Abstract
Artificial intelligence is more present than ever in virtually all sectors of society. This is in large part due to the development of increasingly powerful deep learning models capable of tackling classification problems that were previously untreatable. As a result, there has been a proliferation of scientific articles applying deep learning to a plethora of different problems. The interest in deep learning in agriculture has been continuously growing since the inception of this type of technique in the early 2010s. Soybeans, being one of the most important agricultural commodities, has frequently been the target of efforts in this regard. In this context, it can be challenging to keep track of a constantly evolving state of the art. This review characterizes the current state of the art of deep learning applied to soybean crops, detailing the main advancements achieved so far and, more importantly, providing an in-depth analysis of the main challenges and research gaps that still remain. The ultimate goal is to facilitate the leap from academic research to technologies that actually work under the difficult conditions found in the the field.
1. Introduction
Soybean (Glycine max L.) has become the most important oilseed crop and one of the five most important food crops worldwide [1,2]. Its high protein content makes soybean a prime source of feed for livestock, and soybean oil is used for both human consumption and industrial applications [1]. While the demand for soybeans continues to grows worldwide [3], environmental pressures due to climate change are becoming more widespread and extreme [4]. In order for the soybean yield to keep up with the demand, new solutions to current production limitations are needed. Although extensive breeding efforts have led to the development of varieties quite robust to different conditions, soybean crops are still vulnerable to many factors. Stresses caused by diseases, pests, unfavorable weather, nutrition imbalances, and others, are responsible for losses that can easily surpass 20% of the total world production [2]. Although completely eliminating losses is likely unfeasible, closely monitoring each one of the relevant variables can greatly mitigate the problem [5]. However, continuous monitoring may require too large a workforce, unless some type of automation is employed. In this context, artificial intelligence techniques emerge as powerful aiding tools for farm monitoring and management [6].
One of the possible definitions for artificial intelligence (AI) states that this is “a computational data-driven approach capable of performing tasks that normally require human intelligence to independently detect, track, or classify objects” [7]. Techniques fitting this definition have existed for many decades, including expert systems, neural networks and other types of machine learning algorithms. With the inception of deep learning models in the first half of the 2010s, the application of artificial intelligence has grown steeply both in number and scope. This is certainly true in the case of agriculture, for which applications like plant disease recognition [5], yield estimation [8], plant nutrition status [9], and biomass estimation [10], among many others, have experienced a surge in the number of articles employing artificial intelligence. Among AI techniques, deep learning has been particularly successful and well adapted to difficult classification problems. One of the reasons for this success is that with deep learning, the explicit extraction of features from the data is no longer required [11,12], making the classification process more straightforward, less biased and more robust to different types of conditions [13].
While the leap between academic research and practical solutions has been successfully completed in some cases (e.g., weed detection and control), in most cases, real-world conditions and variability are too challenging for techniques and models that are, more often than not, trained on data that represent only a small fraction of reality [14]. The most direct way to address this problem is to expand the datasets used to train the models. This is by no means a trivial task, especially considering that the variability involved in some classification problems may require a number of images that can reach the order of millions. Increasing the practice of data sharing and exploring citizen science concepts can help reduce the problem, but in many situations, all-encompassing datasets may be unfeasible [14]. If supervised learning is adopted, there is the additional challenge of data annotation, a process that is often expensive, time-consuming and error-prone [15]. New annotation strategies capable of speeding up the process are already being studied [16], but these were still incipient when this article was written.
AI models failing when presented with new data with distinct statistical distribution, a phenomenon often called “covariate shift” [17], is arguably the most important hurdle for the more effective use of artificial intelligence-based technologies in agriculture, but other factors are almost always present. Each application has its own challenges, so systemically understanding how data and technical issues affect the performance of the models is fundamental for the construction of suitable solutions. Many of these challenges have already been experienced in previous studies and reported in the literature, so a proper understanding of the current state of the art is critical to the novelty of new research and to avoid repeating mistakes. Research on deep learning applied to crop management has been extensive across different types of crops, so including all research would make the article somewhat redundant and impractically long. Soybean, being a major agricultural commodity, has received considerable attention from researchers, to the point that it encapsulates most of the approaches adopted across different crops. This was the main motivation for narrowing the scope of the review to research related to this crop only.
The use of deep learning for soybean monitoring started to gain momentum after 2015. Early research was mostly dedicated to disease and pest detection, but soon, applications like phenotyping, seed counting, cultivar identification and yield prediction began being explored. Since the beginning, studies have been focusing on the investigation of different deep learning models and architectures in the context of each different application and domain. Although this type of research has yielded relevant results, there are not many technologies being effectively used in practice. One important exception is weed detection, as machinery from different manufacturers already have the ability to not only detect the weeds but also actuate to eliminate the problem. For most applications, there are still significant challenges that require more suitable solutions. New approaches emphasizing model interpretability and fine tuning are beginning to be explored [18,19,20], but the research gap is still substantial. In this context, the main objective of this review is to properly characterize the current state of the art of deep learning applied to soybean monitoring and management. Special emphasis is given to the main challenges and research gaps reported in the articles, as well to issues that are not usually addressed by the authors but are relevant for the effectiveness of the proposed techniques nonetheless. This article differs from other reviews because it does not focus on findings that are specific to each application, concentrating instead on general trends and challenges that affect most efforts to apply deep learning to solve problems related to the soybean.
The article is organized as follows. Section 2 presents the definitions of some relevant terms used in this review, as well as some acronyms used throughout the text. Section 3 describes the current state of the art of the application of deep learning in the context of soybean crops. Section 4 provides an in-depth discussion about the main challenges and research gaps that still require additional research effort. Finally, Section 5 offers some final remarks.
2. Definitions and Acronyms
Some terms deemed to be of particular importance in the context of this work are defined in this section. Most of the definitions are adapted from [7,21]. A list of acronyms used in this article with the respective meanings is given in Table 1.

Table 1.
Acronyms used in this review.
Artificial intelligence: a computational data-driven approach capable of performing tasks that normally require human intelligence to independently detect, track, or classify objects.
Data annotation: the process of adding metadata to a dataset such as indicating the objects of interest in an image. This is typically performed manually by human specialists.
Deep learning: a special case of machine learning that utilizes artificial neural networks with many layers of processing to implicitly extract features from the data and recognize patterns of interest. Deep learning is appropriate for large datasets with complex features and where there are unknown relationships within the data.
Domain adaptation: techniques that have the objective of adapting the knowledge learned in a source domain to apply it to a different but related target domain.
Image augmentation: process of applying different image processing techniques to alter existing images in order to create more data for training the model.
Machine learning: application of artificial intelligence (AI) algorithms that underpin the ability to learn characteristics of the classes of interest via extraction of features from a dataset. Once the model is developed, it can be used to predict the desired output on test data or unknown images.
Model: a representation of what a machine learning program has learned from the data.
Overfitting: when a model closely predicts the training data but fails to fit the testing data.
Proximal images: images captured in close proximity to the objects of interest.
Segmentation: the process of partitioning a digital image containing the objects of interest into multiple segments of similarity or classes either automatically or manually. In the latter case, the human-powered task is also called image annotation in the context of training AI algorithms.
Semi-supervised learning: a combination of supervised and unsupervised learning in which a small portion of the data is used for a first supervised training, and the remainder of the process is carried out with unlabeled data.
Supervised learning: a machine learning model, based on a known labeled training dataset that is able to predict a class label (classification) or numeric value (regression) for new unknown data.
Transfer learning: machine learning technique that transfers knowledge learned from one domain to other. Weight fine-tuning and domain adaptation are arguably the most employed transfer learning techniques.
