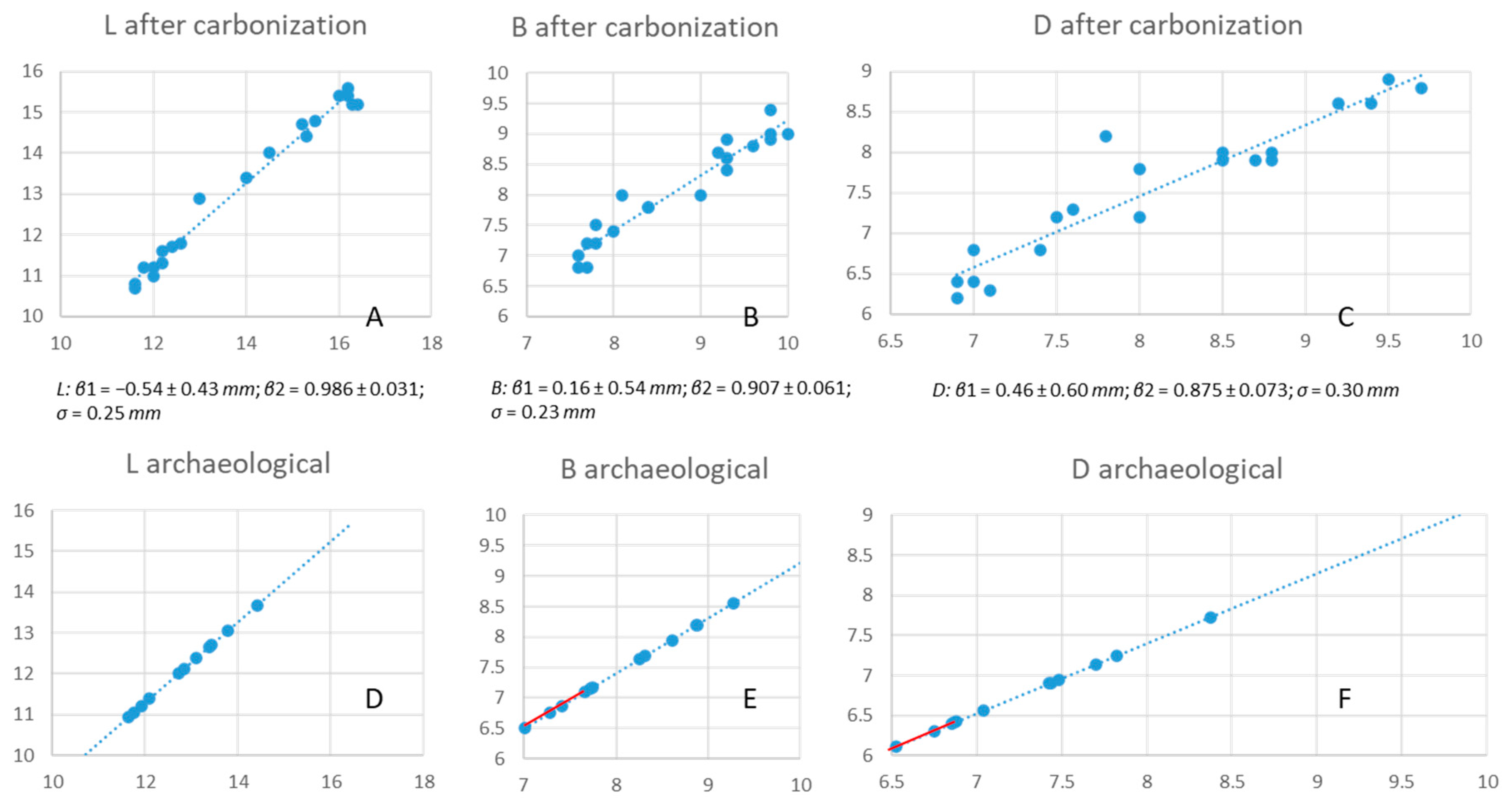
2.3. Morphometric Seed Analysis
2.4. Experimental Carbonization
- y: the dependent variable (outcome or response variable).
- β1: the intercept (the value of y when x = 0).
- β2: the slope coefficient (the change in y for a one-unit increase in x).
- x: the independent variable (predictor or explanatory variable).
- ε: the error term, which represents the unexplained variation in y that is not accounted for by the linear relationship with x. It accounts for randomness, measurement errors, or influences from other variables not included in the model. It ensures that the equation acknowledges real-world data variability rather than assuming a perfect deterministic relationship between x and y.
- Site-level analysis using original, uncorrected data (five aggregated samples).
- Individual seed-level analysis using original, uncorrected data (twelve discrete samples).
- Site-level analysis incorporating carbonization-induced morphological corrections (five aggregated samples).
- Individual seed-level analysis incorporating carbonization-induced morphological corrections (twelve discrete samples).
2.5. Data Analysis
2.5.1. Classification of Samples
- Length (L): 15 intervals, ranging from 4 to ≤60 mm.
- Breadth (B): 9 intervals, ranging from 3 to ≤19 mm.
- Depth (D): 6 intervals, ranging from 0.1 to ≤18 mm.
- Depth-to-breadth ratio (D/B): 6 intervals, ranging from 0.3 to ≤1.5.
- Volumetric dimension (L × B × D): 13 intervals, ranging from 36 to ≤1200 mm3.
- K: Total number of variables
- c ∈ {1, 2,..., C}, where C = 67 represents the descriptive morphological parameters
- Kc: Number of states for each descriptor c (approximately 10 states per descriptor)
- xik, xjk: Specific values of variable k for sampling units i and j, respectively
- xi.: Mean value for sampling unit i Equation (6)
- xj.: Mean value for sampling unit j
- x.k: Mean value for variable k Equation (7)
- x..: Comprehensive overall mean Equation (8)
- dij = 0 indicates complete morphological congruence, signifying that samples i and j are statistically indistinguishable across all analyzed variables
- dij = 1 represents maximal morphological divergence, indicating that samples i and j exhibit complete heterogeneity across the entire suite of descriptive parameters
- Definition of ‘neighborhood’ proximity.
- Algorithmic updating of the dissimilarity matrix.
- Estimation of intercluster edge lengths.
- Objective Function Methodology: Ward’s approach is fundamentally grounded in an optimization principle that minimizes the incremental variance resulting from cluster mergers. This mathematical precision offers a transparent and systematic mechanism for hierarchical clustering, enabling researchers to objectively quantify cluster formation [66].
- Variance Minimization Strategy: By emphasizing the reduction of within-cluster variance, the method preferentially generates compact, spherical clusters. This characteristic is particularly valuable when investigating complex dissimilarity matrices, as it facilitates the identification of inherent data groupings [67].
- Metric Adaptability: Although initially conceived for squared Euclidean distances, Ward’s method demonstrates remarkable versatility, allowing adaptation to diverse distance metrics and dissimilarity matrices [68].
2.5.2. Identification of Samples
- Prior probability p(Hi|I): The initial probabilistic assessment of the hypothesis based on pre-existing knowledge (background information).
- Likelihood p(D|Hi,I): The probability of observing the specific data given the hypothesis and background information.
