Methane Biofiltration Processes: A Summary of Biotic and Abiotic Factors
Abstract
1. Introduction
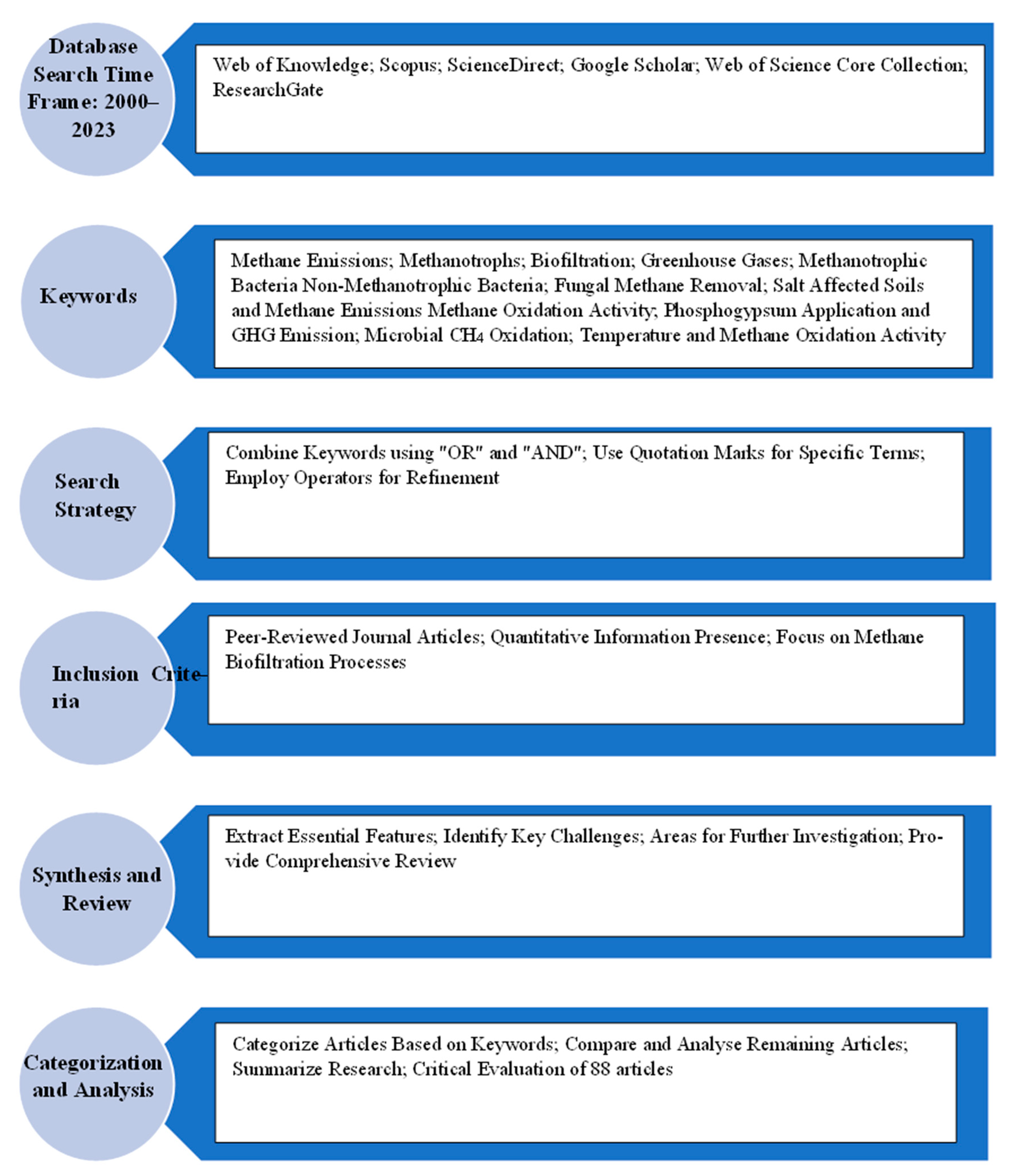
2. Methodology
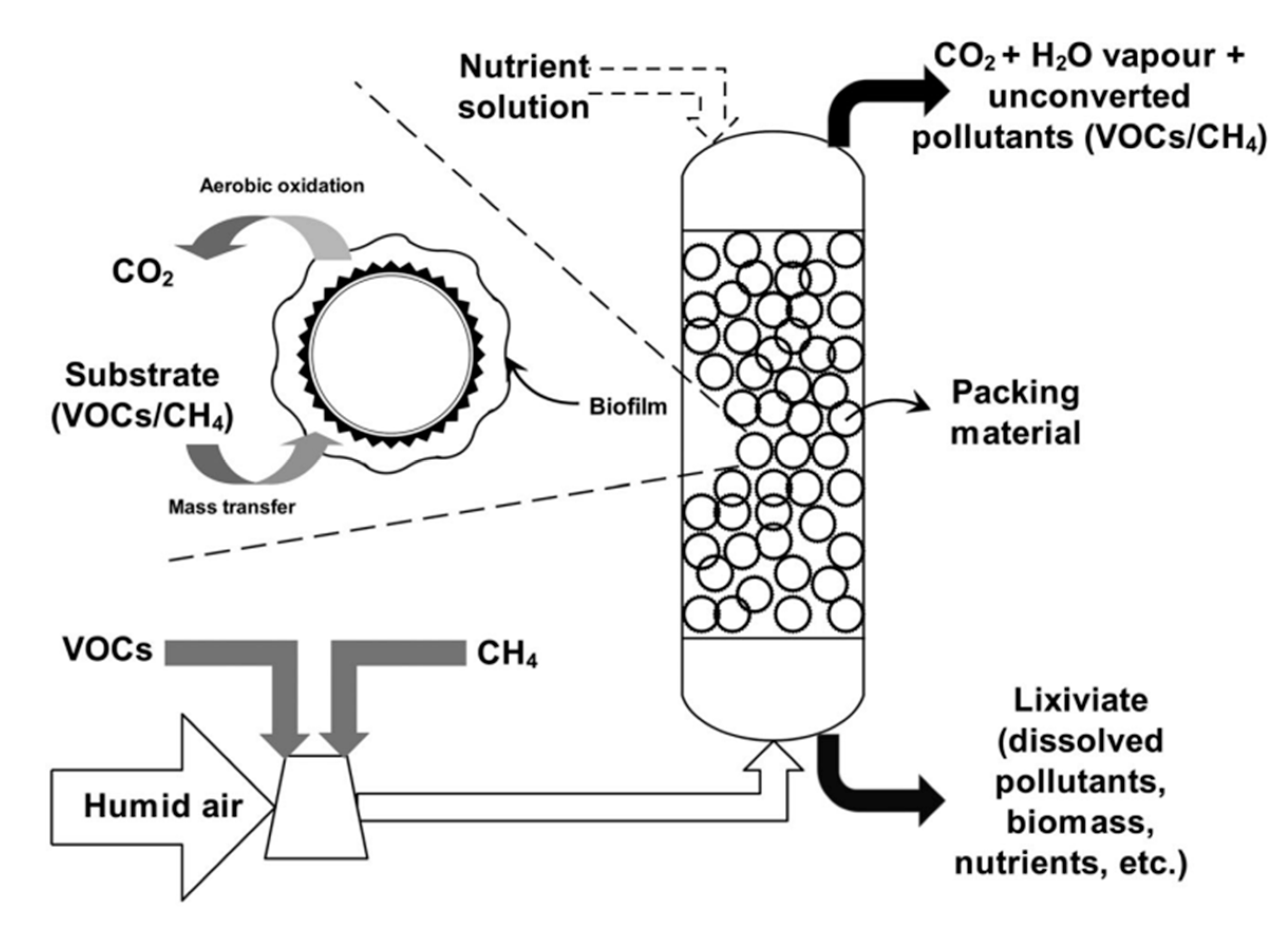
3. Biofiltration Processes
4. Microorganisms
4.1. Methanotrophs
4.2. Non-Methanotrophic Bacteria
4.3. Fungal Methane Removal
5. Influence of Salt Concentration (Sodium and Sulfur) on Methane Oxidation Activity
6. Influence of Temperature on Methane Oxidation Activity
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Duan, Z.; Scheutz, C.; Kjeldsen, P. Mitigation of methane emissions from three Danish landfills using different biocover systems. Waste Manag. 2022, 149, 156–167. [Google Scholar] [CrossRef]
- Lebrero, R.; Pérez, R.; Muñoz, R. Quantification of biofilter pressure drop: Influence of organic and inorganic loading. Bio Technol. 2011, 102, 3081–3087. [Google Scholar]
- Wilms, R.; Sass, H.; Köpke, B.; Cypionka, H.; Engelen, B. Methane and sulfate profiles within the sub-surface of a tidal flat are reflected by the distribution of sulfate-reducing bacteria and methanogenic archaea. FEMS Microbiol Ecol. 2007, 59, 611–621. [Google Scholar] [CrossRef]