Unsupervised learning: machine learning that finds patterns in unlabeled data.
3. Literature Review
The search for articles was carried out in May 2023 on Scopus and Google Scholar, as both encompass virtually all relevant bibliographic databases. The terms used in the search were “deep learning” and “soybean”. The terms were deliberately kept general in order to reduce the likelihood of relevant work being missed. The downside of this strategy is that the filtering process was labor-intensive and time-consuming. Only articles in which soybeans were featured prominently were kept, which means that references in which soybean was only one among several crops were removed. Although high-quality conference papers do exist, in most cases, the review process is either absent or lacks rigor, so this type of publication was also removed. After that, the list of references of the selected literature were inspected for articles that were missed in the original search, leading to the final number of 61 articles.
This section is divided into four subsections according to the type of data used as input for the deep learning models: proximal images, UAV images, satellite data (images and vegetation indices) and other types of data (weather, soil, yield, etc.). Figure 1 summarizes the types of data employed according to the acquisition distance.
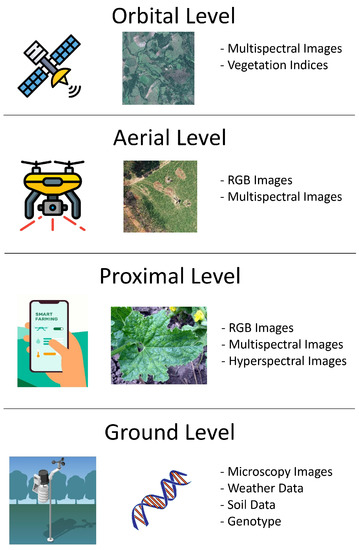
Figure 1.
Types of data used in the articles considered in this review according to the acquisition distance.
3.1. Proximal Images as Main Input Data
In the context of this review, investigations employing proximal images are by far the most common (34 out of 61 articles, Table 2). Applications related to plant health (weed detection, disease recognition, and pest detection) dominate, but other applications, like seed counting and cultivar identification, are also common. Most approaches use common RGB images, but the use of hyperspectral images is growing fast due to their ability to offer a detailed spectral profiling for any object of interest. It can also be observed that while classification accuracies are almost always very high, accurate object detection (localization) is harder to achieve. It is important to emphasize, however, that the high classification accuracies are almost always obtained on data with rather limited variability, so the reported results are not necessarily valid for real practical conditions (this issue is addressed in more depth in the next section).

Table 2.
List of articles employing proximal images and deep learning for soybean monitoring and management.
A problem that affects virtually all research using agricultural images but is rarely mentioned is that the datasets used in the experiments normally only cover a small part of the variability that can be found in practice [35]. This is mostly due to the impracticality of capturing the whole range of conditions and variations found under real uncontrolled conditions [5]. Image augmentation can be a partial solution for the lack of variability, as long as it is applied correctly following the guidelines discussed in Section 4. This type of technique has been used, for example, to increase the size of challenging classes, with encouraging results [1]. It has also been applied in the context of unsupervised learning, also yielding superior results [15]. In any case, as briefly discussed in the introduction, the lack of training data variability is arguably the most important challenge for making deep learning models more broadly applicable in the context of agriculture.
Because of the challenges posed by the variety of characteristics found in the agricultural environment, in many cases, the research is conducted under relatively controlled conditions, which can differ significantly from real field conditions [11,44]. Situations that are common under daily operations, like the presence of stains, debris and other spurious objects, normally are not present in experimental data [30], and the complex backgrounds found in real fields can pose significant challenges to most deep learning architectures [46]. Occlusions can also make it difficult to detect and classify the objects of interest, and suitable solutions may require unconventional sensor configurations or statistical corrections to mitigate this problem [46]. As difficult as they may be, the exclusion of these less than ideal but relatively common conditions can cause the model to become highly biased, making it difficult to anticipate how the model will perform with new data [1]. Interestingly, some authors tried to increase data variety by training the model with both images of attached leaves and severed leaves placed on an alternative background as a means to make the model more robust, but the opposite occurred [1]. The authors speculated that the background may contain information that actually helps with the classification, but more research is needed to reach a more reliable conclusion.
Utilizing fully synthetic images is another possible way to increase datasets without the need for collecting new data or manual annotation [39,42]. Although this type of image may not be an exact representation of real samples, there are some classification problems that have well-defined characteristics, which can be properly captured by the synthetic samples, and generative approaches using architectures like GANs can produce representative synthetic images [41]. Hybrid datasets containing both real and synthetic images have been successfully employed for seed phenotyping purposes [39].
Another difficulty that is often present in classification problems using proximal images is class imbalance. Usually, some classes will naturally occur more often, while others may be much rarer. As a result, the former will tend to have more samples associated than the latter. If the difference in the number of samples across the classes is too severe, the model will likely be biased toward the larger classes and will perform poorly for the underrepresented ones [13]. The most straightforward way to reduce the problem is by either increasing the number of samples of the smaller classes by means of image augmentation, or by removing samples from the larger classes. There are also neural network architectures (e.g., RetinaNet) that are less susceptible to this type of problem [31]. It is worth noting that class imbalance does not seem to always have a negative effect. In the context of unsupervised learning, some results with unbalanced datasets were similar or even better than those obtained with balanced classes [15].
Depending on the characteristics of the problem to be solved, a single data source may not provide enough information for unambiguous answers. For this reason, the field of research called data fusion, which tries to find the most effective ways to combine different types of data, has been growing considerably in the last few years. One of the most common data fusion types is the combination of different types of images [49]. This type of approach has been particularly useful in the context of remote sensing, and especially satellite images [50], but it has also been adopted for proximal images. In Flores et al. [11], the authors combined the information contained in RGB and CIR images for weed detection, with fused images being generated using wavelet transforms.
3.2. UAV Images as Main Input Data
Agricultural monitoring is among the most natural applications for UAVs, as fields tend to be vast, and the probability of accidents with injuries is low due to those areas usually being sparsely populated. As in the case of proximal images, RGB is predominant, but other types of imaging sensors are starting to be employed as technology evolves and costs fall. Weed detection is by far the most common application, with other uses like yield prediction and disease recognition also being explored (Table 3).

Table 3.
List of articles employing UAV images and deep learning for soybean monitoring and management.
Complex backgrounds are often a challenge when images are captured in the field. This is particularly true in the case of UAV-captured images because not only are the conditions not controlled but there is also little room for adjustments of the angle and position of capture. This is particularly problematic in the detection of small objects, although certain models for object localization (e.g., YOLO v5) are particularly well-suited for the detection of small targets [22].
Although there are some precautions that can be taken in order to produce images of good quality, including the careful selection of the most appropriate time of the day and weather for carrying out the UAV flight missions, it is often impossible to prevent the sub-optimal conditions that produce low-quality images. Lighting effects like shadows and specular reflections can be particularly damaging [62]. In cases like these, even highly trained experts can fail to correctly analyze the image [52]. Images with these characteristics should not be used for training, but the model will inevitably have to deal with this type of situation if applied in practice. In order to avoid problems, it is good practice to consider only classifications that reach a certain confidence threshold (most models are capable of associating a probability to each classification) [52].
The issues of data imbalance and lack of data variability were also identified in the case of UAV images [52]. Similarly to what occurs with proximal images, the presence of one or both problems tends to lead to biased models that lack robustness and generalization capability, albeit some architectures being capable of dealing with these problems more effectively than others [55]. Although addressing these problems is not always straightforward, this is a requirement for the development of technologies that work under real conditions. One relatively straightforward way to increase variability is to gather data through multiple different years [59]. Class imbalance, on the other hand, can be counteracted by a number of techniques that include data subsampling, data augmentation and class weighting [62].
