Climate Change during Cretaceous/Paleogene as a Driving Force for the Evolutionary Radiation of Physical Dormancy in Fabaceae
Abstract
1. Introduction
2. Methods and Materials
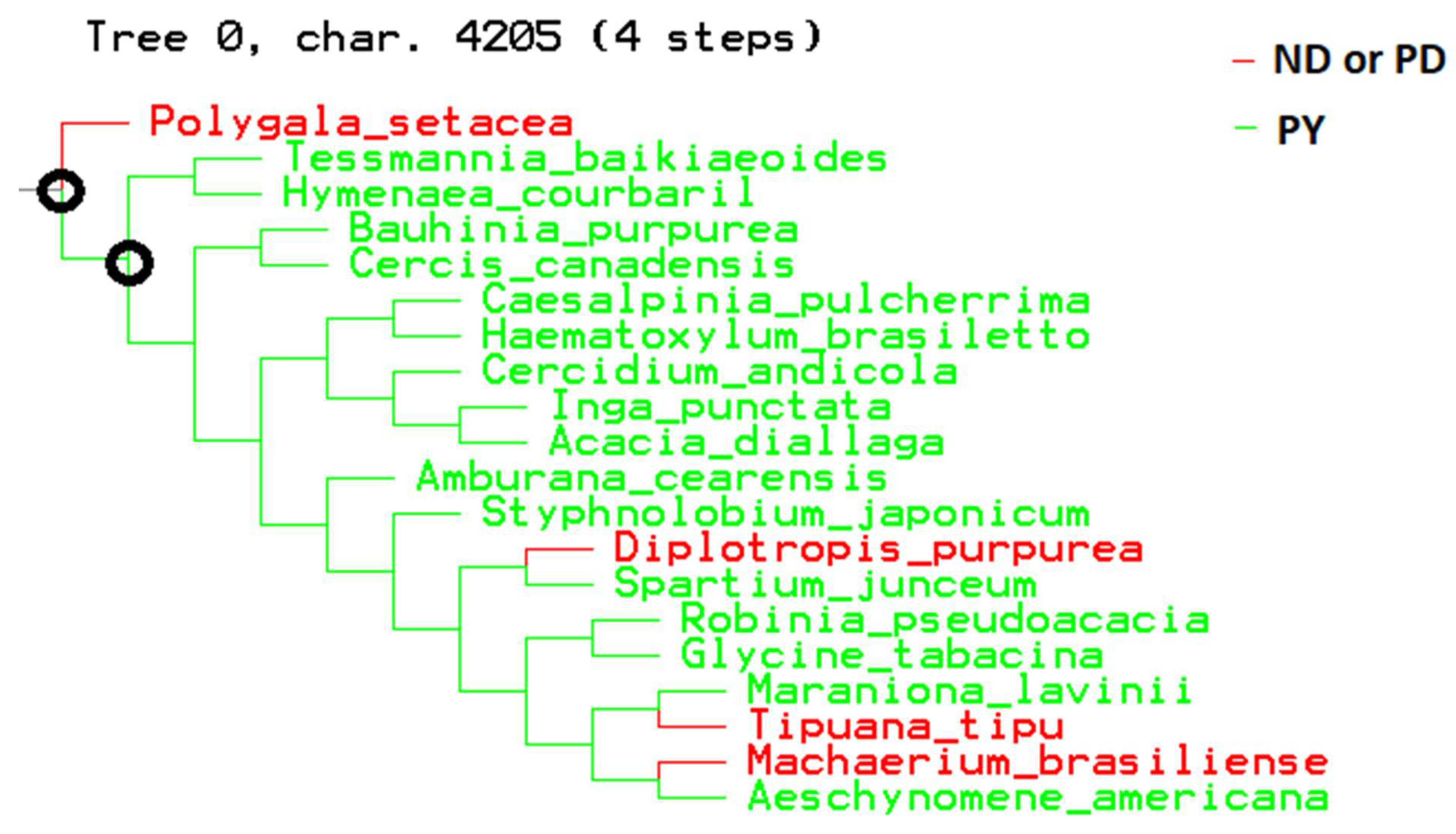
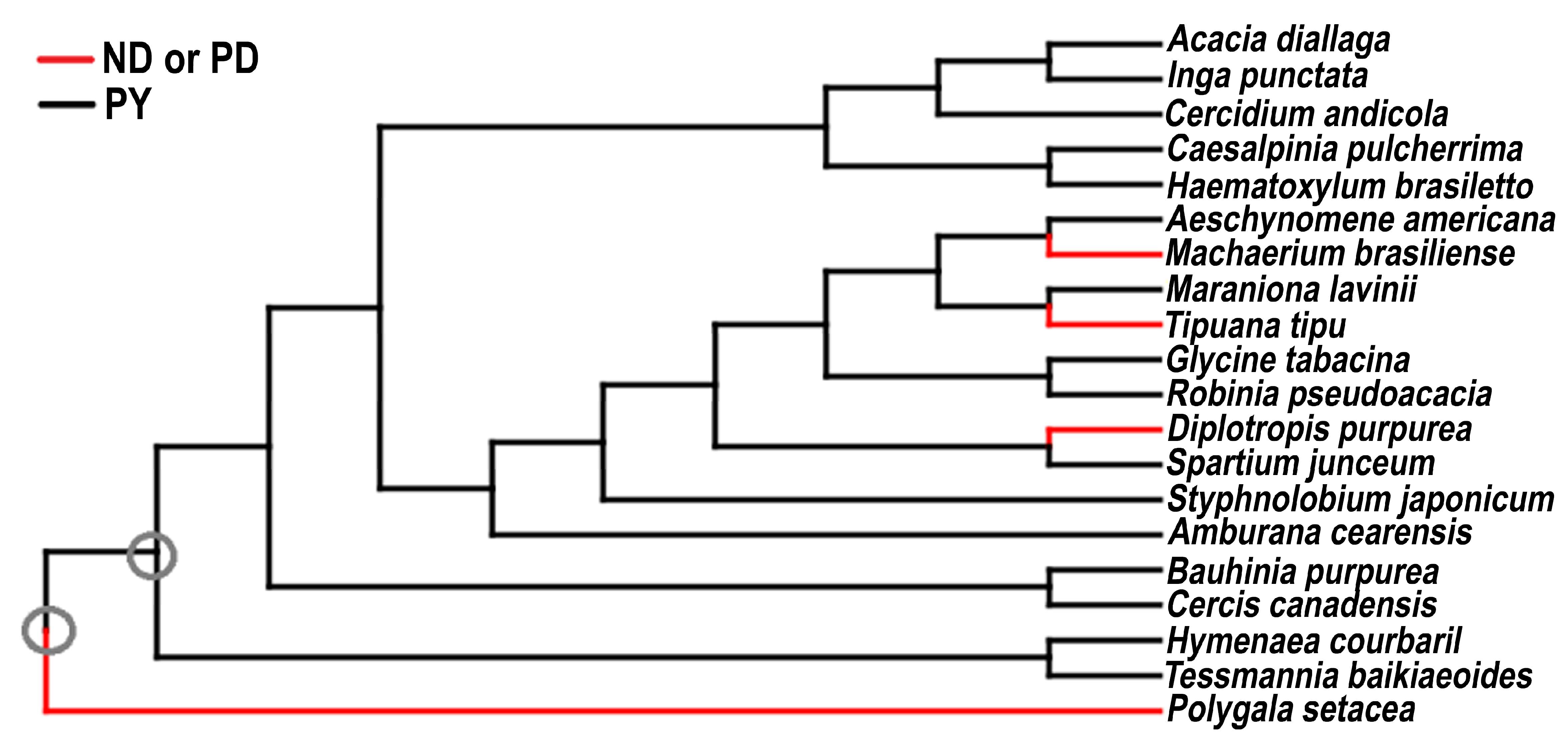
3. Data and Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baskin, C.C.; Baskin, J.M. Seeds: Ecology, Biogeography, and Evolution of Dormancy and Germination, 2nd ed.; Academic Press: Cambridge, MA, USA, 2014. [Google Scholar]
- Baskin, J.M.; Baskin, C.C. The great diversity in kinds of seed dormancy: A revision of the Nikolaeva–Baskin classification system for primary seed dormancy. Seed Sci. Res. 2021, 31, 249–277. [Google Scholar] [CrossRef]
- Willis, C.G.; Baskin, C.C.; Baskin, J.M.; Auld, J.R.; Venable, D.L.; Cavender-Bares, J.; Donohue, K.; Rubio de Casas, R.; Group, N.G.W. The evolution of seed dormancy: Environmental cues, evolutionary hubs, and diversification of the seed plants. New Phytol. 2014, 203, 300–309. [Google Scholar] [CrossRef] [PubMed]
- Graeber, K.; Nakabayashi, K.; Leubner-Metzger, G. Development of Dormancy. In Encyclopedia of Applied Plant Sciences, 2nd ed.; Thomas, B., Murray, B.G., Murphy, D.J., Eds.; Academic Press: Oxford, UK, 2017; pp. 483–489. [Google Scholar]
- Baskin, J.M.; Baskin, C.C. A classification system for seed dormancy. Seed Sci. Res. 2004, 14, 1–16. [Google Scholar] [CrossRef]
- Crocker, W. Mechanics of dormancy in seeds. Am. J. Bot. 1916, 3, 99–120. [Google Scholar] [CrossRef]
- Keeley, J.E.; Pausas, J.G.; Rundel, P.W.; Bond, W.J.; Bradstock, R.A. Fire as an evolutionary pressure shaping plant traits. Trends Plant Sci. 2011, 16, 406–411. [Google Scholar] [CrossRef] [PubMed]
- Temple, S.A. Plant-animal mutualism: Coevolution with dodo leads to near extinction of plant. Science 1977, 197, 885–886. [Google Scholar] [CrossRef]