- Normalization constant p(D|I): A probabilistic scaling factor that ensures the total probability across all hypotheses equals unity, expressed by the marginal probability of data (D) given the background information (I).
- Principle of Indifference: Uniform probability distribution across 24 taxonomic units, with p(Hi) = 1/24 for each hypothesis. The Principle of Indifference (alternatively termed the Principle of Insufficient Reason)—a fundamental probabilistic heuristic. When confronted with N mutually exclusive and exhaustive propositions (H) and an absence of discriminating evidence, this principle prescribes an equiprobable distribution, such that each proposition is assigned an equal probability p(Hi) = 1/N.
- Empirical Frequency-Based Priors: Alternatively, one can establish prior probabilities based on the observed frequency distribution of taxonomic groups within the comprehensive seed ensemble utilized in the analysis (Table 1).
- ni denotes the number of seeds associated with taxonomic unit ti
- N represents the total number of seeds in the analytical ensemble
- p(Hi) quantifies the prior probability as the frequency-weighted representation of each taxonomic group
3. Results
3.1. Key Diagnostic Characters
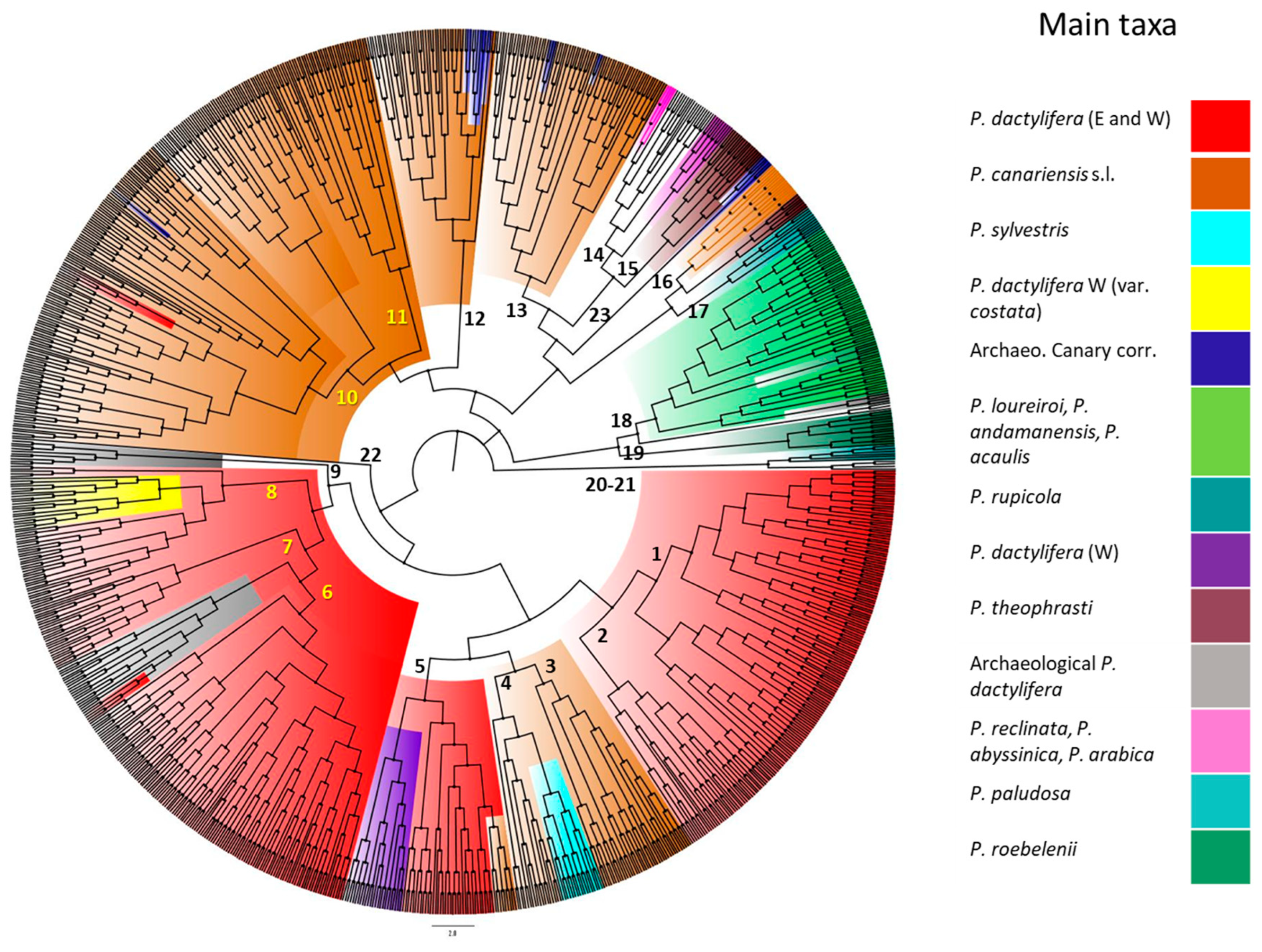
3.2. Morphotypes and Species
3.3. Identification of Phoenix Archaeological Seed Samples from the Canary Islands
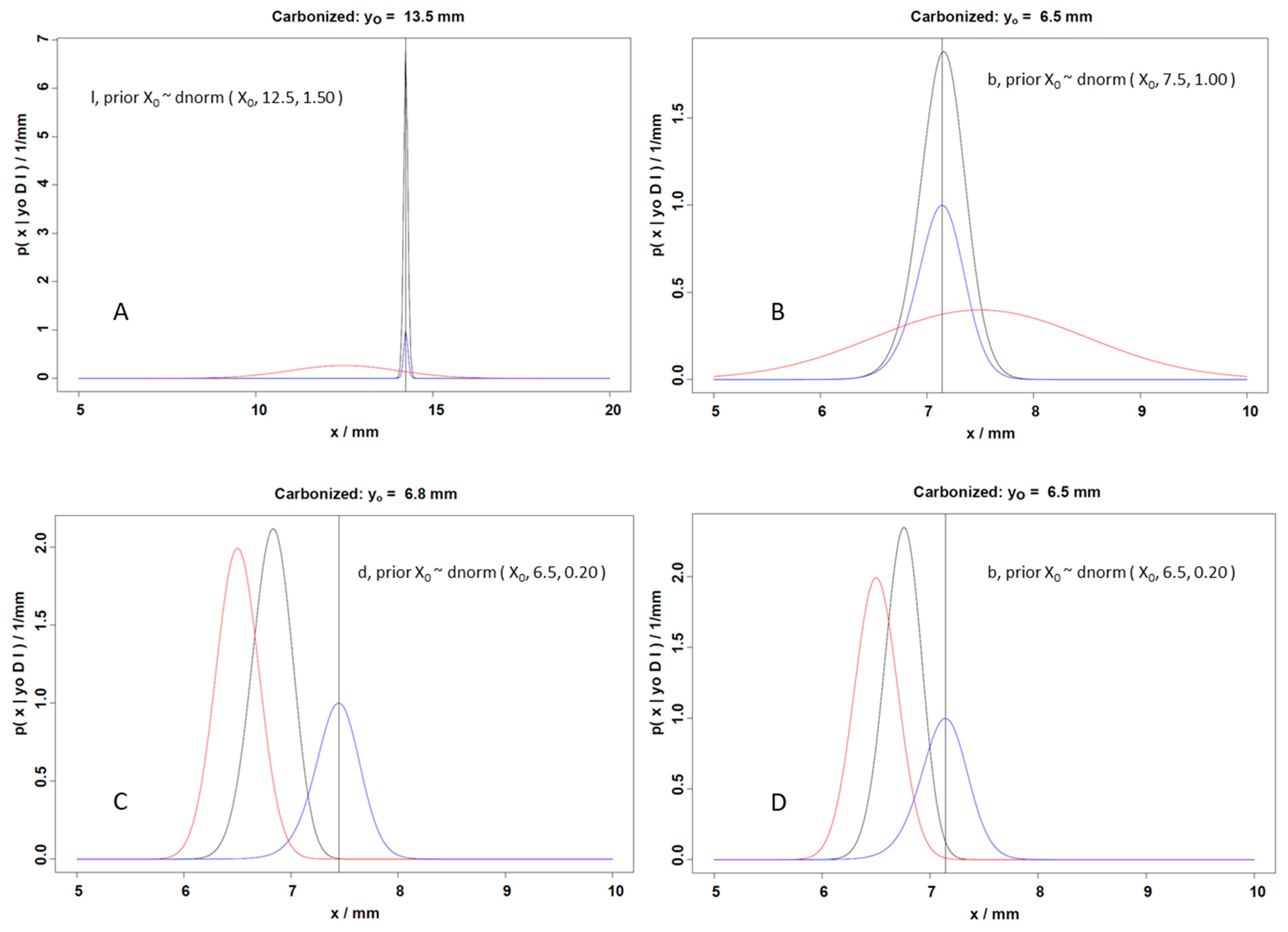
- Panel A: Displays a predictive distribution for length (l), given a dnorm(X0, 12.5, 1.50) prior. With a prior centered around X0 = 12.5 mm and a relatively high standard deviation (1.50), the y-axis represents probability density, and the x-axis represents length (x) in millimeters. The posterior (black curve) aligns closely with the observed data, indicating that the likelihood strongly influences the posterior due to the narrow spread of the likelihood function (blue).
- Panel B: Represents the posterior distribution of breadth (b), given a dnorm(X0, 7.5, 1.00) prior. With a prior centered around X0 = 7.5 mm and moderate standard deviation (1.00), the posterior distribution, where the carbonized seed dimension (Y0) is 6.5 mm, shifts significantly compared to the prior, reflecting an interplay between prior assumptions and observed data.
- Panel C: Depicts the posterior distribution of depth (d), given a dnorm(X0, 6.5, 0.20) prior. With a prior centered around X0 = 6.5 mm and a low standard deviation (0.20), the narrow prior strongly influences the posterior, where the carbonized seed dimension (Y0) is 6.8 mm, which aligns closely with the prior distribution.