Most studies adopt fully supervised learning [60], as the process is more controllable and the results tend to be better. However, as mentioned in the introduction, the annotation process is expensive and time-consuming, so unsupervised approaches can become very attractive if the considerable challenges can be overcome. One study dedicated to weed detection obtained some encouraging results and led to interesting remarks, like the fact that using a number of data clusters much higher than the number of classes actually improves classification, and that cluster granularity can play an important role in improving models arising from unsupervised learning [15].
Semi-supervised learning is often adopted as a relatively straightforward way to combine the advantages of both supervised and unsupervised learning. In order to reduce the need for manual labeling, the study described in Menezes et al. [54] introduced a pseudo-labeling process that automatically annotates a large dataset based on a small set of pre-labeled samples. Although this automatic labeling process is not perfect, the additional samples used for training led to significantly higher accuracies.
Data fusion has also been applied in the context of UAV images. In Maimaitijiang et al. [53], thermal information was fused with spectral and visual features for yield estimation across different soybean phenotypes. The combination of all three types of data yielded results that were superior to those obtained with any single sensor or pair of sensors. In addition, the deep learning model greatly outperformed other machine learning algorithms in terms of adaptability to different soybean varieties and to spatial variations.
One peculiarity of UAV images is that there is a trade-off between flight height and spatial resolution [60]. In many cases, an altitude believed to provide enough resolution for the problem at hand is selected, and no more thought is given to the matter. However, with low flights, one of the most attractive advantages of UAVs, which is the fast sweep of large areas, is reduced. The ideal approach is to perform several flights at different altitudes and investigate the influence of this factor on the accuracy. A quicker and cheaper alternative is to carry out a low-altitude mission and then simulate different altitudes by undersampling the images [56]. This is not ideal because image distortions may vary depending on the altitude, and the interference between neighboring pixels is partially lost by simply reducing the number of pixels in the image, but the results should be reliable enough to determine the flight height that provides the best balance between object resolvability and area coverage [62]. It should also be considered that multispectral and hyperspectral sensors can provide valuable information about the spectral responses of the objects of interest, but they usually have lower spatial resolutions (especially the latter) and may require considerably lower flights [61].
There are other consequences attached to flying low other than image resolution. If rotary-wing UAVs are used, the airflow through the blades can disturb plants and soil, affecting the quality of the information present in the images [65]. In addition, the likelihood of collisions increase, and time for recovery is smaller. Higher flight altitudes can be possible with the use of higher-resolution cameras [35], but these tend to be considerably more expensive.
3.3. Satellite Images as Main Input Data
Satellite images are very convenient for agricultural monitoring, as they cover large areas and many satellites have relatively short revisit times. However, despite the notable evolution of the spatial resolutions offered by images taken from orbit, these are still not enough for many applications. The two main problems associated with insufficient spatial resolutions are the instances in which objects of interest cannot be resolved, and the presence of mixed pixels containing information pertaining to multiple classes [66]. Thus, it is not a surprise that applications which can be enabled with relatively coarse spatial resolutions prevail, like yield prediction and crop mapping (Table 4). All articles selected in this review employ either multispectral images or derived features (especially vegetation indices).

Table 4.
List of articles employing other types of data and deep learning for soybean monitoring and management.
One advantage of satellite images that is beginning to be explored more effectively is the possibility to extract temporal information [71]. Although in principle, it is possible to build time series using proximal and UAV images, this is much easier in the case of satellites, as their revisits are predefined and time series are naturally generated, and also because of the difficulty in guaranteeing consistency of reflectance values and spatial alignment over time when using UAV images [60]. The incorporation of temporal information can provide valuable cues that lead to better models [67]. The authors remarked, however, that the collection frequency of satellite imagery may not correspond to the development of diseases, which can lead to delayed responses and significant losses. Also, too-short time series can lead to overfitting [71].
Extreme weather events are relatively rare but do have a big impact on the satellite data time series. Including data captured during those rare events is very important to teach the model to recognize those types of situations, especially considering that predictions become more challenging when conditions are not typical. With climate change, extreme events tend to become more common across the globe, which emphasizes even more the need for the training data to be as complete and comprehensive as possible [50].
Methods heavily reliant on temporal information can suffer greatly when the time series is broken by the removal of spurious pixels, normally due to the presence of clouds and shadows [71]. Data imputation by interpolation algorithms can mitigate the problem, but these are not perfect, and the estimated pixels can cause misclassifications. Once again, building models carefully designed with robustness in mind is the best option to deal with imperfect data [71].
Severe class imbalance was also observed for satellite images. In Bi et al. [67], the authors observed that there were many more instances of healthy areas than diseased ones. Instead of increasing the number of samples of the smaller class by image augmentation or decreasing the number of samples of the larger class, the authors opted for an approach in which the costs for the misclassification of the smaller class were increased, thus producing a more balanced model. The authors observed that this approach leads to a trade-off between precision and recall: the larger the weights given to the smaller class, the fewer associated false negatives there will be, but the number of false positives inevitably increases. Ultimately, the ideal class weight will depend on the application and the type of error deemed more damaging.
Training deep neural networks from scratch can be very time-consuming, especially if the amount of training data is high. For this reason, it is common practice to freeze the weights in the backbone of pretrained networks and update only the weights in the last few layers of the architecture, a procedure commonly called transfer learning. There are several standard CNN models that were pre-trained using the 1000-class ImageNet dataset [73], including AlexNet, GoogleNet, Xception and ResNet, among others. Although using these general pre-trained models usually works well, transfer learning can be applied in a more targeted way. Using satellite images for yield prediction, the authors in Khaki et al. [68] showed that a model trained from scratch for a given crop can be successfully transferred to other crops without significant accuracy loss. The choice between training from scratch and transfer learning will ultimately depend on the time available for the experiments and on the perceived importance of having a model fully tuned to the training data.
Historically, spectral indices extracted from satellite images have been used more frequently than the images themselves. Although those indices have been shown to produce good results for a variety of applications, they can limit the transferability of machine learning models over space and time and usually require careful selection of the features used to feed the models [66]. With the inherent ability of deep learning architectures to implicitly extract appropriate features from the data, some studies have adopted a more direct approach by using either raw [50,66] or slightly preprocessed images [20,68,71]. This allows a rich variety of nonlinear, hierarchical and complex features to be learned from the data itself [66]. Despite the shortcomings of using spectral indices, it is worth considering that the combination of carefully selected VIs with raw images can lead to superior results [66].
While obtaining satellite images for classification purposes is relatively easy, the same cannot be said about the reference ground data. Some of the challenges involved in obtaining ground data were discussed in Xu et al. [71]: field surveys and censuses are expensive, labor-intensive, and time-consuming; visual comparison with high-resolution RGB photos from GPS records and Google Earth require considerable effort and are not appropriate for large-scale monitoring; and government-sponsored national-scale surveys are available in only a few countries. Transferring models trained on regions with rich ground reference data to target regions seem to be one of the most viable options [71], but generating models with this level of generalization capability is far from trivial as discussed in Section 4.
3.4. Other Types of Data as Main Input for the Models
Only five articles selected in this review used data other than proximal, UAV or satellite images (Table 5). From those, one employed microscopy images, two used genetic data, and two adopted a variety of data sources. Phenotyping applications, which include yield prediction, are the most common in this case.

Table 5.
List of articles employing satellite images and deep learning for soybean monitoring and management.