- Paulsen, T.R.; Colville, L.; Daws, M.I.; Eliassen, S.; Högstedt, G.; Kranner, I.; Thompson, K.; Vandvik, V. The crypsis hypothesis explained: A reply to Jayasuriya et al. (2015). Seed Sci. Res. 2015, 25, 402–408. [Google Scholar] [CrossRef]
- Paulsen, T.; Colville, L.; Kranner, I.; Daws, M.; Högstedt, G.; Vandvik, V.; Thompson, K. Physical dormancy in seeds: A game of hide and seek? New Phytol. 2013, 198, 496–503. [Google Scholar] [CrossRef]
- Brancalion, P.H.S.; Novembre, A.D.L.C.; Rodrigues, R.R.; Filho, J.M. Dormancy as exaptation to protect mimetic seeds against deterioration before dispersal. Ann. Bot. 2010, 105, 991–998. [Google Scholar] [CrossRef]
- Jaganathan, G.K. Crypsis hypothesis as an explanation for evolution of impermeable coats in seeds is anecdotal. Ecol. Res. 2018, 33, 857–861. [Google Scholar] [CrossRef]
- Jaganathan, G.; Yule, K.; Liu, B. On the evolutionary and ecological value of breaking physical dormancy by endozoochory. Perspect. Plant Ecol. Evol. Syst. 2016, 22, 11–22. [Google Scholar] [CrossRef]
- Jaganathan, G.K. Are wildfires an adapted ecological cue breaking physical dormancy in the Mediterranean basin? Seed Sci. Res. 2015, 25, 120–126. [Google Scholar] [CrossRef]
- Jayasuriya, K.; Athugala, Y.S.; Wijayasinghe, M.M.; Baskin, J.M.; Baskin, C.C.; Mahadevan, N. The crypsis hypothesis: A stenopic view of the selective factors in the evolution of physical dormancy in seeds. Seed Sci. Res. 2015, 25, 127–137. [Google Scholar] [CrossRef]
- Van Staden, J.; Manning, J.C.; Kelly, K.M. Legumes seeds: The structure function equation. In Advances in Legume Biology; Stirton, C.H., Zarucchi, J.L., Eds.; Missouri Bot Garden: St. Louis, MO, USA, 1989; pp. 417–450. [Google Scholar]
- Baskin, J.M.; Baskin, C.C.; Li, X. Taxonomy, Anatomy and Evolution of Physical Dormancy in Seeds. Plant Species Biol. 2000, 15, 139–152. [Google Scholar] [CrossRef]
