Ectopic Expression of AtYUC8 Driven by GL2 and TT12 Promoters Affects the Vegetative Growth of Arabidopsis
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Plant Morphology Measurement
2.3. Statistical Analysis
3. Results
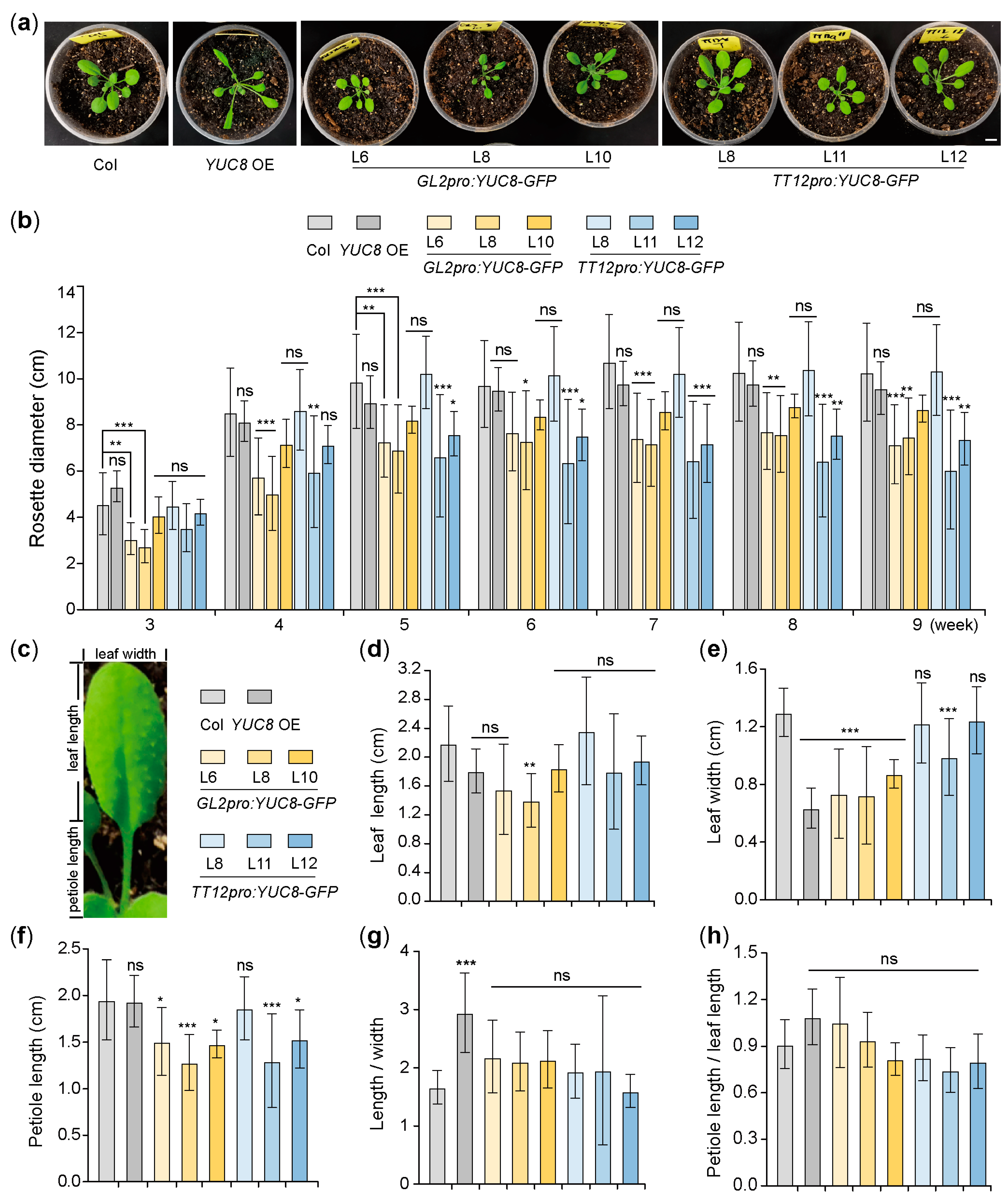
3.1. Ectopic Expression of the YUC8 under the Control of GL2 and TT12 Promoters Affects the Size of Arabidopsis Rosette
3.2. Ectopic Expression of the YUC8 under the Control of GL2 and TT12 Promoters Affects the Development of Arabidopsis Leaves
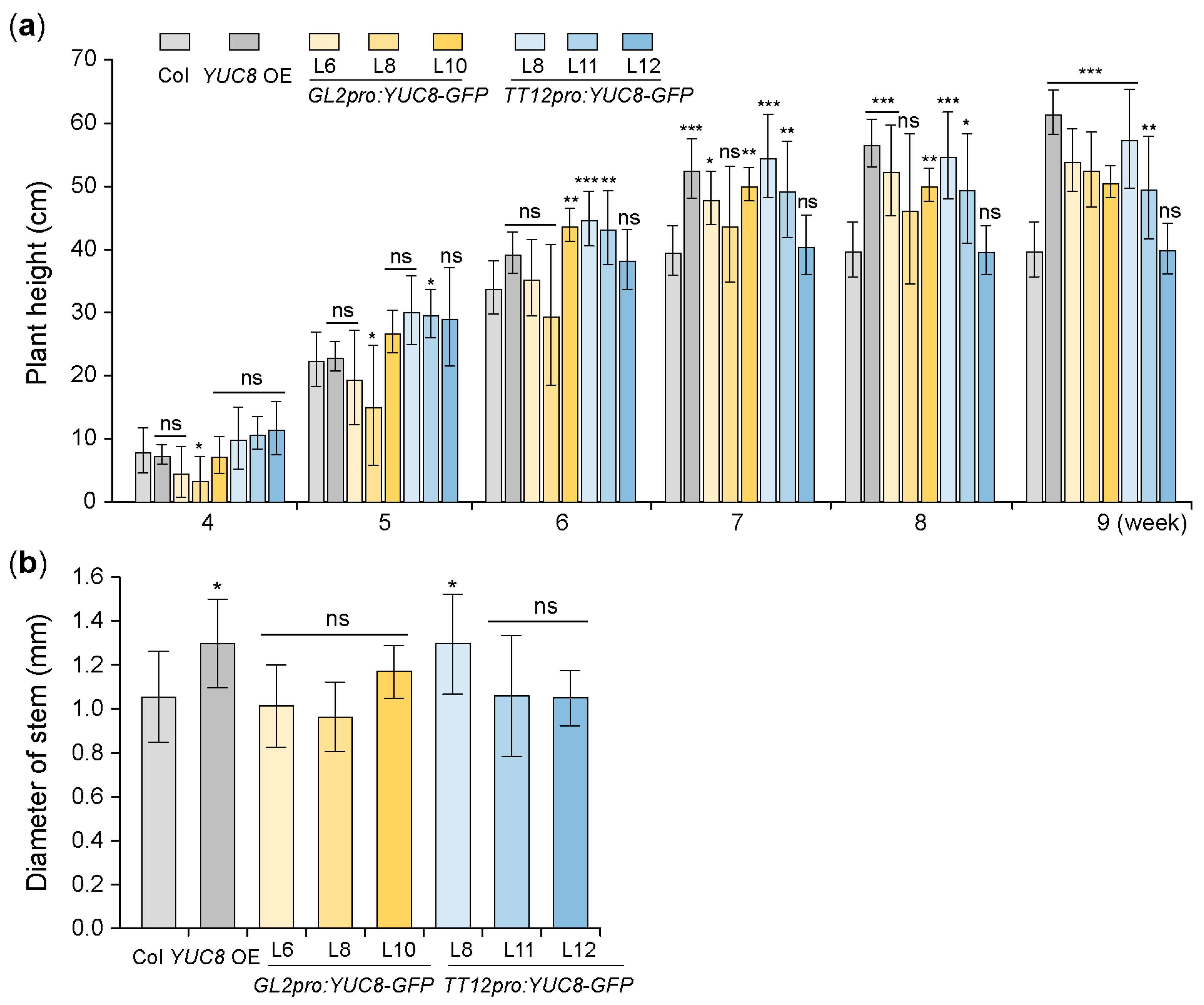
3.3. Ectopic Expression of the YUC8 under the Control of GL2 and TT12 Promoters Affects the Arabidopsis Height but Does Not Affect the Stem Thickness
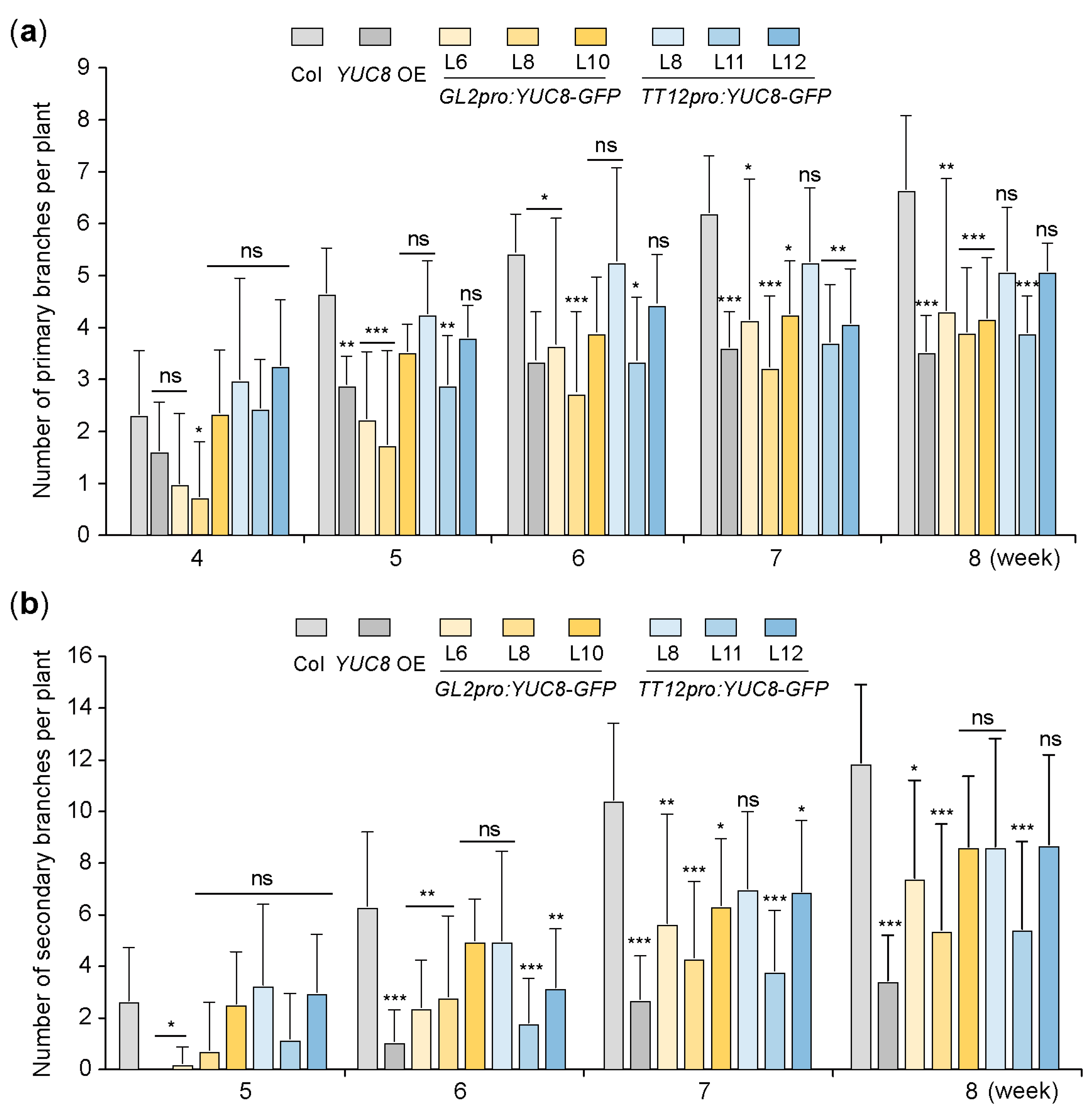
3.4. Ectopic Expression of the YUC8 under the Control of GL2 and TT12 Promoters Inhibits the Branching of Arabidopsis Plants
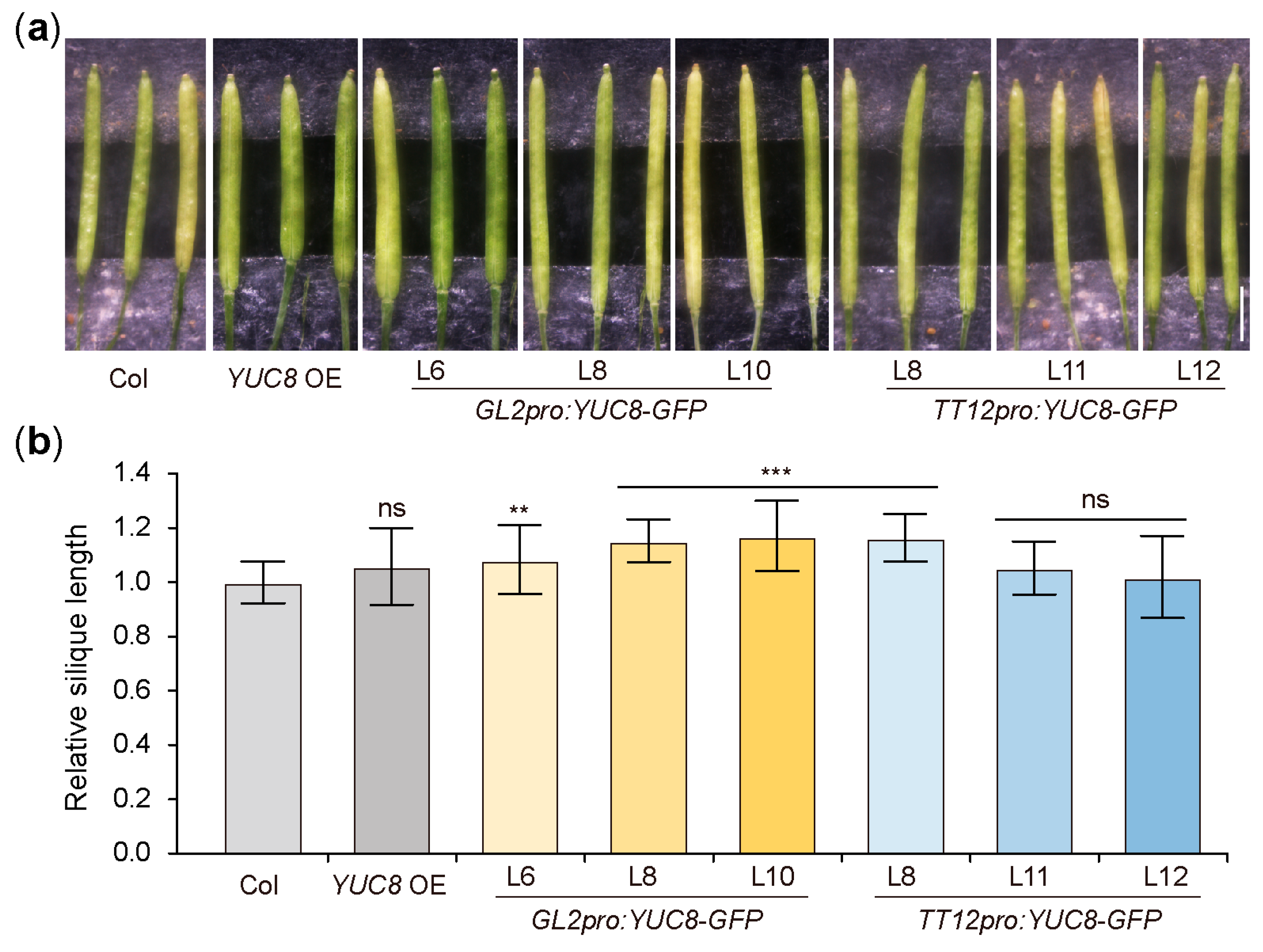
3.5. Ectopic Expression of the YUC8 under the Control of GL2 and TT12 Produce Longer Siliques
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ljung, K. Auxin metabolism and homeostasis during plant development. Development 2013, 140, 943–950. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y. Essential roles of local auxin biosynthesis in plant development and in adaptation to environmental changes. Annu. Rev. Plant Biol. 2018, 69, 417–435. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.D.; Slovin, J.P.; Hendrickson, A.M. Two genetically discrete pathways convert tryptophan to auxin: More redundancy in auxin biosynthesis. Trends Plant Sci. 2003, 8, 197–199. [Google Scholar] [CrossRef] [PubMed]
- Morffy, N.; Strader, L.C. Old Town Roads: Routes of auxin biosynthesis across kingdoms. Curr. Opin. Plant Biol. 2020, 55, 21–27. [Google Scholar] [CrossRef]