The challenges identified in the case of microscopy images are often similar to those found when other types of images are used. The variability of the data, especially regarding the visual appearance of the objects of interest, is a problem that seems to always be present when it comes to agricultural data [74]. Images that are cluttered with both the objects of interest and spurious debris also pose significant challenges.
Digital images usually have a tridimensional data structure, with two spatial dimensions and one spectral dimension with length varying from 3 (RGB) to several hundreds (hyperspectral). Other types of data are often unidimensional, requiring networks with structures that are slightly different from those employed with images. In Khaki et al. [75], a hybrid CNN-RNN model was adopted to extract information from variables related to soil, weather, yield and crop management. This approach, despite being rather sensitive to some variables, showed good generalization capabilities.
The performance of artificial intelligence models depend not only on the quality of the data collected, but also on the quality of the annotations and reference data used to teach the model. This is especially true when the reference values are quantitative, and data like yield and weather variables can contain inconsistencies from a variety of causes that include filling errors and sensor malfunction. Prediction models are particularly susceptible to those inconsistencies, so a solution that is frequently applied is to filter the reference data in order to remove outliers. However, as argued in Li et al. [69], if the filtering criteria are not carefully designed, atypical data that are actually caused by uncommon events may also be removed, which ultimately may lead to models that lack robustness to abnormal conditions. Thus, keeping all the data tends to be the best strategy in many cases [69].
Studies dealing with genotypes have some specific characteristics that are not usually found for other types of data [18,76]. Genotype matrices usually have missing values, so it is common practice to fill those values using imputation methods that estimate those values from a template population. However, some studies employing deep learning have reported better results with no imputation, probably because imputation fills most missing values with reference alleles, thus deflating the effects of different genotypes [76]. Indeed, deep learning models show remarkable robustness to noisy inputs [18], so imputing data that may not represent well the missing values will often have an adverse effect.
4. Discussion
High data variability and the associated phenomenon of covariate shift are omnipresent in agricultural applications. Many articles considered in this review do not even acknowledge this issue, but many recognize that the data used in the respective experiments do not cover the whole variability expected for that application, and that more data collection and research efforts are needed in order to increase the robustness of the models [35,62]. With the number of publicly shared datasets growing, the variability of available data also tends to grow [60]. However, it is unlikely that more data collection alone will solve the problem, as the number and depth of factors that introduce variability are usually very high. In this context, domain adaptation techniques arise as a suitable way to make models more adaptable to different conditions. Domain adaptation aims at adapting a given classifier to data with statistical distributions that differ from the data used for training [77]. This type of technique is frequently applied in the context of satellite images, but its use is quickly being extended to proximal and UAV images as well.
Data augmentation can be valuable to artificially increase the number and variety of samples and produce more robust models. However, this technique is not always applied correctly, and one particularly egregious mistake is to apply augmentation prior to division into training and test sets. The problem with this procedure is that many augmentation operations produce new images that are only slightly modified versions of the original image. Thus, there will exist several almost identical copies of the same image. This is not a problem if augmentation is applied only to the training set but causes severely biased and unrealistic results if applied prior to the set division because very similar images will be present in both the training and test sets, which in practice is equivalent to using the exact same images for training and testing. Unfortunately this is a problem that has become widespread [14], and the situation is not better in the literature considered in this review. Both authors and reviewers should be aware of this problem to avoid the publication of reports containing invalid results. It is also worth mentioning that while it is often claimed that image augmentation decreases overfitting, the opposite can easily happen because the new images generated are still highly correlated to the original ones [60]. This has led some authors to argue that employing the dropout technique during training is often a more suitable option [60]. In any case, the only way to properly evaluate the effectiveness of augmentation and alternative techniques is by carrying out experiments with independent datasets, which is rarely done.
The division into training, test and validation sets is usually performed randomly. Although this is indeed the best option in most cases, there is always a chance that the statistical distributions in one or more of the sets is biased, causing the results to become unreliable. One way to avoid this problem is the adoption of cross validation, in which experiments are repeated a certain number of times (folds) using different data partitions, that is, the sets are always different in each repetition. Unfortunately, only a small percentage of the studies adopt this strategy, probably because of the added training effort. The absence of cross validation, together with the improper use of image augmentation, causes many of the results reported in the literature to be unreliable and, in some cases, misleading.
Data annotation is a demanding process no matter the application, not only because of the time and cost involved but also due to possible inconsistencies arising from its inherent subjectivity [60,62] and from measurement inaccuracies [39]. This is a problem because the generated labels are used as reference for both training and evaluation of the models, and if those are not consistent, both the models and the associated results could be biased [62]. These challenges are even more pronounced in the case of pixel-wise classification. This approach, which requires that each pixel in an image be correctly labeled, is often adopted for applications such as disease severity estimation and crop mapping. Considering that deep learning models usually require large amounts of data for proper training and that there are very few available datasets with this kind of annotation [25], this is a hurdle difficult to overcome. Semi-automatic labeling can be helpful in cases like this, but further research is needed for more suitable solutions.
Despite the annotation challenges posed by pixel-wise classification, if individual pixels indeed carry enough information to enable such a fine-grained classification, a large amount of training samples can be obtained even if only a few images are available. In most cases, this does not solve the lack of variability problem, as just a few images will almost certainly not be representative enough, even under moderate variability. However, having a large number of samples coming from just a few images can be advantageous in cases for which the variability is rather low, like, for example, in the analysis of seeds using flatbed hyperspectral imaging systems. Hyperspectral images are particularly suitable for pixel-wise classification because each pixel carries a wealth of information about the spectral characteristics of that specific point in space [47].
The number of parameters used in deep learning models can vary greatly, a fact that has motivated many authors to investigate the computational load associated to different models [1,13,58]. While those differences can indeed have a great impact on the time spent training the models [69], it is worth pointing out that, once trained, even the larger models can run at reasonable times in most devices [21], although memory usage can become a problem under more restrictive conditions [55]. Thus, unless computational resources are scarce and real-time operation is required, the size of the model tends to not have a large impact on its usability. When real-time operation is required, architectures especially designed to be lightweight are usually employed, especially if the model is coupled with some kind of actuator [32].
In classification problems, experiments usually try to maximize accuracy, which often results in relatively similar values for precision and recall, that is, the number of false negatives and false positives tend to balance out. However, for some applications, one type of error may be much more damaging than the other [62]. The case of weed detection is a good example. If other objects are recognized as weeds (false positives), there will be a waste of herbicide but the crop will still be protected [32]. On the other hand, if the model fails to detect the weeds, they will remain in the field and can potentially spread, causing losses. It is worth pointing out that most studies do address this point and offer some useful comments on the subject. This also stresses the importance of employing the right evaluation metrics in order to fully characterize the methods being proposed.
One aspect of machine learning models that has been attracting considerable attention is the issue of interpretability [20]. In many instances, the relationship between input data and the answer provided by the model is not clear, making it difficult to gain scientific insights and increasing the risk of wrong conclusions. Some studies have been conducting experiments specifically designed to investigate and increase interpretability [18,19,20], but this is still an open problem that will require considerable research effort to be solved.
It is common practice to reduce the size of the images inputted to the models, either because a pretrained model with standard input size is being used, or due to computer memory and processing constraints. This reduction inevitably leads to loss of information, which in turn may result in lower accuracy [56]. In order to preserve all the information originally captured, properly selecting patches of appropriate size and processing them separately can yield better results [63], even if the object of interest is broken up in the process [78]. Although patch selection is not always a trivial task, there are a few simple techniques that can be used to select potential candidates with relative ease [78]. Some of the studies considered in this review adopted this strategy [29,56,74].