- Wang, Y.; Fan, G.; Liu, Y.; Sun, F.; Shi, C.; Liu, X.; Peng, J.; Chen, W.; Huang, X.; Cheng, S.; et al. The sacred lotus genome provides insights into the evolution of flowering plants. Plant J. 2013, 76, 557–567. [Google Scholar] [CrossRef]
- Shi, T.; Rahmani, R.; Gugger, P.; Wang, M.; Li, H.; Zhang, Y.; Li, Z.-Z.; Van de Peer, Y.; Marchal, K.; Chen, J. Distinct Expression and Methylation Patterns for Genes with Different Fates following a Single Whole-Genome Duplication in Flowering Plants. Mol. Biol. Evol. 2020, 37, 2394–2413. [Google Scholar] [CrossRef]
- Wu, S.; Han, B.; Jiao, Y. Genetic Contribution of Paleopolyploidy to Adaptive Evolution in Angiosperms. Mol. Plant 2020, 13, 59–71. [Google Scholar] [CrossRef]
- Bolingue, W.; Vu, B.; Leprince, O.; Buitink, J. Characterization of dormancy behaviour in seeds of the model legume Medicago truncatula. Seed Sci. Res. 2010, 20, 97–107. [Google Scholar] [CrossRef]
- Smýkal, P.; Vernoud, V.; Blair, M.W.; Soukup, A.; Thompson, R.D. The role of the testa during development and in establishment of dormancy of the legume seed. Front. Plant Sci. 2014, 5, 351. [Google Scholar]
- Chen, H.; Chu, P.; Zhou, Y.-L.; Ding, Y.; Li, Y.; Liu, J.; Jiang, L.-W.; Huang, S.-Z. Ectopic expression of NnPER1, a Nelumbo nucifera 1-cysteine peroxiredoxin antioxidant, enhances seed longevity and stress tolerance in Arabidopsis. Plant J. 2016, 88, 608–619. [Google Scholar] [CrossRef]
- Al-Mssallem, I.S.; Hu, S.; Zhang, X.; Lin, Q.; Liu, W.; Tan, J.; Yu, X.; Liu, J.; Pan, L.; Zhang, T.; et al. Genome sequence of the date palm Phoenix dactylifera L. Nat. Commun. 2013, 4, 2274. [Google Scholar] [CrossRef] [PubMed]
- Sano, N.; Rajjou, L.; North, H.; Debeaujon, I.; Marion-Poll, A.; Seo, M. Staying Alive: Molecular Aspects of Seed Longevity. Plant Cell Physiol. 2015, 57, pcv186. [Google Scholar] [CrossRef]