- Casanova-Sáez, R.; Voß, U. Auxin metabolism controls developmental decisions in land plants. Trends Plant Sci. 2019, 24, 741–754. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y. Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 2010, 61, 49–64. [Google Scholar] [CrossRef]
- Wright, A.D.; Sampson, M.B.; Neuffer, M.G.; Michalczuk, L.; Slovin, J.P.; Cohen, J.D. Indole-3-Acetic acid biosynthesis in the mutant maize orange pericarp, a tryptophan auxotroph. Science 1991, 254, 998–1000. [Google Scholar] [CrossRef]
- Normanly, J.; Cohen, J.D.; Fink, G.R. Arabidopsis thaliana auxotrophs reveal a tryptophan-independent biosynthetic pathway for indole-3-acetic acid. Proc. Natl. Acad. Sci. USA 1993, 90, 10355–10359. [Google Scholar] [CrossRef]
- Wang, B.; Chu, J.; Yu, T.; Xu, Q.; Sun, X.; Yuan, J.; Xiong, G.; Wang, G.; Wang, Y.; Li, J. Tryptophan-independent auxin biosynthesis contributes to early embryogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2015, 112, 4821–4826. [Google Scholar] [CrossRef]
- Zhao, Y.; Hull, A.K.; Gupta, N.R.; Goss, K.A.; Alonso, J.; Ecker, J.R.; Normanly, J.; Chory, J.; Celenza, J.L. Trp-dependent auxin biosynthesis in Arabidopsis: Involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev. 2002, 16, 3100–3112. [Google Scholar] [CrossRef]
- Crane, R.A.; Cardénas, V.M.; Castaneda, N.; Jackson, C.L.; Riley, C.J.; Mostafa, I.; Kong, W.; Chhajed, S.; Chen, S.; Brusslan, J.A. Negative regulation of age-related developmental leaf senescence by the IAOx Pathway, PEN1, and PEN3. Front. Plant Sci. 2019, 10, 1202. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Dai, X.; Aoi, Y.; Takebayashi, Y.; Yang, L.; Guo, X.; Zeng, Q.; Yu, H.; Kasahara, H.; Zhao, Y. Two homologous INDOLE-3-ACETAMIDE (IAM) HYDROLASE genes are required for the auxin effects of IAM in Arabidopsis. J. Genet. Genom. 2020, 47, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Mano, Y.; Nemoto, K. The pathway of auxin biosynthesis in plants. J. Exp. Bot. 2012, 63, 2853–2872. [Google Scholar] [CrossRef]
- Stepanova, A.N.; Robertson-Hoyt, J.; Yun, J.; Benavente, L.M.; Xie, D.Y.; Dolezal, K.; Schlereth, A.; Jürgens, G.; Alonso, J.M. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 2008, 133, 177–191. [Google Scholar] [CrossRef]
- Tao, Y.; Ferrer, J.L.; Ljung, K.; Poger, F.; Hong, F.; Long, J.A.; Li, L.; Moreno, J.E.; Bowman, M.E.; Ivans, L.J.; et al. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 2008, 133, 164–176. [Google Scholar] [CrossRef] [PubMed]
- Mashiguchi, K.; Tanaka, K.; Sakai, T.; Sugawara, S.; Kawaide, H.; Natsume, M.; Hanada, A.; Yaeno, T.; Shirasu, K.; Yao, H.; et al. The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 18512–18517. [Google Scholar] [CrossRef] [PubMed]
- Won, C.; Shen, X.; Mashiguchi, K.; Zheng, Z.; Dai, X.; Cheng, Y.; Kasahara, H.; Kamiya, Y.; Chory, J.; Zhao, Y. Conversion of tryptophan to indole-3-acetic acid by tryptophan aminotransferases of Arabidopsis and YUCCAs in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 18518–18523. [Google Scholar] [CrossRef]
- Stepanova, A.N.; Yun, J.; Robles, L.M.; Novak, O.; He, W.; Guo, H.; Ljung, K.; Alonso, J.M. The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell 2011, 23, 3961–3973. [Google Scholar] [CrossRef]
- Zhao, Y. Auxin biosynthesis: A simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol. Plant 2012, 5, 334–338. [Google Scholar] [CrossRef]