When training from scratch is adopted, all weights of the network need to be initialized and updated as training progresses. Frequently, those weights are initialized randomly, but under certain conditions, unfavorable initial weights may lead to excessive training times and, more importantly, the network may end up in a local minima that can be far from the best possible results. For this reason, it may be a better approach to employ some pre-initialization method capable of selecting more appropriate initial weights [34]. It is also worth mentioning that the strategy (algorithms) used for training can have a big impact on the results [35].
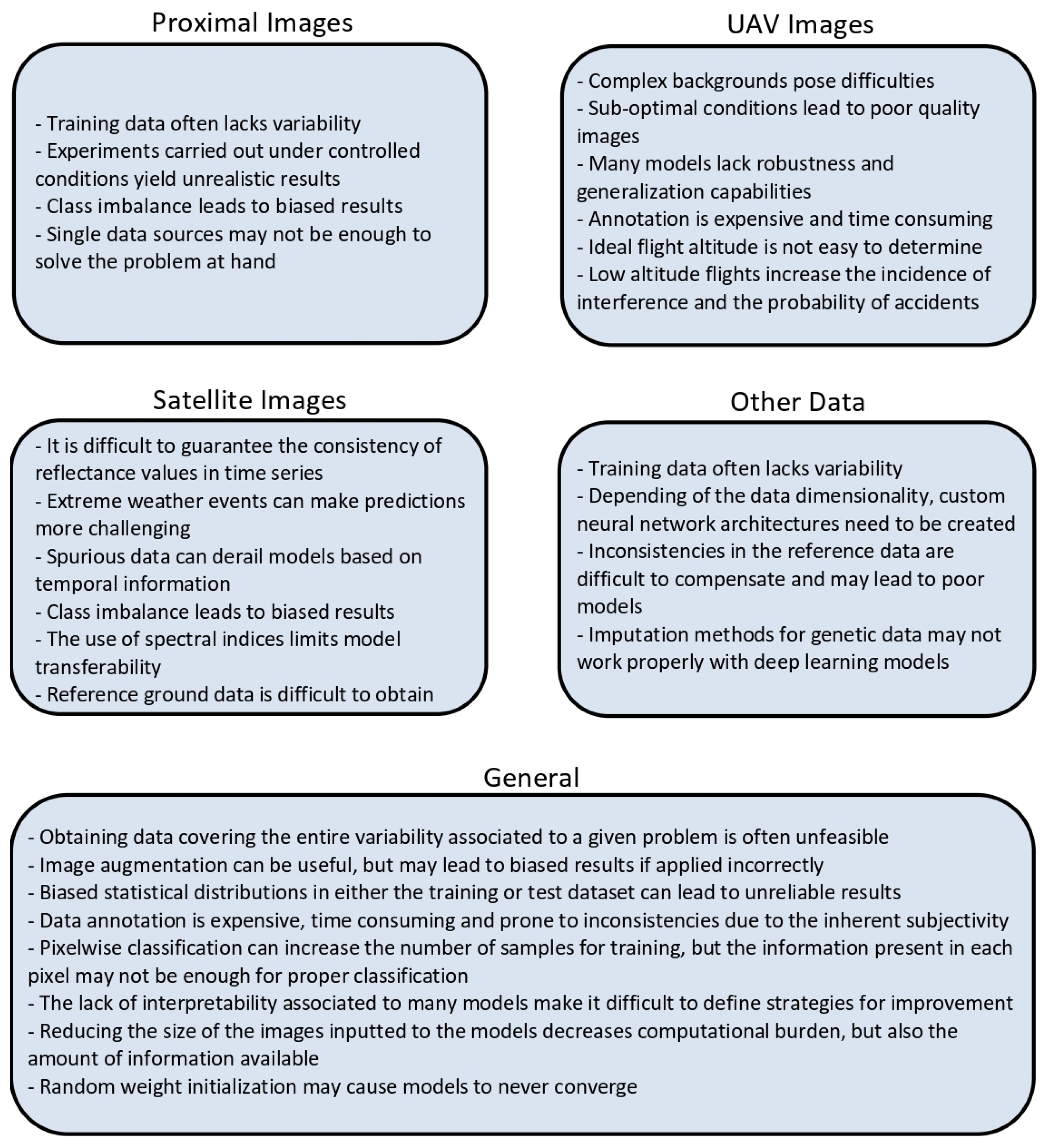
Figure 2 summarizes all challenges and weaknesses identified in this review.
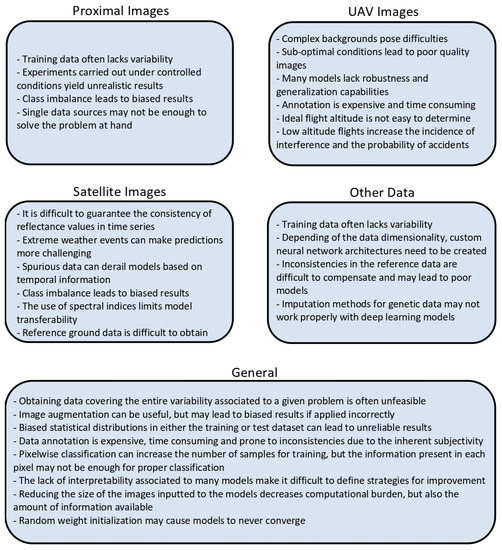
Figure 2.
Challenges and weaknesses identified in this review for each type of data.
5. Conclusions
This review explored the current state of the art of deep learning applied to problems attached to soybean crops. The number of articles dedicated to the subject has been growing steadily, and significant progress has been achieved not only in terms of accuracy but also in understanding how the models arrive at their answers. Despite this progress, there are still many challenges and research gaps that lack suitable solutions. Many of those challenges were identified and discussed in this article, and potential solutions were proposed whenever possible. Among the tendencies for future work that could be inferred, some seem to be quickly gaining momentum, including the fusion of different types of data, the attempt to increase interpretability and untangle the inner workings of deep learning models, and the incorporation of temporal information whenever appropriate. The leap from academic research to practical solutions has already been completed in a few cases, but there is still much to be done in order for artificial intelligence and deep learning to realize their full potential in soybean crop management and monitoring. It is our hope that the discussion developed in this review will help achieve this goal in a timely manner.
As for future perspectives, based on the way artificial intelligence and crop management have evolved so far, some tendencies are likely to prevail in the near future. Artificial intelligence and deep learning techniques should continue to evolve at a fast pace, continuously expanding the range of applications related to crop management that can benefit from this type of technique. At the same time, improved interpretability and a better understanding of the way deep learning architectures work will likely make it feasible to design models that are both lighter and more robust. As technical and technological hurdles are removed, the number of technologies based on deep learning ready to be used under more realistic conditions should grow. On the other hand, limitations related to data representativeness and model generalization will continue to exist, but these will tend to become less intense as sensors and data gathering techniques continue to evolve. It is also worth pointing out that the rapid development of other branches of AI can also have an impact that is difficult to foresee as exemplified by ChatGPT’s repercussion across society [79].
Funding
This research was funded by FAPESP, Proc. 2022/09319-9.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data sharing not applicable.
Conflicts of Interest
The author declares no conflict of interest.