- Koenen, E.; Ojeda Alayon, D.; Bakker, F.; Wieringa, J.; Kidner, C.; Hardy, O.; Pennington, R.; Herendeen, P.; Bruneau, A.; Hughes, C. The Origin of the Legumes is a Complex Paleopolyploid Phylogenomic Tangle Closely Associated with the Cretaceous–Paleogene (K–Pg) Mass Extinction Event. Syst. Biol. 2021, 70, 508–526. [Google Scholar] [CrossRef] [PubMed]
- Marques, A.; Buijs, G.; Ligterink, W.; Hilhorst, H. Evolutionary ecophysiology of seed desiccation sensitivity. Funct. Plant Biol. 2018, 45, 1083–1095. [Google Scholar] [CrossRef] [PubMed]
- Lavin, M.; Herendeen, P.S.; Wojciechowski, M.F. Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the tertiary. Syst. Biol. 2005, 54, 575–594. [Google Scholar] [CrossRef]
- Lyson, T.R.; Miller, I.M.; Bercovici, A.D.; Weissenburger, K.; Fuentes, A.J.; Clyde, W.C.; Hagadorn, J.W.; Butrim, M.J.; Johnson, K.R.; Fleming, R.F. Exceptional continental record of biotic recovery after the Cretaceous–Paleogene mass extinction. Science 2019, 366, 977–983. [Google Scholar] [CrossRef]
- Centeno-González, N.K.; Martínez-Cabrera, H.I.; Porras-Múzquiz, H.; Estrada-Ruiz, E. Late Campanian fossil of a legume fruit supports Mexico as a center of Fabaceae radiation. Commun. Biol. 2021, 4, 41. [Google Scholar] [CrossRef]
- Nascimento, F.F.; Reis, M.D.; Yang, Z. A biologist’s guide to Bayesian phylogenetic analysis. Nat. Ecol. Evol. 2017, 1, 1446–1454. [Google Scholar] [CrossRef]
- Smith, S.D. Using phylogenetics to detect pollinator-mediated floral evolution. New Phytol. 2010, 188, 354–363. [Google Scholar] [CrossRef]
- Wickett, N.J.; Mirarab, S.; Nguyen, N.; Warnow, T.; Carpenter, E.; Matasci, N.; Ayyampalayam, S.; Barker, M.S.; Burleigh, J.G.; Gitzendanner, M.A. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc. Natl. Acad. Sci. USA 2014, 111, E4859–E4868. [Google Scholar] [CrossRef]