- Zhao, Y.; Christensen, S.K.; Fankhauser, C.; Cashman, J.R.; Cohen, J.D.; Weigel, D.; Chory, J. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 2001, 291, 306–309. [Google Scholar] [CrossRef]
- Dai, X.; Mashiguchi, K.; Chen, Q.; Kasahara, H.; Kamiya, Y.; Ojha, S.; DuBois, J.; Ballou, D.; Zhao, Y. The biochemical mechanism of auxin biosynthesis by an Arabidopsis YUCCA flavin-containing monooxygenase. J. Biol. Chem. 2013, 288, 1448–1457. [Google Scholar] [CrossRef]
- Cheng, Y.; Dai, X.; Zhao, Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006, 20, 1790–1799. [Google Scholar] [CrossRef]
- Cheng, Y.; Dai, X.; Zhao, Y. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 2007, 19, 2430–2439. [Google Scholar] [CrossRef] [PubMed]
- Hentrich, M.; Böttcher, C.; Düchting, P.; Cheng, Y.; Zhao, Y.; Berkowitz, O.; Masle, J.; Medina, J.; Pollmann, S. The jasmonic acid signaling pathway is linked to auxin homeostasis through the modulation of YUCCA8 and YUCCA9 gene expression. Plant J. 2013, 74, 626–637. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Dai, X.; De-Paoli, H.; Cheng, Y.; Takebayashi, Y.; Kasahara, H.; Kamiya, Y.; Zhao, Y. Auxin overproduction in shoots cannot rescue auxin deficiencies in Arabidopsis roots. Plant Cell Physiol. 2014, 55, 1072–1079. [Google Scholar] [CrossRef]
- Larsson, E.; Vivian-Smith, A.; Offringa, R.; Sundberg, E. Auxin homeostasis in Arabidopsis ovules is anther-dependent at maturation and changes dynamically upon fertilization. Front. Plant Sci. 2017, 8, 1735. [Google Scholar] [CrossRef]
- Liu, H.; Luo, Q.; Tan, C.; Song, J.; Zhang, T.; Men, S. Biosynthesis- and transport-mediated dynamic auxin distribution during seed development controls seed size in Arabidopsis. Plant J. 2023, 113, 1259–1277. [Google Scholar] [CrossRef]