References
- Bevers, N.; Sikora, E.J.; Hardy, N.B. Soybean disease identification using original field images and transfer learning with convolutional neural networks. Comput. Electron. Agric. 2022, 203, 107449. [Google Scholar] [CrossRef]
- Savary, S.; Willocquet, L.; Pethybridge, S.; Esker, P.; McRoberts, N.; Nelson, A. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 2019, 3, 430–439. [Google Scholar] [CrossRef]
- Gui, J.; Xu, H.; Fei, J. Non-Destructive Detection of Soybean Pest Based on Hyperspectral Image and Attention-ResNet Meta-Learning Model. Sensors 2023, 23, 678. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Jin, N.; Yu, Q. Impacts of climate change and crop management practices on soybean phenology changes in China. Sci. Total Environ. 2020, 707, 135638. [Google Scholar] [CrossRef] [PubMed]
- Barbedo, J.G.A. Impact of dataset size and variety on the effectiveness of deep learning and transfer learning for plant disease classification. Comput. Electron. Agric. 2018, 153, 46–53. [Google Scholar] [CrossRef]
- Jha, K.; Doshi, A.; Patel, P.; Shah, M. A comprehensive review on automation in agriculture using artificial intelligence. Artif. Intell. Agric. 2019, 2, 1–12. [Google Scholar] [CrossRef]
- Bock, C.H.; Pethybridge, S.J.; Barbedo, J.G.A.; Esker, P.D.; Mahlein, A.K.; Ponte, E.M.D. A phytopathometry glossary for the twenty-first century: Towards consistency and precision in intra- and inter-disciplinary dialogues. Trop. Plant Pathol. 2022, 47, 14–24. [Google Scholar] [CrossRef]
- Koirala, A.; Walsh, K.B.; Wang, Z.; McCarthy, C. Deep learning—Method overview and review of use for fruit detection and yield estimation. Comput. Electron. Agric. 2019, 162, 219–234. [Google Scholar] [CrossRef]
- Li, D.; Li, C.; Yao, Y.; Li, M.; Liu, L. Modern imaging techniques in plant nutrition analysis: A review. Comput. Electron. Agric. 2020, 174, 105459. [Google Scholar] [CrossRef]
- Castro, W.; Marcato Junior, J.; Polidoro, C.; Osco, L.P.; Gonçalves, W.; Rodrigues, L.; Santos, M.; Jank, L.; Barrios, S.; Valle, C.; et al. Deep Learning Applied to Phenotyping of Biomass in Forages with UAV-Based RGB Imagery. Sensors 2020, 20, 4802. [Google Scholar] [CrossRef]
- Flores, P.; Zhang, Z.; Igathinathane, C.; Jithin, M.; Naik, D.; Stenger, J.; Ransom, J.; Kiran, R. Distinguishing seedling volunteer corn from soybean through greenhouse color, color-infrared, and fused images using machine and deep learning. Ind. Crops Prod. 2021, 161, 113223. [Google Scholar] [CrossRef]
- Uzal, L.; Grinblat, G.; Namías, R.; Larese, M.; Bianchi, J.; Morandi, E.; Granitto, P. Seed-per-pod estimation for plant breeding using deep learning. Comput. Electron. Agric. 2018, 150, 196–204. [Google Scholar] [CrossRef]
- Ahmad, A.; Saraswat, D.; Aggarwal, V.; Etienne, A.; Hancock, B. Performance of deep learning models for classifying and detecting common weeds in corn and soybean production systems. Comput. Electron. Agric. 2021, 184, 106081. [Google Scholar] [CrossRef]
- Barbedo, J.G.A. Deep learning applied to plant pathology: The problem of data representativeness. Trop. Plant Pathol. 2022, 47, 85–94. [Google Scholar] [CrossRef]
- dos Santos Ferreira, A.; Freitas, D.M.; da Silva, G.G.; Pistori, H.; Folhes, M.T. Unsupervised deep learning and semi-automatic data labeling in weed discrimination. Comput. Electron. Agric. 2019, 165, 104963. [Google Scholar] [CrossRef]
- Meir, Y.; Barbedo, J.G.A.; Keren, O.; Godoy, C.V.; Amedi, N.; Shalom, Y.; Geva, A.B. Using Brainwave Patterns Recorded from Plant Pathology Experts to Increase the Reliability of AI-Based Plant Disease Recognition System. Sensors 2023, 23, 4272. [Google Scholar] [CrossRef]
- Sugiyama, M.; Nakajima, S.; Kashima, H.; Buenau, P.; Kawanabe, M. Direct Importance Estimation with Model Selection and Its Application to Covariate Shift Adaptation. In Proceedings of the Advances in Neural Information Processing Systems, Vancouver, BC, Canada, 3–6 December 2007; Platt, J., Koller, D., Singer, Y., Roweis, S., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2007; Volume 20. [Google Scholar]
- Shook, J.; Gangopadhyay, T.; Wu, L.; Ganapathysubramanian, B.; Sarkar, S.; Singh, A.K. Crop yield prediction integrating genotype and weather variables using deep learning. PLoS ONE 2021, 16, e0252402. [Google Scholar] [CrossRef]
- da Silva, L.A.; Bressan, P.O.; Gonçalves, D.N.; Freitas, D.M.; Machado, B.B.; Gonçalves, W.N. Estimating soybean leaf defoliation using convolutional neural networks and synthetic images. Comput. Electron. Agric. 2019, 156, 360–368. [Google Scholar] [CrossRef]
- Xu, J.; Yang, J.; Xiong, X.; Li, H.; Huang, J.; Ting, K.; Ying, Y.; Lin, T. Towards interpreting multi-temporal deep learning models in crop mapping. Remote Sens. Environ. 2021, 264, 112599. [Google Scholar] [CrossRef]
- Barbedo, J.G.A. A Review on the Use of Computer Vision and Artificial Intelligence for Fish Recognition, Monitoring, and Management. Fishes 2022, 7, 335. [Google Scholar] [CrossRef]
- Barnhart, I.H.; Lancaster, S.; Goodin, D.; Spotanski, J.; Dille, J.A. Use of open-source object detection algorithms to detect Palmer amaranth (Amaranthus palmeri) in soybean. Weed Sci. 2022, 70, 648–662. [Google Scholar] [CrossRef]
- Chandel, N.S.; Chakraborty, S.K.; Rajwade, Y.A.; Dubey, K.; Tiwari, M.K.; Jat, D. Identifying crop water stress using deep learning models. Neural Comput. Appl. 2021, 33, 5353–5367. [Google Scholar] [CrossRef]
- Falk, K.G.; Jubery, T.Z.; Mirnezami, S.V.; Parmley, K.A.; Sarkar, S.; Singh, A.; Ganapathysubramanian, B.; Singh, A.K. Computer vision and machine learning enabled soybean root phenotyping pipeline. Plant Methods 2020, 16, 5. [Google Scholar] [CrossRef]
- Gonçalves, J.P.; Pinto, F.A.; Queiroz, D.M.; Villar, F.M.; Barbedo, J.G.; Ponte, E.M.D. Deep learning architectures for semantic segmentation and automatic estimation of severity of foliar symptoms caused by diseases or pests. Biosyst. Eng. 2021, 210, 129–142. [Google Scholar] [CrossRef]
- Guo, Y.; Gao, Z.; Zhang, Z.; Li, Y.; Hu, Z.; Xin, D.; Chen, Q.; Zhu, R. Automatic and Accurate Acquisition of Stem-Related Phenotypes of Mature Soybean Based on Deep Learning and Directed Search Algorithms. Front. Plant Sci. 2022, 13, 906751. [Google Scholar] [CrossRef]