- Berry, K. The first plants to recolonize western North America following the Cretaceous/Paleogene mass extinction event. Int. J. Plant Sci. 2020, 182, 19–27. [Google Scholar] [CrossRef]
- Berry, K. Seed traits linked to differential survival of plants during the Cretaceous/Paleogene impact winter. Acta Palaeobot. 2020, 60, 307–322. [Google Scholar] [CrossRef]
- Bello, M.A.; Bruneau, A.; Forest, F.; Hawkins, J.A. Elusive Relationships within Order Fabales: Phylogenetic Analyses Using matK and rbcL Sequence Data. Syst. Bot. 2009, 34, 102–114. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Goloboff, P.A.; Catalano, S.A. GB-to-TNT: Facilitating creation of matrices from GenBank and diagnosis of results in TNT. Cladistics 2012, 28, 503–513. [Google Scholar] [CrossRef]
- Goloboff, P.A.; Catalano, S.A. TNT version 1.5, including a full implementation of phylogenetic morphometrics. Cladistics 2016, 32, 221–238. [Google Scholar] [CrossRef]
- Bouckaert, R.; Vaughan, T.G.; Barido-Sottani, J.; Duchêne, S.; Fourment, M.; Gavryushkina, A.; Heled, J.; Jones, G.; Kühnert, D.; De Maio, N. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019, 15, e1006650. [Google Scholar] [CrossRef]
- Koenen, E.; Ojeda, D.; Steeves, R.; Migliore, J.; Bakker, F.; Wieringa, J.; Kidner, C.; Hardy, O.; Pennington, R.; Bruneau, A. Large-scale genomic sequence data resolve the deepest divergences in the legume phylogeny and support a near-simultaneous evolutionary origin of all six subfamilies. New Phytol. 2020, 225, 1355–1369. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J. TreeView v. 1.7.2 and TreeAnnotator v. 2.6.4; Institute of Evolutionary Biology, University of Edinburgh: Edinburgh, UK, 2021; Available online: https://beast.community/treeannotator (accessed on 6 August 2022).
- Maddison, W.P.; Maddison, D.R. Mesquite: A Modular System for Evolutionary Analysis. Available online: https://www.mesquiteproject.org (accessed on 6 August 2022).