- Panel D: Also pertains to breadth (b) and thus shows the posterior distribution of breadth, given a dnorm(X0, 6.5, 0.20) prior. With a prior centered around X0 = 6.5 mm and a much narrower prior standard deviation (0.20), the prior significantly restricts the posterior’s range, where the carbonized seed dimension (Y0) is 6.5 mm.
4. Discussion
| Taxa | Alto Garajonay | Sobrado de los Gomeros | Cueva Pintada | Lomo de los Gatos | Guayedra |
|---|---|---|---|---|---|
| P. canariensis var. canariensis | 0.709 | 0.590 | 0.81 | 0.66 | 0.66 |
| P. canariensis var. porphyrococca | 0 | 0.017 | 0 | 0.03 | 0.03 |
| P. canariensis Wildpret’s Large Date Group | 0.166 | 0.080 | 0.19 | 0.03 | 0.03 |
| P. canariensis Subtotal | 0.875 | 0.688 | 1.00 | 0.72 | 0.72 |
| P. dactylifera s.l. Subtotal | 0 | 0.045 | 0 | 0.08 | 0.08 |
| P. farinifera | 0 | 0 | 0 | 0.05 | 0.05 |
| P. roebelenii | 0.021 | 0 | 0 | 0.07 | 0.07 |
| P. theophrasti | 0.104 | 0.015 | 0 | 0.03 | 0.03 |
| Others Subtotal | 0.125 | 0.102 | 0 | 0.19 | 0.19 |
| Probability ratio P. canariensis vs. others | 7 | 6.74 | ꝏ | 3.78 | 3.78 |
| Probability ratio P. canariensis vs. P. dactylifera s.l. | ꝏ | 15.28 | ꝏ | 8.25 | 8.25 |
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Barrow, S. A monograph of Phoenix L. (Palmae: Coryphoideae). Kew Bull. 1998, 53, 513–575. [Google Scholar] [CrossRef]
- Beccari, O. Rivista monografica delle specie del genere. Phoenix L. Malesia 1890, 3, 345–416. [Google Scholar]
- Carreño, E. Diversidad Genética en Especies del Género Phoenix. Ph.D. Thesis, Escuela Politécnica Superior, Universidad Miguel Hernández, Orihuela, Spain, 2017. Available online: https://dialnet.unirioja.es/servlet/tesis?codigo=135027 (accessed on 6 February 2025).
- IPNI. Phoenix in International Plant Names Index (IPNI). Available online: https://www.ipni.org/?q=Phoenix%2CPhoenix (accessed on 6 February 2025).
- Rivera, D.; Obón, C.; García, J.; Egea, T.; Alcaraz, F.; Laguna, E.; Carreño, E.; Johnson, D.; Krueger, R.; Delgadillo, J.; et al. Carpological analysis of Phoenix (Arecaceae): Contributions to the taxonomy and evolutionary history of the genus. Bot. J. Linn. Soc. 2014, 175, 74–122. [Google Scholar] [CrossRef]
- Rivera, D.; Abellán, J.; Palazón, J.A.; Obón, C.; Alcaraz, F.; Carreño, E.; Laguna, E.; Ruiz, A.; Johnson, D. Modelling ancient areas for date palms (Phoenix species: Arecaceae): Bayesian analysis of biological and cultural evidence. Bot. J. Linn. Soc. 2020, 193, 228–262. [Google Scholar] [CrossRef]
- Martínez-Rico, M. El género Phoenix en Jardinería y Paisajismo: El Caso de Phoenix canariensis. Ph.D. Thesis, Universidad Miguel Hernández de Elche, Orihuela, Spain, 2017. Available online: https://dialnet.unirioja.es/servlet/dctes?codigo=134995 (accessed on 6 February 2025).