- He, H.; Ma, X.; Guan, H.; Wang, F.; Shen, P. Recognition of soybean pods and yield prediction based on improved deep learning model. Front. Plant Sci. 2023, 13, 1096619. [Google Scholar] [CrossRef]
- Karlekar, A.; Seal, A. SoyNet: Soybean leaf diseases classification. Comput. Electron. Agric. 2020, 172, 105342. [Google Scholar] [CrossRef]
- Li, Y.; Jia, J.; Zhang, L.; Khattak, A.M.; Sun, S.; Gao, W.; Wang, M. Soybean Seed Counting Based on Pod Image Using Two-Column Convolution Neural Network. IEEE Access 2019, 7, 64177–64185. [Google Scholar] [CrossRef]
- de Castro Pereira, R.; Hirose, E.; Ferreira de Carvalho, O.L.; da Costa, R.M.; Borges, D.L. Detection and classification of whiteflies and development stages on soybean leaves images using an improved deep learning strategy. Comput. Electron. Agric. 2022, 199, 107132. [Google Scholar] [CrossRef]
- Rairdin, A.; Fotouhi, F.; Zhang, J.; Mueller, D.S.; Ganapathysubramanian, B.; Singh, A.K.; Dutta, S.; Sarkar, S.; Singh, A. Deep learning-based phenotyping for genome wide association studies of sudden death syndrome in soybean. Front. Plant Sci. 2022, 13, 966244. [Google Scholar] [CrossRef]
- Sabóia, H.d.S.; Mion, R.L.; Silveira, A.d.O.; Mamiya, A.A. Real-time selective spraying for viola rope control in soybean and cotton crops using deep learning. Eng. Agríc. 2022, 42, e20210163. [Google Scholar] [CrossRef]
- Srilakshmi, A.; Geetha, K. A novel framework for soybean leaves disease detection using DIM-U-net and LSTM. Multimed. Tools Appl. 2023, 82, 28323–28343. [Google Scholar] [CrossRef]
- Tang, J.; Wang, D.; Zhang, Z.; He, L.; Xin, J.; Xu, Y. Weed identification based on K-means feature learning combined with convolutional neural network. Comput. Electron. Agric. 2017, 135, 63–70. [Google Scholar] [CrossRef]
- Tetila, E.C.; Machado, B.B.; Astolfi, G.; de Souza Belete, N.A.; Amorim, W.P.; Roel, A.R.; Pistori, H. Detection and classification of soybean pests using deep learning with UAV images. Comput. Electron. Agric. 2020, 179, 105836. [Google Scholar] [CrossRef]
- Wang, B.; Li, H.; You, J.; Chen, X.; Yuan, X.; Feng, X. Fusing deep learning features of triplet leaf image patterns to boost soybean cultivar identification. Comput. Electron. Agric. 2022, 197, 106914. [Google Scholar] [CrossRef]
- Wu, Q.; Zhang, K.; Meng, J. Identification of Soybean Leaf Diseases via Deep Learning. J. Inst. Eng. (India) Ser. A 2019, 100, 659–666. [Google Scholar] [CrossRef]
- Xiang, S.; Wang, S.; Xu, M.; Wang, W.; Liu, W. YOLO POD: A fast and accurate multi-task model for dense Soybean Pod counting. Plant Methods 2023, 19, 8. [Google Scholar] [CrossRef]
- Yang, S.; Zheng, L.; He, P.; Wu, T.; Sun, S.; Wang, M. High-throughput soybean seeds phenotyping with convolutional neural networks and transfer learning. Plant Methods 2021, 17, 50. [Google Scholar] [CrossRef]
- Yoosefzadeh-Najafabadi, M.; Tulpan, D.; Eskandari, M. Using Hybrid Artificial Intelligence and Evolutionary Optimization Algorithms for Estimating Soybean Yield and Fresh Biomass Using Hyperspectral Vegetation Indices. Remote Sens. 2021, 13, 2555. [Google Scholar] [CrossRef]
- Yu, X.; Chen, C.; Gong, Q.; Lu, L. Application of Data Enhancement Method Based on Generative Adversarial Networks for Soybean Leaf Disease Identification. Am. J. Biochem. Biotechnol. 2022, 18, 417–427. [Google Scholar] [CrossRef]
- Zhang, K.; Wu, Q.; Chen, Y. Detecting soybean leaf disease from synthetic image using multi-feature fusion faster R-CNN. Comput. Electron. Agric. 2021, 183, 106064. [Google Scholar] [CrossRef]
- Zhang, X.; Cui, J.; Liu, H.; Han, Y.; Ai, H.; Dong, C.; Zhang, J.; Chu, Y. Weed Identification in Soybean Seedling Stage Based on Optimized Faster R-CNN Algorithm. Agriculture 2023, 13, 175. [Google Scholar] [CrossRef]
- Zhang, C.; Lu, X.; Ma, H.; Hu, Y.; Zhang, S.; Ning, X.; Hu, J.; Jiao, J. High-Throughput Classification and Counting of Vegetable Soybean Pods Based on Deep Learning. Agronomy 2023, 13, 1154. [Google Scholar] [CrossRef]
- Zhao, G.; Quan, L.; Li, H.; Feng, H.; Li, S.; Zhang, S.; Liu, R. Real-time recognition system of soybean seed full-surface defects based on deep learning. Comput. Electron. Agric. 2021, 187, 106230. [Google Scholar] [CrossRef]
- Zhao, J.; Kaga, A.; Yamada, T.; Komatsu, K.; Hirata, K.; Kikuchi, A.; Hirafuji, M.; Ninomiya, S.; Guo, W. Improved Field-Based Soybean Seed Counting and Localization with Feature Level Considered. Plant Phenomics 2023, 5, 0026. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Zhou, L.; Zhang, C.; Bao, Y.; Wu, B.; Chu, H.; Yu, Y.; He, Y.; Feng, L. Identification of Soybean Varieties Using Hyperspectral Imaging Coupled with Convolutional Neural Network. Sensors 2019, 19, 4065. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Zhang, J.; Chao, M.; Xu, X.; Song, P.; Zhang, J.; Huang, Z. A Rapid and Highly Efficient Method for the Identification of Soybean Seed Varieties: Hyperspectral Images Combined with Transfer Learning. Molecules 2020, 25, 152. [Google Scholar] [CrossRef]
- Barbedo, J.G.A. Data Fusion in Agriculture: Resolving Ambiguities and Closing Data Gaps. Sensors 2022, 22, 2285. [Google Scholar] [CrossRef]
- Sun, J.; Di, L.; Sun, Z.; Shen, Y.; Lai, Z. County-Level Soybean Yield Prediction Using Deep CNN-LSTM Model. Sensors 2019, 19, 4363. [Google Scholar] [CrossRef] [PubMed]
- Babu, V.; Ram, N. Deep residual CNN with contrast limited adaptive histogram equalization for weed detection in soybean crops. Trait. Signal 2020, 39, 717–722. [Google Scholar] [CrossRef]
- dos Santos Ferreira, A.; Freitas, D.M.; da Silva, G.G.; Pistori, H.; Folhes, M.T. Weed detection in soybean crops using ConvNets. Comput. Electron. Agric. 2017, 143, 314–324. [Google Scholar] [CrossRef]
- Maimaitijiang, M.; Sagan, V.; Sidike, P.; Hartling, S.; Esposito, F.; Fritschi, F.B. Soybean yield prediction from UAV using multimodal data fusion and deep learning. Remote Sens. Environ. 2020, 237, 111599. [Google Scholar] [CrossRef]