- Escobar, D.F.; Silveira, F.A.; Morellato, L.P.C. Timing of seed dispersal and seed dormancy in Brazilian savanna: Two solutions to face seasonality. Ann. Bot. 2018, 121, 1197–1209. [Google Scholar] [CrossRef]
- Travlos, I.; Economou, G.; Karamanos, A. Seed germination and seedling emergence of Spartium junceum L. in response to heat and other pre-sowing treatments. J. Agron. 2007, 6, 152. [Google Scholar]
- Galindez, G.; Ceccato, D.V.; Malagrina, G.M.; Pidal, B.; Chilo, G.N.; Bach, H.G.; Fortunato, R.H.; Ortega Baes, F.P. Physical seed dormancy in native legume species of Argentina. Bol. Soc. Argent. Bot. 2016, 51, 73–78. [Google Scholar]
- Rodríguez-Arévalo, I.; Mattana, E.; García, L.; Liu, U.; Lira, R.; Dávila, P.; Hudson, A.; Pritchard, H.W.; Ulian, T. Conserving seeds of useful wild plants in Mexico: Main issues and recommendations. Genet. Resour. Crop Evol. 2017, 64, 1141–1190. [Google Scholar] [CrossRef]
- Cannon, S.B.; Ilut, D.; Farmer, A.D.; Maki, S.L.; May, G.D.; Singer, S.R.; Doyle, J.J. Polyploidy did not predate the evolution of nodulation in all legumes. PLoS ONE 2010, 5, e11630. [Google Scholar] [CrossRef] [PubMed]
- Cannon, S.B.; McKain, M.R.; Harkess, A.; Nelson, M.N.; Dash, S.; Deyholos, M.K.; Peng, Y.; Joyce, B.; Stewart, C.N., Jr.; Rolf, M.; et al. Multiple Polyploidy Events in the Early Radiation of Nodulating and Nonnodulating Legumes. Mol. Biol. Evol. 2014, 32, 193–210. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, R.; Jiang, K.-W.; Qi, J.; Hu, Y.; Guo, J.; Zhu, R.; Zhang, T.; Egan, A.N.; Yi, T.-S. Nuclear phylotranscriptomics and phylogenomics support numerous polyploidization events and hypotheses for the evolution of rhizobial nitrogen-fixing symbiosis in Fabaceae. Mol. Plant 2021, 14, 748–773. [Google Scholar] [CrossRef]
- Kreplak, J.; Madoui, M.-A.; Cápal, P.; Novák, P.; Labadie, K.; Aubert, G.; Bayer, P.E.; Gali, K.K.; Syme, R.A.; Main, D.; et al. A reference genome for pea provides insight into legume genome evolution. Nat Genet. 2019, 51, 1411–1422. [Google Scholar] [CrossRef]
- Huegele, I.; Manchester, S. An Early Paleocene Carpoflora from the Denver Basin of Colorado, USA, and Its Implications for Plant-Animal Interactions and Fruit Size Evolution. Int. J. Plant Sci. 2020, 181, 6. [Google Scholar] [CrossRef]
- Schulte, P.; Alegret, L.; Arenillas, I.; Arz, J.A.; Barton, P.J.; Al, E. The Chicxulub Asteroid Impact and Mass Extinction at the Cretaceous-Paleogene Boundary. Science 2010, 327, 1214–1218. [Google Scholar] [CrossRef]
- Chiarenza, A.A.; Mannion, P.D.; Lunt, D.J.; Farnsworth, A.; Jones, L.A.; Kelland, S.J.; Allison, P.A. Ecological niche modelling does not support climatically-driven dinosaur diversity decline before the Cretaceous/Paleogene mass extinction. Nat. Commun. 2019, 10, 1091. [Google Scholar] [CrossRef]
- Chiarenza, A.A.; Farnsworth, A.; Mannion, P.D.; Lunt, D.J.; Valdes, P.J.; Morgan, J.V.; Allison, P.A. Asteroid impact, not volcanism, caused the end-Cretaceous dinosaur extinction. Proc. Natl. Acad. Sci. USA 2020, 117, 17084–17093. [Google Scholar] [CrossRef]
- Tabor, C.R.; Bardeen, C.G.; Otto-Bliesner, B.L.; Garcia, R.R.; Toon, O.B. Causes and Climatic Consequences of the Impact Winter at the Cretaceous-Paleogene Boundary. Geophys. Res. Lett. 2020, 47, e60121. [Google Scholar] [CrossRef]
- Roberts, E.H. Predicting the Storage Life of Seeds. Seed Sci. Technol. 1973, 1, 499–514. [Google Scholar]
- Gould, S.J.; Vrba, E.S. Exaptation; a missing term in the science of form. Paleobiology 1982, 8, 4–15. [Google Scholar] [CrossRef]