- Rivera, D.; Obón, C.; Alcaraz, F.; Egea, T.; Carreño, E.; Laguna, E.; Saro, I.; Sosa, P.A. The date palm with blue dates Phoenix senegalensis André (Arecaceae): A horticultural enigma is solved. Sci. Hortic. 2014, 180, 236–242. [Google Scholar] [CrossRef]
- Rivera, D.; Obón, C.; Verde, A.; Fajardo, J.; Valdés, A.; Alcaraz, F.; Carreño, E.; Heinrich, M.; Martínez-Rico, M.; Ríos, S.; et al. La palmera datilera y la palmera canaria en la fitoterapia tradicional de España. Rev. Fitoter. 2014, 14, 67–81. [Google Scholar]
- Spennemann, D. Canary Date Palms (Phoenix canariensis) as ornamental plants. The first thirty years of the horticultural trade. Huntia 2019, 17, 79–102. [Google Scholar]
- Zona, S. The Horticultural History of the Canary Island date palm (Phoenix canariensis). Gard. Hist. 2008, 36, 301–309. Available online: https://www.jstor.org/stable/40649462 (accessed on 22 March 2025).
- Castilla-Beltrán, A.; Fernández-Palacios, E.; Vrydaghs, L.; Mallol, C.; Fernández-Palacios, J.M.; de Nascimento, L. Phytoliths from modern plants in the Canary Islands as a reference for the reconstruction of long-term vegetation change and culture-environment interactions. Veg. Hist. Archaeobotany 2024, 33, 705–723. [Google Scholar] [CrossRef]
- Saro, I.; González-Pérez, M.; García-Verdugo, C.; Sosa, P. Patterns of genetic diversity in Phoenix canariensis, a widespread oceanic palm species endemic from the Canarian archipelago. Tree Genet. Genomes 2015, 11, 815. [Google Scholar] [CrossRef]
- Saro, I.; García-Verdugo, C.; González-Pérez, M.; Naranjo, A.; Santana, A.; Sosa, P.A. Genetic structure of the Canarian palm tree (Phoenix canariensis) at the island scale, Does the “island within islands” concept apply to species with high colonization abilities? Plant Biol. 2019, 21, 101–109. [Google Scholar] [CrossRef]
- Vidal-Matutano, P.; Delgado-Darias, T.; López-Dos Santos, N.; Henríquez-Valido, P.; Velasco-Vázquez, J.; Alberto-Barroso, V. Use of decayed wood for funerary practices, Archaeobotanical analysis of funerary wooden artefacts from Prehispanic (ca. 400–1500 CE) Gran Canaria (Canary Islands, Spain). Quat. Int. 2021, 593, 384–398. [Google Scholar] [CrossRef]
- Montesino, J. Los paisajes y la gente de La Gomera. In La Gomera. Entre Bosques y Taparuchas. Actas XI Semana Científica Telesforo Bravo; Afonso, J., Ed.; Instituto de Estudios Hispánicos de Canarias: Puerto de la Cruz, Spain, 2016; pp. 39–100. [Google Scholar]
- Sosa, P.A.; Naranjo, A.; Marques, M.; Escandell, A.; González-Pérez, M. Atlas de los Palmerales de Gran Canaria; Obra Social de la Caja de Canarias: Las Palmas, Spain, 2007. [Google Scholar]
- Sosa, P.A.; Saro, I.; Gil, J.; Rivera, D.; Alcaraz, F.J.; Obón, C. Biología, distribución y genética de la palmera canaria. Quercus 2018, 387, 45–52. [Google Scholar]
- Obón, C.; Rivera, D.; Alcaraz, F.; Carreño, E.; Ríos, S.; Laguna, E.; Sánchez-Balibrea, J.; del Arco, M.; Bergmeier, E.; Johnson, D. What are palm groves of Phoenix? Conservation of Phoenix palm groves in the European Union. Biodivers. Conserv. 2018, 27, 1905–1922. [Google Scholar] [CrossRef]
- González-Pérez, M.; Caujapé-Castells, J.; Sosa, P.A. Molecular evidence of hybridisation between the endemic Phoenix canariensis and the widespread P. dactylifera with Random Amplified Polymorphic DNA (RAPD) markers. Plant Syst. Evol. 2004, 247, 165–175. [Google Scholar] [CrossRef]
- Saro, I.; Rodríguez-Rodríguez, P.; Rivera, D.; Obón, C.; Aberlenc, F.; Díaz-Pérez, A.; Zehdi-Azouzi, S.; Curbelo, L.; Sosa, P.A. The Genetic Characterization of the Canarian Endemic Palm (Phoenix canariensis) by Simple Sequence Repeats and Chloroplast Markers: A Tool for the Molecular Traceability of Phoenix Hybridization. Diversity 2024, 16, 411. [Google Scholar] [CrossRef]
- Purugganan, M. What is domestication? Trends Ecol. Evol. 2022, 37, 663–671. [Google Scholar] [CrossRef]
- Thompson, J. Specific Hypotheses on the Geographic Mosaic of Coevolution. Am. Nat. 1999, 153, S1–S14. [Google Scholar]
- Archetti, M. Evidence from the domestication of apple for the maintenance of autumn colours by coevolution. Proc. R. Soc. B 2009, 276, 2575–2580. [Google Scholar] [CrossRef]
- Slatkin, M.; Maynard-Smith, J. Models of Coevolution. Q. Rev. Biol. 1979, 54, 233–263. [Google Scholar] [CrossRef]
- Jackson, F. The Coevolutionary Relationship of Humans and Domesticated Plants. Yearb. Phys. Anthropol. 1996, 39, 161–176. [Google Scholar] [CrossRef]
- Morales, J.; Gil, J. Gathering in a new environment, the use of wild food plants during the first colonization of the Canary Islands, Spain (3–2nd BC to 15th AD). In Plants and People, Choices and Diversity Through Time; Chevalier, A., Marinova, E., Peña-Chocarro, L., Eds.; EARTH Series; Oxbow Books: Oxford, UK, 2014; Volume 1, pp. 216–227. [Google Scholar]
- Serrano, J.G.; Ordóñez, A.C.; Santana, J.; Sánchez-Cañadillas, E.; Arnay, M.; Rodríguez-Rodríguez, A.; Morales, J.; Velasco-Vázquez, J.; Alberto-Barroso, V.; Delgado-Darias, T.; et al. The genomic history of the Indigenous people of the Canary Islands. Nat. Commun. 2023, 14, 4641. [Google Scholar] [CrossRef]
- Santana, J.; Del Pino, M.; Morales, J.; Fregel, R.; Hagenblad, J.; Morquecho, A.; Brito-Mayor, A.; Henríquez, P.; Jiménez, J.; Serrano, J.; et al. The chronology of the human colonization of the Canary Islands. Proc. Nat. Acad. Sci. USA 2024, 121, e2302924121. [Google Scholar] [CrossRef]
- Abreu-Galindo, J. Historia de la Conquista de las Siete Islas de Canaria; Goya: Santa Cruz de Tenerife, Spain, 1977. [Google Scholar]
- Frutuoso, G. Las Islas Canarias, Saudades da terra. In Fontes Rerum Canariarum XII; Serra, E., Régulo, J., Pestana, S., Eds.; Instituto de Estudios Canarios: La Laguna, Spain, 1964. [Google Scholar]
- Morales-Padrón, F. Canarias. Crónicas de su Conquista; Cabildo Insular de Gran Canaria: Las Palmas de Gran Canaria, Spain, 1993. [Google Scholar]
- Morales, J.; Speciale, C.; Rodríguez-Rodríguez, A.; Henríquez-Valido, P.; Marrero-Salas, E.; Hernández-Marrero, J.C.; López, R.; López, R.; Delgado-Darias, T.; Hagenblad, J.; et al. Agriculture and crop dispersal in the western periphery of the Old World, the Amazigh/Berber settling of the Canary Islands (ca. 2nd–15th centuries CE). Veg. Hist. Archaeobot. 2023, 1–15. [Google Scholar] [CrossRef]
- De-Nascimento, L.; Willis, K.J.; Fernández-Palacios, J.M.; Criado, C.; Whittaker, R.J. The long-term ecology of the lost forests of La Laguna, Tenerife (Canary Islands). J. Biogeogr. 2009, 36, 499–514. [Google Scholar] [CrossRef]
- De-Nascimento, L.; Nogué, S.; Criado, C.; Ravazzi, C.; Whittaker, R.J.; Willis, K.J.; Fernández-Palacios, J.M. Reconstructing Holocene vegetation on the island of Gran Canaria before and after human colonization. Holocene 2015, 26, 113–125. [Google Scholar] [CrossRef]
- Arco, M.C. Recursos Vegetales en la Prehistoria de Canarias; Serie Museo Arqueológico: La Laguna, Spain, 1993. [Google Scholar]
- Mireles-Betancor, F.; Olmo, S.; Rodríguez, A. El poblado prehispánico de Tufia (Telde, Gran Canaria). Intervenciones arqueológicas 1997–1999. Mus. Canar. 2006, 61, 13–63. Available online: https://www.elmuseocanario.com/images/documentospdf/revistaelmuseo/Revistas/2006.pdf (accessed on 25 March 2025).