- Menezes, G.K.; Astolfi, G.; Martins, J.A.C.; Tetila, E.C.; da Silva Oliveira Junior, A.; Gonçalves, D.N.; Marcato Junior, J.; Silva, J.A.; Li, J.; Gonçalves, W.N.; et al. Pseudo-label semi-supervised learning for soybean monitoring. Smart Agric. Technol. 2023, 4, 100216. [Google Scholar] [CrossRef]
- Razfar, N.; True, J.; Bassiouny, R.; Venkatesh, V.; Kashef, R. Weed detection in soybean crops using custom lightweight deep learning models. J. Agric. Food Res. 2022, 8, 100308. [Google Scholar] [CrossRef]
- Veeranampalayam Sivakumar, A.N.; Li, J.; Scott, S.; Psota, E.; Jhala, A.; Luck, J.D.; Shi, Y. Comparison of Object Detection and Patch-Based Classification Deep Learning Models on Mid- to Late-Season Weed Detection in UAV Imagery. Remote Sens. 2020, 12, 2136. [Google Scholar] [CrossRef]
- Teodoro, P.E.; Teodoro, L.P.R.; Baio, F.H.R.; da Silva Junior, C.A.; dos Santos, R.G.; Ramos, A.P.M.; Pinheiro, M.M.F.; Osco, L.P.; Gonçalves, W.N.; Carneiro, A.M.; et al. Predicting Days to Maturity, Plant Height, and Grain Yield in Soybean: A Machine and Deep Learning Approach Using Multispectral Data. Remote Sens. 2021, 13, 4632. [Google Scholar] [CrossRef]
- Tetila, E.C.; Machado, B.B.; Menezes, G.K.; Da Silva Oliveira, A.; Alvarez, M.; Amorim, W.P.; De Souza Belete, N.A.; Da Silva, G.G.; Pistori, H. Automatic Recognition of Soybean Leaf Diseases Using UAV Images and Deep Convolutional Neural Networks. IEEE Geosci. Remote Sens. Lett. 2020, 17, 903–907. [Google Scholar] [CrossRef]
- Tian, F.; Vieira, C.C.; Zhou, J.; Zhou, J.; Chen, P. Estimation of Off-Target Dicamba Damage on Soybean Using UAV Imagery and Deep Learning. Sensors 2023, 23, 3241. [Google Scholar] [CrossRef] [PubMed]
- Trevisan, R.; Pérez, O.; Schmitz, N.; Diers, B.; Martin, N. High-Throughput Phenotyping of Soybean Maturity Using Time Series UAV Imagery and Convolutional Neural Networks. Remote Sens. 2020, 12, 3617. [Google Scholar] [CrossRef]
- Yang, Q.; She, B.; Huang, L.; Yang, Y.; Zhang, G.; Zhang, M.; Hong, Q.; Zhang, D. Extraction of soybean planting area based on feature fusion technology of multi-source low altitude unmanned aerial vehicle images. Ecol. Inform. 2022, 70, 101715. [Google Scholar] [CrossRef]
- Zhang, Z.; Khanal, S.; Raudenbush, A.; Tilmon, K.; Stewart, C. Assessing the efficacy of machine learning techniques to characterize soybean defoliation from unmanned aerial vehicles. Comput. Electron. Agric. 2022, 193, 106682. [Google Scholar] [CrossRef]
- Zhang, S.; Feng, H.; Han, S.; Shi, Z.; Xu, H.; Liu, Y.; Feng, H.; Zhou, C.; Yue, J. Monitoring of Soybean Maturity Using UAV Remote Sensing and Deep Learning. Agriculture 2023, 13, 110. [Google Scholar] [CrossRef]
- Zhou, J.; Zhou, J.; Ye, H.; Ali, M.L.; Chen, P.; Nguyen, H.T. Yield estimation of soybean breeding lines under drought stress using unmanned aerial vehicle-based imagery and convolutional neural network. Biosyst. Eng. 2021, 204, 90–103. [Google Scholar] [CrossRef]
- Delavarpour, N.; Koparan, C.; Nowatzki, J.; Bajwa, S.; Sun, X. A Technical Study on UAV Characteristics for Precision Agriculture Applications and Associated Practical Challenges. Remote Sens. 2021, 13, 1204. [Google Scholar] [CrossRef]
- Sagan, V.; Maimaitijiang, M.; Bhadra, S.; Maimaitiyiming, M.; Brown, D.R.; Sidike, P.; Fritschi, F.B. Field-scale crop yield prediction using multi-temporal WorldView-3 and PlanetScope satellite data and deep learning. ISPRS J. Photogramm. Remote Sens. 2021, 174, 265–281. [Google Scholar] [CrossRef]
- Bi, L.; Hu, G.; Raza, M.M.; Kandel, Y.; Leandro, L.; Mueller, D. A Gated Recurrent Units (GRU)-Based Model for Early Detection of Soybean Sudden Death Syndrome through Time-Series Satellite Imagery. Remote Sens. 2020, 12, 3621. [Google Scholar] [CrossRef]
- Khaki, S.; Pham, H.; Wang, L. Simultaneous corn and soybean yield prediction from remote sensing data using deep transfer learning. Sci. Rep. 2021, 11, 11132. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zeng, H.; Zhang, M.; Wu, B.; Zhao, Y.; Yao, X.; Cheng, T.; Qin, X.; Wu, F. A county-level soybean yield prediction framework coupled with XGBoost and multidimensional feature engineering. Int. J. Appl. Earth Obs. Geoinf. 2023, 118, 103269. [Google Scholar] [CrossRef]
- Schwalbert, R.A.; Amado, T.; Corassa, G.; Pott, L.P.; Prasad, P.; Ciampitti, I.A. Satellite-based soybean yield forecast: Integrating machine learning and weather data for improving crop yield prediction in southern Brazil. Agric. For. Meteorol. 2020, 284, 107886. [Google Scholar] [CrossRef]
- Xu, J.; Zhu, Y.; Zhong, R.; Lin, Z.; Xu, J.; Jiang, H.; Huang, J.; Li, H.; Lin, T. DeepCropMapping: A multi-temporal deep learning approach with improved spatial generalizability for dynamic corn and soybean mapping. Remote Sens. Environ. 2020, 247, 111946. [Google Scholar] [CrossRef]
- Zhang, S.; Ban, X.; Xiao, T.; Huang, L.; Zhao, J.; Huang, W.; Liang, D. Identification of Soybean Planting Areas Combining Fused Gaofen-1 Image Data and U-Net Model. Agronomy 2023, 13, 863. [Google Scholar] [CrossRef]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. ImageNet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar] [CrossRef]
- Akintayo, A.; Tylka, G.L.; Singh, A.K.; Ganapathysubramanian, B.; Singh, A.; Sarkar, S. A deep learning framework to discern and count microscopic nematode eggs. Sci. Rep. 2018, 8, 9145. [Google Scholar] [CrossRef]
- Khaki, S.; Wang, L.; Archontoulis, S.V. A CNN-RNN Framework for Crop Yield Prediction. Front. Plant Sci. 2020, 10, 1750. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, D.; He, F.; Wang, J.; Joshi, T.; Xu, D. Phenotype Prediction and Genome-Wide Association Study Using Deep Convolutional Neural Network of Soybean. Front. Genet. 2019, 10, 1091. [Google Scholar] [CrossRef]
- Tuia, D.; Persello, C.; Bruzzone, L. Domain Adaptation for the Classification of Remote Sensing Data: An Overview of Recent Advances. IEEE Geosci. Remote Sens. Mag. 2016, 4, 41–57. [Google Scholar] [CrossRef]
- Barbedo, J.G.A. Plant disease identification from individual lesions and spots using deep learning. Biosyst. Eng. 2019, 180, 96–107. [Google Scholar] [CrossRef]
- Stokel-Walker, C.; Noorden, R.V. The promise and peril of generative AI. Nature 2023, 614, 214–216. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).