- Gould, S.J. The Structure of Evolutionary Theory; Belknap Press: Cambridge, MA, USA, 2002. [Google Scholar]
- Vanneste, K.; Maere, S.; Peer, Y. Tangled up in two: A burst of genome duplications at the end of the Cretaceous and the consequences for plant evolution. Philos. Trans. R. Soc. B Biol. Sci. 2019, 369, 5042–5050. [Google Scholar] [CrossRef]
- Leebens-Mack, J.; Wickett, N.; Deyholos, M.K.; Degironimo, L.; Pires, J.C. One thousand plant transcriptomes and the phylogenomics of green plants. Nature 2019, 574, 679–685. [Google Scholar]
- Van de Peer, Y.; Ashman, T.-L.; Soltis, P.; Soltis, D. Polyploidy: An evolutionary and ecological force in stressful times. Plant Cell 2021, 33, 11–26. [Google Scholar] [CrossRef]
- Darwin, C. On the Origin of Species; John Murray: London, UK, 1859. [Google Scholar]
- Linkies, A.; Graeber, K.; Knight, C.; Leubner-Metzger, G. The evolution of seeds. New Phytol. 2010, 186, 817–831. [Google Scholar] [CrossRef]
- Li, H.-T.; Yi, T.-S.; Gao, L.-M.; Ma, P.-F.; Zhang, T.; Yang, J.-B.; Gitzendanner, M.A.; Fritsch, P.W.; Cai, J.; Luo, Y. Origin of angiosperms and the puzzle of the Jurassic gap. Nat. Plants 2019, 5, 461–470. [Google Scholar] [CrossRef]
- Sauquet, H.; Magallón, S. Key questions and challenges in angiosperm macroevolution. New Phytol. 2018, 219, 1170–1187. [Google Scholar] [CrossRef]
- Jacobs, B.F.; Herendeen, P.S. Eocene dry climate and woodland vegetation in tropical Africa reconstructed from fossil leaves from northern Tanzania. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2004, 213, 115–123. [Google Scholar] [CrossRef]
- Morley, R.J. Origin and Evolution of Tropical Rain Forests; John Wiley & Sons: Hoboken, NJ, USA, 2000. [Google Scholar]
- Turner, S.K. Constraints on the onset duration of the Paleocene–Eocene Thermal Maximum. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2018, 376, 20170082. [Google Scholar] [CrossRef] [PubMed]
- Tosso, F.; Hardy, O.J.; Doucet, J.-L.; Daïnou, K.; Kaymak, E.; Migliore, J. Evolution in the Amphi-Atlantic tropical genus Guibourtia (Fabaceae, Detarioideae), combining NGS phylogeny and morphology. Mol. Phylogenet. Evol. 2018, 120, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Cabrera, H.I.; Estrada-Ruiz, E. Wood anatomy reveals high theoretical hydraulic conductivity and low resistance to vessel implosion in a Cretaceous fossil forest from northern Mexico. PLoS ONE 2014, 9, e108866. [Google Scholar] [CrossRef] [PubMed]
- Rubio de Casas, R.; Willis, C.G.; Pearse, W.D.; Baskin, C.C.; Baskin, J.M.; Cavender-Bares, J. Global biogeography of seed dormancy is determined by seasonality and seed size: A case study in the legumes. New Phytol. 2017, 214, 1527–1536. [Google Scholar] [CrossRef] [PubMed]
- Manchester, S. Fruits and Seeds of the Middle Eocene Nut Beds Flora, Clarno Formation, Oregon. Palaeontogr. Am. 1994, 58, 205. [Google Scholar]
- Muller, J. Fossil pollen records of extant angiosperms. Bot. Rev. 1981, 47, 1. [Google Scholar] [CrossRef]
- Endress, P.K. Reproductive structures and phylogenetic significance of extant primitive angiosperms. Plant Syst. Evol. 1986, 152, 1–28. [Google Scholar] [CrossRef]
- Chai, M.; Zhou, C.; Molina, I.; Fu, C.; Nakashima, J.; Li, G.; Zhang, W.; Park, J.; Tang, Y.; Jiang, Q.; et al. A class II KNOX gene, KNOX4, controls seed physical dormancy. Proc. Natl. Acad. Sci. USA 2016, 113, 6997–7002. [Google Scholar] [CrossRef]