- Rodríguez-Santana, C. El trabajo de las fibras vegetales entre los antiguos canarios. Pajar 2002, 12, 4–10. [Google Scholar]
- González, P.; Moreno, M.; Jiménez, A. El Yacimiento Arqueológico de La Cerera. Un modelo de Ocupación en la Isla de Gran Canaria; Cabildo de Gran Canaria: Las Palmas, Spain, 2009. [Google Scholar]
- Hernández-Marrero, J.; Navarro, J.; Trujillo, J.; Cancel, S.; Machado, C.; País, J.; Morales, J.; Rando, J.C. An approach to prehistoric shepherding in La Gomera (Canary Islands) through the study of domestic spaces. Quat. Int. 2016, 414, 337–349. [Google Scholar] [CrossRef]
- Morales, J.; Navarro, J.; Rodríguez, A. Plant offerings to the Gods, seed remains from a pre-Hispanic sacrificial altar in La Gomera island (Canary Islands, Spain). In Windows on the African Past; Fahmy, A.G., Kahlheber, S., D’Andrea, A., Eds.; Africa Magna Verlag: Frankfurt, Germany, 2011; pp. 67–78. [Google Scholar]
- Morales, J.; Rodríguez, A.; Henríquez, P. Agricultura y recolección vegetal en la arqueología prehispánica de las Islas Canarias (siglos III–XV d.C.), la contribución de los estudios carpológicos. In Miscelanea en Homenaje a Lydia Zapata Peña (1965–2015); Fernández-Eraso, J., Mugica, J.A., Arrizabalaga, A., Garcia-Diez, M., Eds.; Universidad del País Vasco: Bilbao, Spain, 2017; pp. 191–220. [Google Scholar]
- del Arco, M.M.; Hernández, C.; Adrián, M.; Armas, E.; del Arco, M.C. El menceyato de Icod en el poblamiento de Tenerife, D. Gaspar, Las Palomas y Los Guanches. Sobre el poblamiento y las estrategias de alimentación vegetal entre los guanches. Eres. Arqueol./Bioantropología 2000, 9, 67–129. [Google Scholar]
- Soler, J.; Pérez, F.; Rodríguez, T. Excavaciones en la Memoria. Estudio Historiográfico del Barranco del Agua de Dios y de la Comarca de Tegueste (Tenerife); Gobierno de Canarias: Santa Cruz de Tenerife, Spain, 2011. [Google Scholar]
- Fontugne, M.; García, A.; Hatté, C.; Núñez, M.; Olmo, S.; Onrubia, J.; García, A.; Pérez-Jordà, G.; Rodríguez, C.; Sáenz, J.I.; et al. Parque arqueológico Cueva Pintada (Gáldar, Gran Canaria). Programa de intervenciones e investigaciones arqueológicas. Avance de los trabajos efectuados entre los años 1995–1997. Investig. Arqueol. Canar. 1999, 6, 489–561. [Google Scholar]
- Morales, J. El Uso de las Plantas en la Prehistoria de Gran Canaria, Alimentación, Agricultura y Ecología; Cabildo de Gran Canaria: Gran Canaria, Spain, 2010. [Google Scholar]
- Martín de Guzmán, C. Estructuras habitacionales del Valle de Guayedra. Not. Arqueol. Hispánico 1982, 10, 301–318. [Google Scholar]
- Navarro, J.; Borges, E.; Barro, A.; Alberto, V.; Hernández, C.; Hernández, J. El Diezmo a Orahan, Pireos o aras de sacrificio en la prehistoria de La Gomera (Islas Canarias). Tabona 2001, 10, 91–126. [Google Scholar]
- De-Nascimento, L.; Nogue, S.; Naranjo, A.; Criado, C.; McGlone, M.; Fernandez-Palacios, E.; Fernandez-Palacios, J.M. Human impact and ecological changes during prehistoric settlement on the Canary Islands. Quat. Sci. Rev. 2020, 239, 106332. [Google Scholar] [CrossRef]
- Phoenix-Spain. Phoenix Spain. Colección Nacional. Available online: http://www.phoenix-spain.org/ (accessed on 6 February 2025).
- Elaigwu, S.; Greenway, G. Microwave-assisted hydrothermal carbonization of rapeseed husk, a strategy for improving its solid fuel properties. Fuel Process. Technol. 2016, 149, 305–312. [Google Scholar] [CrossRef]
- Guiotoku, M.; Rambo, C.; Hansel, F.; Magalhaes, W.; Hotza, D. Microwave-assisted hydrothermal carbonization of lignocellulosic materials. Mater. Lett. 2009, 63, 2707–2709. [Google Scholar] [CrossRef]
- Perrier, X.; Flori, A.; Bonnot, F. Data analysis methods. In Genetic Diversity of Cultivated Tropical Plants; Hamon, P., Seguin, M., Perrier, X., Glaszmann, J., Eds.; Science Publishers, Enfield: London, UK, 2003; pp. 43–76. [Google Scholar]
- Perrier, X.; Jacquemoud-Collet, J. DARwin Software (6.0.021, 2019-04-26). Available online: https://darwin.cirad.fr/ (accessed on 6 February 2025).