- Blakeslee, J.J.; Spatola Rossi, T.; Kriechbaumer, V. Auxin biosynthesis: Spatial regulation and adaptation to stress. J. Exp. Bot. 2019, 70, 5041–5049. [Google Scholar] [CrossRef]
- Kong, Y.; Zhu, Y.; Gao, C.; She, W.; Lin, W.; Chen, Y.; Han, N.; Bian, H.; Zhu, M.; Wang, J. Tissue-specific expression of SMALL AUXIN UP RNA41 differentially regulates cell expansion and root meristem patterning in Arabidopsis. Plant Cell Physiol. 2013, 54, 609–621. [Google Scholar] [CrossRef]
- Song, Y.; Xu, Z. Ectopic overexpression of an AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) gene OsIAA4 in rice induces morphological changes and reduces responsiveness to Auxin. Int. J. Mol. Sci. 2013, 14, 13645–13656. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.Z.; Jin, S.H.; Li, P.; Jiang, X.Y.; Li, Y.J.; Hou, B.K. Ectopic expression of UGT84A2 delayed flowering by indole-3-butyric acid-mediated transcriptional repression of ARF6 and ARF8 genes in Arabidopsis. Plant Cell Rep. 2017, 36, 1995–2006. [Google Scholar] [CrossRef]
- Zhang, G.Z.; Jin, S.H.; Jiang, X.Y.; Dong, R.R.; Li, P.; Li, Y.J.; Hou, B.K. Ectopic expression of UGT75D1, a glycosyltransferase preferring indole-3-butyric acid, modulates cotyledon development and stress tolerance in seed germination of Arabidopsis thaliana. Plant Mol. Biol. 2016, 90, 77–93. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Hou, B.; Zhang, G. The ectopic expression of Arabidopsis glucosyltransferase UGT74D1 affects leaf positioning through modulating indole-3-acetic acid homeostasis. Sci. Rep. 2021, 11, 1154. [Google Scholar] [CrossRef] [PubMed]