- Lomax, B.H.; Hilton, J.; Bateman, R.M.; Upchurch, G.R.; Lake, J.A.; Leitch, I.J.; Cromwell, A.; Knight, C.A. Reconstructing relative genome size of vascular plants through geological time. New Phytol. 2014, 201, 636–644. [Google Scholar] [CrossRef]
- Clark, J.; Donoghue, P. Whole-Genome Duplication and Plant Macroevolution. Trends Plant Sci. 2018, 23, 933–945. [Google Scholar] [CrossRef]
- Fawcett, J.A.; Maere, S.; Van de Peer, Y. Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proc. Natl. Acad. Sci. USA 2009, 106, 5737–5742. [Google Scholar] [CrossRef]
- Gasper, A.L.; Almeida, T.; Dittrich, V.; Smith, A.; Salino, A. Molecular phylogeny of the fern family Blechnaceae (Polypodiales) with a revised genus-level treatment. Cladistics 2017, 33, 429–446. [Google Scholar] [CrossRef]
- Li, Z.-Z. Evolution by Ancient Gene and Genome Duplication in Hexapods and Land Plants. Ph.D. Dissertation, University of Arizona, Tucson, AZ, USA, 2020. [Google Scholar]
- Luo, M.-C.; You, F.M.; Li, P.; Wang, J.-R.; Zhu, T.; Dandekar, A.M.; Leslie, C.A.; Aradhya, M.; McGuire, P.E.; Dvorak, J. Synteny analysis in Rosids with a walnut physical map reveals slow genome evolution in long-lived woody perennials. BMC Genom. 2015, 16, 707. [Google Scholar] [CrossRef]
- Wolfe, J.A.; Upchurch, G.R. Leaf assemblages across the Cretaceous-Tertiary boundary in the Raton Basin, New Mexico and Colorado. Proc. Natl. Acad. Sci. USA 1987, 84, 5096–5100. [Google Scholar] [CrossRef]
- Shimada, N. A cluster of genes encodes the two types of chalcone isomerase involved in the biosynthesis of general flavonoids and legume-specific 5-deoxy(iso)flavonoids in Lotus japonicus. Plant Physiol. 2003, 131, 941–951. [Google Scholar] [CrossRef]
- Przysiecka, Ł.; Książkiewicz, M.; Wolko, B.; Naganowska, B. Structure, expression profile and phylogenetic inference of chalcone isomerase-like genes from the narrow-leafed lupin (Lupinus angustifolius L.) genome. Front. Plant Sci. 2015, 6, 268. [Google Scholar] [CrossRef]
- Cheng, A.-X.; Zhang, X.; Han, X.-J.; Zhang, Y.-Y.; Gao, S.; Liu, C.-J.; Lou, H.-X. Identification of chalcone isomerase in the basal land plants reveals an ancient evolution of enzymatic cyclization activity for synthesis of flavonoids. New Phytol. 2018, 217, 909–924. [Google Scholar] [CrossRef]
- Ni, R.; Zhu, T.-T.; Zhang, X.-S.; Wang, P.-Y.; Sun, C.-J.; Qiao, Y.-N.; Lou, H.-X.; Cheng, A.-X. Identification and evolutionary analysis of chalcone isomerase fold proteins in ferns. J. Exp. Bot. 2019, 71, 290–304. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jaganathan, G.K.; Berry, K. Climate Change during Cretaceous/Paleogene as a Driving Force for the Evolutionary Radiation of Physical Dormancy in Fabaceae. Seeds 2023, 2, 309-317. https://doi.org/10.3390/seeds2030023
Jaganathan GK, Berry K. Climate Change during Cretaceous/Paleogene as a Driving Force for the Evolutionary Radiation of Physical Dormancy in Fabaceae. Seeds. 2023; 2(3):309-317. https://doi.org/10.3390/seeds2030023
Chicago/Turabian StyleJaganathan, Ganesh K., and Keith Berry. 2023. "Climate Change during Cretaceous/Paleogene as a Driving Force for the Evolutionary Radiation of Physical Dormancy in Fabaceae" Seeds 2, no. 3: 309-317. https://doi.org/10.3390/seeds2030023
APA StyleJaganathan, G. K., & Berry, K. (2023). Climate Change during Cretaceous/Paleogene as a Driving Force for the Evolutionary Radiation of Physical Dormancy in Fabaceae. Seeds, 2(3), 309-317. https://doi.org/10.3390/seeds2030023