- Perrier, X.; Jacquemoud-Collet, J. Darwin. Dissimilarity Analysis and Representation for Windows, version 6; CIRAD: Montpellier, France, 2014. [Google Scholar]
- Hsu, C.N.; Huang, H.J.; Wong, T. Implications of the Dirichlet assumption for discretization of continuous variables in naive Bayesian classifiers. Mach. Learn. 2003, 53, 235–263. [Google Scholar] [CrossRef]
- Clarke, E.J.; Barton, B.A. Entropy and MDL discretization of continuous variables for Bayesian belief networks. Int. J. Intell. Syst. 2000, 15, 61–92. [Google Scholar] [CrossRef]
- Galli, S. Data Discretization in Machine Learning (4 July 2022). Available online: https://www.blog.trainindata.com/data-discretization-in-machine-learning/ (accessed on 6 February 2025).
- Lustgarten, J.L.; Gopalakrishnan, V.; Grover, H.; Visweswaran, S. Improving classification performance with discretization on biomedical datasets. AMIA Annu. Symp. Proc. 2008, 2008, 445–449. Available online: https://pmc.ncbi.nlm.nih.gov/articles/PMC2656082/ (accessed on 6 February 2025). [PubMed] [PubMed Central]
- Fayyad, U.; Irani, K. Multi-Interval Discretization of Continuous-Valued Attributes for Classification Learning. In Proceedings of the 13th International Joint Conference on Artificial Intelligence, Chambéry, France, 28 August–3 September 1993; IJCAI Organization, Ed.; Morgan Kaufmann: San Mateo, CA, USA, 1993; Volume 2, pp. 1022–1027. Available online: https://www.ijcai.org/Proceedings/93-2/Papers/022.pdf (accessed on 6 February 2025).
- Dougherty, J.; Kohavi, R.; Sahami, M. Supervised and unsupervised discretization of continuous features. In Proceedings of the 12th International Conference on Machine Learning, Tahoe City, CA, USA, 9–12 July 1995; Prieditis, A., Russel, S., Eds.; Morgan Kaufmann: San Francisco, CA, USA, 1995; pp. 194–202. [Google Scholar] [CrossRef]
- Hong, S. Use of contextual information for feature ranking and discretization. IEEE Trans. Knowl. Data Eng. 1997, 9, 718–730. [Google Scholar] [CrossRef]
- IBM Support. How is the Chi-Square Distance in Proximities and Cluster Defined? Available online: https://www.ibm.com/support/pages/how-chi-square-distance-proximities-and-cluster-defined (accessed on 3 February 2025).
- Cardona, L.; Vargas-Cardona, H.; Navarro González, P.; Cardenas-Peña, D.; Orozco Gutiérrez, Á. Classification of categorical data based on the chi-square dissimilarity and t-sne. Computation 2020, 8, 104. [Google Scholar] [CrossRef]
- Ward, J.H. Hierarchical Grouping to Optimize an Objective Function. J. Am. Stat. Assoc. 1963, 58, 236–244. [Google Scholar] [CrossRef]
- Murtagh, F.; Legendre, P. Ward’s Hierarchical Clustering Method: Clustering Criterion and Agglomerative Algorithm. arXiv 2011, arXiv:1111.6285. Available online: https://arxiv.org/pdf/1111.6285 (accessed on 3 February 2025).
- Anselin, L.; GeoDa, An Introduction to Spatial Data Science, Cluster Analysis (2). Hierarchical Clustering Methods [11/02/2020 (Latest Update)]. Available online: https://geodacenter.github.io/workbook/7bh_clusters_2a/lab7bh.html (accessed on 3 February 2025).
- Strauss, T.; von Maltitz, M.J. Generalising Ward’s Method for Use with Manhattan Distances. PLoS ONE 2017, 12, e0168288. [Google Scholar] [CrossRef] [PubMed]
- Murtagh, F.; Legendre, P. Ward’s Hierarchical Agglomerative Clustering Method: Which Algorithms Implement Ward’s Criterion? J. Classif. 2014, 31, 274–295. [Google Scholar] [CrossRef]
- Székely, G.J.; Rizzo, M.L. Hierarchical Clustering via Joint Between-Within Distances: Extending Ward’s Minimum Variance Method. J. Classif. 2005, 22, 151–183. [Google Scholar] [CrossRef]
- Ran, X.; Xi, Y.; Lu, Y.; Wang, X.; Lu, Z. Comprehensive survey on hierarchical clustering algorithms and the recent developments. Artif. Intell. Rev. 2023, 56, 8219–8264. [Google Scholar] [CrossRef]
- Rambaut, A.; Fig Tree Drawing Tool Version 1.4.2. Edinburgh, Institute of Evolutionary Biology, University of Edinburgh, 2006–2014. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 3 February 2025).
- Microsoft Corporation. Microsoft PowerPoint [Software], Version 16.0; Microsoft Corporation: Redmond, WA, USA, 2021. [Google Scholar]
- Zellner, A. An Introduction to Bayesian Inference in Econometrics; John Wiley and Sons: New York, NY, USA, 1971. [Google Scholar]
- Zellner, A. Bayesian and non-Bayesian analysis of the regression model with multivariate Student-t error terms. J. Am. Stat. Assoc. 1976, 71, 400–405. [Google Scholar] [CrossRef]
- Zellner, A.; Ando, T. Bayesian and non-Bayesian analysis of the seemingly unrelated regression model with Student-t errors, and its application for forecasting. Int. J. Forecast. 2010, 26, 413–434. [Google Scholar] [CrossRef]
- Zellner, A. Bayesian Analysis in Econometrics and Statistics, The Zellner View and Papers; Edward Elgar Pub: Cheltenham, UK, 1997. [Google Scholar]
- Wessel, P.; Luis, J.F.; Uieda, L.; Scharroo, R.; Wobbe, F.; Smith, W.; Tian, D. GMT 6, The Generic Mapping Tools Version 6. Geochem. Geophys. Geosystems 2019, 20, 5556–5564. [Google Scholar] [CrossRef]
- Microsoft Corporation. Microsoft Excel [Software], version 16.0; Microsoft Corporation: Redmond, WA, USA, 2021. [Google Scholar]
- Rivera, D.; Ferrer-Gallego, P.P.; Obón, C.; Alcaraz, F.; Valera, J.; Goncharov, N.P.; Laguna, E.; Kislev, M. Fossil or non-fossil? A best-practice guide for archaeobotanical taxa. Taxon 2024, 73, 425–435. [Google Scholar] [CrossRef]
- Rivera, D.; Ferrer-Gallego, P.P.; Obón, C.; Alcaraz, F.; Laguna, E.; Goncharov, N.P. Exploring the Origins of Hexaploid Wheats: Typification of Archaeological Triticum vulgare var. antiquorum and Description of Modern Triticum sphaerococcum subsp. antiquorum (Poaceae: Triticeae). Taxonomy 2024, 4, 780–794. [Google Scholar] [CrossRef]
- Boardman, S.; Jones, G. Experiments on the effects of charring on cereal plant components. J. Archaeol. Sci. 1990, 17, 1–11. [Google Scholar] [CrossRef]
- Braadbaart, F. Carbonisation and morphological changes in modern dehusked and husked Triticum dicoccum and Triticum aestivum grains. Veg. Hist. Archaeobotany 2008, 17, 155–166. [Google Scholar] [CrossRef]
- Ruas, M.P.; Bouby, L. Carbonization, Preservation and Deformation of Carpological Remains. Palethnologie 2010, 2, 8509. [Google Scholar] [CrossRef]
- Ruas, M.P.; Bouby, L. Carbonisation, conservation et déformation des restes carpologiques. Palethnologie 2010, 2, 8442. [Google Scholar] [CrossRef]
- Antolín, F. Experimental archaeology as a resource for approaching formation processes of seed assemblages. First results and future perspectives. Estrat Crític 2012, 6, 35–48. Available online: https://ddd.uab.cat/pub/estcri/estcri_a2012n6/estcri_a2012n6p35.pdf (accessed on 3 February 2025).