- Park, S.C.; Kim, H.S.; Lee, H.U.; Kim, Y.H.; Kwak, S.S. Overexpression of Arabidopsis YUCCA6 enhances environment stress tolerance and inhibits storage root formation in sweetpotato. Plant Biotechnol. Rep. 2019, 13, 345–352. [Google Scholar] [CrossRef]
- Wang, X.; Yu, R.; Wang, J.; Lin, Z.; Han, X.; Deng, Z.; Fan, L.; He, H.; Deng, X.W.; Chen, H. The asymmetric expression of SAUR genes mediated by ARF7/19 promotes the gravitropism and phototropism of plant hypocotyls. Cell Rep. 2020, 31, 107529. [Google Scholar] [CrossRef] [PubMed]
- Stadler, R.; Lauterbach, C.; Sauer, N. Cell-to-cell movement of green fluorescent protein reveals post-phloem transport in the outer integument and identifies symplastic domains in Arabidopsis seeds and embryos. Plant Physiol. 2005, 139, 701–712. [Google Scholar] [CrossRef]
- Sun, B.; Shang, L.; Li, Y.; Zhang, Q.; Chu, Z.; He, S.; Yang, W.; Ding, X. Ectopic expression of OsJAZs alters plant defense and development. Int. J. Mol. Sci. 2022, 23, 4581. [Google Scholar] [CrossRef]
- Lloret, A.; Quesada-Traver, C.; Conejero, A.; Arbona, V.; Gómez-Mena, C.; Petri, C.; Sánchez-Navarro, J.A.; Zuriaga, E.; Leida, C.; Badenes, M.L.; et al. Regulatory circuits involving bud dormancy factor PpeDAM6. Hortic. Res. 2021, 8, 261. [Google Scholar] [CrossRef]
- Chen, K.Q.; Tang, X.G.; Song, M.R.; Guo, Y.G.; Liu, L.F.; Xue, H.; Dai, H.Y.; Zhang, Z.H. Functional identification of MdMYB5 involved in secondary cell wall formation in apple. Fruit Res. 2021, 1, 6. [Google Scholar] [CrossRef]
- Yuan, J.; Yu, Z.; Li, Y.; Shah, S.H.A.; Xiao, D.; Hou, X.; Li, Y. Ectopic expression of BrIQD35 promotes drought stress tolerance in Nicotiana benthamiana. Plant Biol. 2022, 24, 887–896. [Google Scholar] [CrossRef]