- Milon, J.; Bouchaud, C.; Viot, C.; Lemoine, M.; Cucchi, T. Exploring the carbonization effect on the interspecific identification of cotton (Gossypium spp.) seeds using classical and 2D geometric morphometrics. J. Archaeol. Sci. Rep. 2023, 49, 104007. [Google Scholar] [CrossRef]
- White, C.; Toro, F.; White, J. Rice carbonization and the archaeobotanical record: Experimental results from the Ban Chiang ethnobotanical collection, Thailand. Archaeol. Anthropol. Sci. 2019, 11, 6501–6513. [Google Scholar] [CrossRef]
- Braadbaart, F.; Wright, P.J.; van der Horst, J.; Boon, J.J. A laboratory simulation of the carbonization of sunflower achenes and seeds. J. Anal. Appl. Pyrolysis 2007, 78, 316–327. [Google Scholar] [CrossRef]
- Sallon, S.; Cherif, E.; Chabrillange, N.; Solowey, E.; Gros-Balthazard, M.; Ivorra, S.; Terral, J.F.; Egli, M.; Aberlenc, F. Origins and insights into the historic Judean date palm based on genetic analysis of germinated ancient seeds and morphometric studies. Sci. Adv. 2020, 6, eaax0384. [Google Scholar] [CrossRef]
- Costantini, L.; Audisio, P. Plant and insect remains from the Bronze Age site of Ra’s al-Jinz (RJ-2), Sultanate of Oman. Paléorient 2000, 26, 143–156. [Google Scholar] [CrossRef]
- Gros-Balthazard, M.; Newton, C.; Ivorra, S.; Pierre, M.H.; Pintaud, J.C.; Terral, J.F. The Domestication Syndrome in Phoenix dactylifera Seeds: Toward the Identification of Wild Date Palm Populations. PLoS ONE 2016, 11, e0152394. [Google Scholar] [CrossRef]
- Terral, J.F.; Newton, C.; Ivorra, S.; Gros-Balthazard, M.; de Morais, C.T.; Picq, S.; Tengberg, M.; Pintaud, J.C. Insights into the historical biogeography of the date palm (Phoenix dactylifera L.) using geometric morphometry of modern and ancient seeds. J. Biogeogr. 2012, 39, 929–941. [Google Scholar] [CrossRef]
- Goor, A. The History of the Date through the Ages in the Holy Land. Econ. Bot. 1967, 21, 320–340. [Google Scholar] [CrossRef]
- Carpenter, J.B.; Elmer, H.S. Pest and Diseases of the Date Palm (Agriculture Handbook 527); U.S. Department of Agriculture, Science and Education Administration: Hyattsville, MD, USA, 1978; pp. 1–42. [Google Scholar]
- Rivera, D.; Johnson, D.; Delgadillo, J.; Carrillo, M.H.; Obón, C.; Krueger, R.; Alcaraz, F.; Ríos, S.; Carreño, E. Historical evidence of the Spanish introduction of date palm (Phoenix dactylifera L.; Arecaceae) into the Americas. Genet. Resour. Crop Evol. 2013, 60, 1433–1452. [Google Scholar] [CrossRef]
- Rivera, D.; Obón, C.; Alcaraz, F.; Egea, T.; Carreño, E.; Laguna, E.; Santos, A.; Wildpret, W. A review of the nomenclature and typification of the Canary Islands endemic palm, Phoenix canariensis (Arecaceae). Taxon 2013, 62, 1275–1282. [Google Scholar] [CrossRef]
- Obón, C.; Sosa, P.; Alcaraz, F.; Saro, I.; Martínez-Rico, M.; Laguna, E.; Ferrer-Gallego, P.P.; Johnson, D.; Pérez de Paz, P.; Rivera, D. Phoenix × arehuquensis nov. hybr. (Arecaceae): The hybrid of P. canariensis × P. reclinata in garden and forest. S. Afr. J. Bot. 2024, 168, 124–129. [Google Scholar] [CrossRef]
- Rivera, D.; Obón, C.; Alcaraz, F.; Egea, T.; Martínez-Rico, M.; Carreño, E.; Laguna, E.; Johnson, D.; Saro, I.; Sosa, P.; et al. Nomenclature and typification of Phoenix senegalensis and Fulchironia senegalensis (Arecaceae). Taxon 2019, 68, 370–378. [Google Scholar] [CrossRef]
- Benítez, N.; Wildpret, H. Catálogo de las Plantas que contiene el Jardín de Aclimatación de la Orotava, en Tenerife; Jardín de Aclimatación: La Orotava, Spain, 1879. [Google Scholar]
- Kislev, M.E. Triticum parvicoccum sp. nov., the oldest naked wheat. Isr. J. Plant Sci. 1980, 28, 95–107. [Google Scholar]