- Chuong, N.N.; Hoang, X.L.T.; Nghia, D.H.T.; Nguyen, N.C.; Thao, D.T.T.; Tran, T.B.; Ngoc, T.T.M.; Thu, N.B.A.; Nguyen, Q.T.; Thao, N.P. Ectopic expression of GmHP08 enhances resistance of transgenic Arabidopsis toward drought stress. Plant Cell Rep. 2021, 40, 819–834. [Google Scholar] [CrossRef] [PubMed]
- Dong, G.; Xiong, H.; Zeng, W.; Li, J.; Du, D. Ectopic expression of the rice grain-size-affecting gene GS5 in maize affects kernel size by regulating endosperm starch synthesis. Genes 2022, 13, 1542. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Ren, T.; Li, J.; Hu, W.; Zhang, J.; Yan, J.; Li, X.; Cong, R.; Guo, S.; Lu, J. Nutrition-mediated cell and tissue-level anatomy triggers the covariation of leaf photosynthesis and leaf mass per area. J. Exp. Bot. 2020, 71, 6524–6537. [Google Scholar] [CrossRef] [PubMed]
- Koyama, K.; Kikuzawa, K. Is whole-plant photosynthetic rate proportional to leaf area? A test of scalings and a logistic equation by leaf demography census. Am. Nat. 2009, 173, 640–649. [Google Scholar] [CrossRef]
- Zheng, M.; Peng, C.; Liu, H.; Tang, M.; Yang, H.; Li, X.; Liu, J.; Sun, X.; Wang, X.; Xu, J.; et al. Genome-wide association study reveals candidate genes for control of plant height, branch initiation height and branch number in rapeseed (Brassica napus L.). Front. Plant Sci. 2017, 8, 1246. [Google Scholar] [CrossRef]
- Figueiredo, D.D.; Batista, R.A.; Roszak, P.J.; Köhler, C. Auxin production couples endosperm development to fertilization. Nat. Plants 2015, 1, 15184. [Google Scholar] [CrossRef]
- Brumos, J.; Robles, L.M.; Yun, J.; Vu, T.C.; Jackson, S.; Alonso, J.M.; Stepanova, A.N. Local auxin biosynthesis is a key regulator of plant development. Dev. Cell 2018, 47, 306–318. [Google Scholar] [CrossRef]
- Romano, C.P.; Robson, P.R.; Smith, H.; Estelle, M.; Klee, H. Transgene-mediated auxin overproduction in Arabidopsis: Hypocotyl elongation phenotype and interactions with the hy6-1 hypocotyl elongation and axr1 auxin-resistant mutants. Plant Mol. Biol. 1995, 27, 1071–1083. [Google Scholar] [CrossRef]
- Ceccato, L.; Masiero, S.; Sinha, D.; Bencivenga, S.; Roig-Villanova, I.; Ditengou, F.A.; Palme, K.; Simon, R.; Colombo, L. Maternal control of PIN1 is required for female gametophyte development in Arabidopsis. PLoS ONE 2013, 8, e66148. [Google Scholar] [CrossRef]
- Wang, J.; Guo, X.; Xiao, Q.; Zhu, J.; Cheung, A.; Yuan, L.; Vierling, E.; Xu, S. Auxin efflux controls orderly nucellar degeneration and expansion of the female gametophyte in Arabidopsis. New Phytol. 2021, 230, 2261–2274. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, C.; Song, J.; Zhang, T.; Liang, M.; Li, S.; Liu, H.; Men, S. Ectopic Expression of AtYUC8 Driven by GL2 and TT12 Promoters Affects the Vegetative Growth of Arabidopsis. Seeds 2023, 2, 278-289. https://doi.org/10.3390/seeds2030021
Tan C, Song J, Zhang T, Liang M, Li S, Liu H, Men S. Ectopic Expression of AtYUC8 Driven by GL2 and TT12 Promoters Affects the Vegetative Growth of Arabidopsis. Seeds. 2023; 2(3):278-289. https://doi.org/10.3390/seeds2030021
Chicago/Turabian StyleTan, Chao, Jia Song, Tan Zhang, Mengxiao Liang, Suxin Li, Huabin Liu, and Shuzhen Men. 2023. "Ectopic Expression of AtYUC8 Driven by GL2 and TT12 Promoters Affects the Vegetative Growth of Arabidopsis" Seeds 2, no. 3: 278-289. https://doi.org/10.3390/seeds2030021
APA StyleTan, C., Song, J., Zhang, T., Liang, M., Li, S., Liu, H., & Men, S. (2023). Ectopic Expression of AtYUC8 Driven by GL2 and TT12 Promoters Affects the Vegetative Growth of Arabidopsis. Seeds, 2(3), 278-289. https://doi.org/10.3390/seeds2030021