- Loredo, J. From Laplace to Supernova SN 1987A, Bayesian Inference in Astrophysics. In Maximum Entropy and Bayesian Methods; Fougere, P.F., Ed.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1990; pp. 81–142. [Google Scholar]
- Kolmogorov, A.N. Foundations of The Theory of Probability, Second English Edition; Translation Edited by Nathan Morrison with an Added Bibliography by A.T. Bharucha-Reid; University of Oregon Chelsea Publishing Company: New York, NY, USA, 1956; p. 5, (Remark 2). [Google Scholar]





| Taxa | Seeds Analyzed | Prior Probability p(Hi|I) |
|---|---|---|
| P. canariensis H.Wildpret var. canariensis | 2808 | 0.301 |
| P. canariensis var. porphyrococca Vasc. & Franco | 135 | 0.014 |
| P. canariensis Wildpret’s Large Date Group | 928 | 0.099 |
| P. × arehuquensis (P. canariensis × P. reclinata hybrid) (*) | 57 | 0.006 |
| P. canariensis Subtotal | 3928 | 0.421 |
| P. dactylifera Eastern group | 867 | 0.093 |
| P. dactylifera hybrids | 233 | 0.025 |
| P. dactylifera Western group (**) | 2749 | 0.295 |
| P. dactylifera Subtotal | 3849 | 0.413 |
| P. ‘Palmer’ | 59 | 0.006 |
| P. abyssinica Drude. | 15 | 0.002 |
| P. acaulis Roxb. | 61 | 0.007 |
| P. andamanensis auct. | 32 | 0.003 |
| P. arabica Burret | 44 | 0.005 |
| P. caespitosa Chiov. | 15 | 0.002 |
| P. farinifera Roxb. | 47 | 0.005 |
| P. loureiroi Kunth | 294 | 0.032 |
| P. loureiroi var. hanceana | 78 | 0.008 |
| P. paludosa Roxb. | 62 | 0.007 |
| P. pusilla Gaertn. | 5 | 0.001 |
| P. reclinata Jacq. | 161 | 0.017 |
| P. roebelenii O’Brien | 156 | 0.017 |
| P. rupicola T.Anderson | 80 | 0.009 |
| P. rupicola ʽMedipalmʼ | 8 | 0.001 |
| P. sylvestris (L.)Roxb. | 178 | 0.019 |
| P. theophrasti Greuter | 264 | 0.028 |
| P. other Subtotal | 1559 | 0.166 |
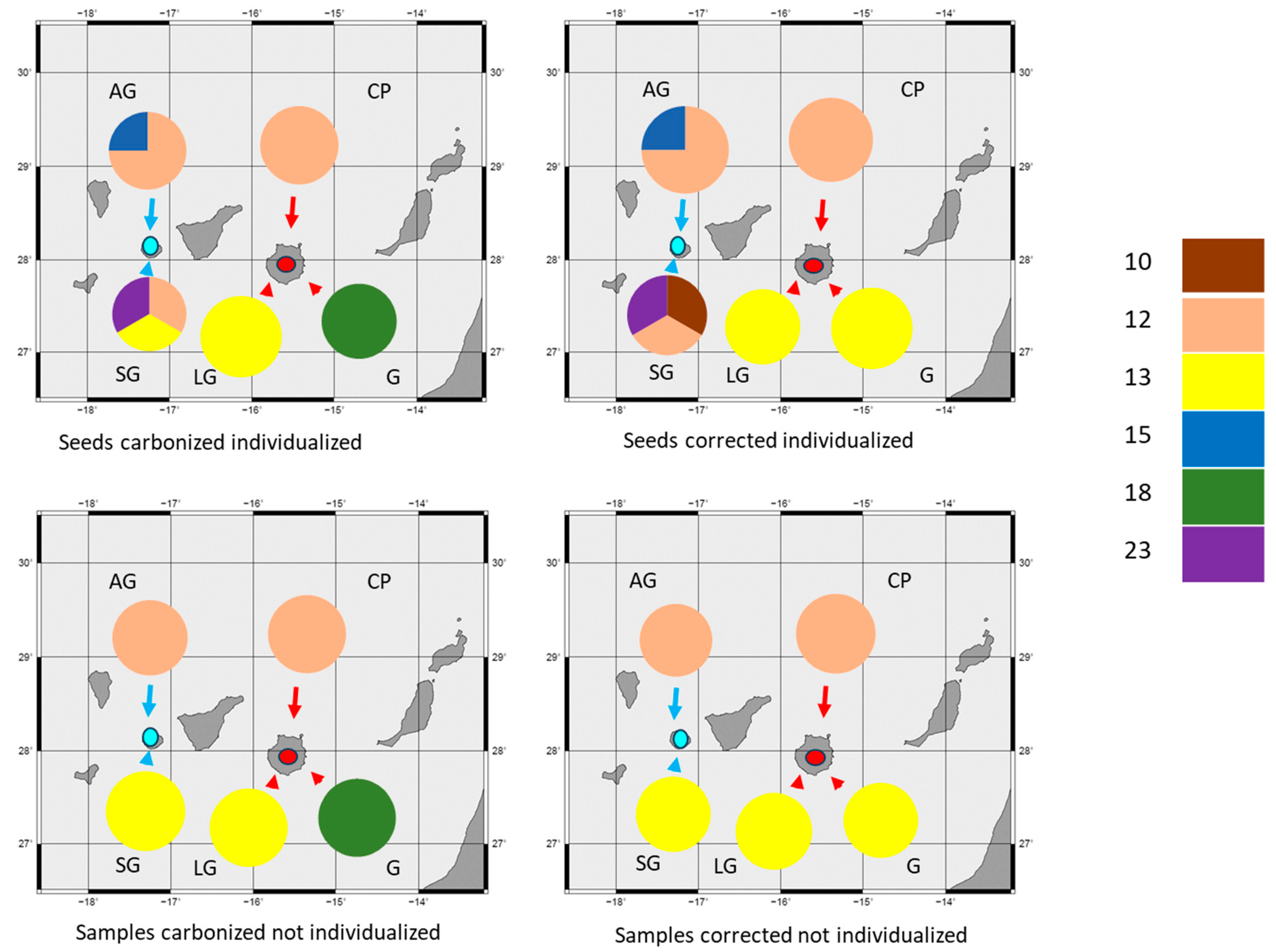
| Sites | Island | Seed Numbers | 10 | 12 | 13 | 15 | 23 |
|---|---|---|---|---|---|---|---|
| Alto del Garajonay | La Gomera | 4 | - | 0.875 | - | 0.125 | - |
| Sobrado de los Gomeros | La Gomera | 3 | 0.167 | 0.167 | 0.5 | - | 0.167 |
| Cueva Pintada | Gran Canaria | 2 | - | 1 | - | - | - |
| Lomo de los Gatos | Gran Canaria | 2 | - | - | 1 | - | - |
| Guayedra | Gran Canaria | 1 | - | - | 1 | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rivera, D.; Martínez-Rico, M.; Morales, J.; Alcaraz, F.; Valera, J.; Johnson, D.; Sosa, P.A.; Abellán, J.; Palazón, J.A.; Rivera-Obón, D.J.; et al. Bayesian Morphometric Analysis for Archaeological Seed Identification: Phoenix (Arecaceae) Palms from the Canary Islands (Spain). Seeds 2025, 4, 19. https://doi.org/10.3390/seeds4020019
Rivera D, Martínez-Rico M, Morales J, Alcaraz F, Valera J, Johnson D, Sosa PA, Abellán J, Palazón JA, Rivera-Obón DJ, et al. Bayesian Morphometric Analysis for Archaeological Seed Identification: Phoenix (Arecaceae) Palms from the Canary Islands (Spain). Seeds. 2025; 4(2):19. https://doi.org/10.3390/seeds4020019
Chicago/Turabian StyleRivera, Diego, Manuel Martínez-Rico, Jacob Morales, Francisco Alcaraz, Javier Valera, Dennis Johnson, Pedro A. Sosa, Javier Abellán, Jose Antonio Palazón, Diego José Rivera-Obón, and et al. 2025. "Bayesian Morphometric Analysis for Archaeological Seed Identification: Phoenix (Arecaceae) Palms from the Canary Islands (Spain)" Seeds 4, no. 2: 19. https://doi.org/10.3390/seeds4020019
APA StyleRivera, D., Martínez-Rico, M., Morales, J., Alcaraz, F., Valera, J., Johnson, D., Sosa, P. A., Abellán, J., Palazón, J. A., Rivera-Obón, D. J., Laguna, E., & Obón, C. (2025). Bayesian Morphometric Analysis for Archaeological Seed Identification: Phoenix (Arecaceae) Palms from the Canary Islands (Spain). Seeds, 4(2), 19. https://doi.org/10.3390/seeds4020019