- Strong, P.J.; Laycock, B.; Mahamud, S.N.S.; Jensen, P.D.; Lant, P.A.; Tyson, G.; Pratt, S. The opportunity for high-performance biomaterials from methane. Microorganisms 2016, 4, 11. [Google Scholar] [CrossRef]
- Wilshusen, J.H.; Hettiaratchi, J.P.A.; De Visscher, A.; Saint-Fort, R. Methane oxidation and formation of EPS in compost: Effect of oxygen concentration. Environ. Pollut. 2004, 129, 305–314. [Google Scholar] [CrossRef]
- Rose, J.L.; Mahler, C.F.; Izzo, R.L.D. Comparison of the methane oxidation rate in four media. Rev. Bras. Ciênc. Solo 2012, 36, 803–812. [Google Scholar] [CrossRef][Green Version]
- Duan, Z.; Hansen, P.O.C.R.; Scheutz, C.; Kjeldsen, P. Mitigation of methane and trace gas emissions through a large-scale active biofilter system at Glatved landfill, Denmark. Waste Manag. 2021, 126, 367–376. [Google Scholar] [CrossRef]
- Woertz, J.; van Heiningen, W.; van Eekert, M.; Kraakman, N.; Kinney, K.; van Groenestijn, J. Dynamic bioreactor operation: Effects of packing material and mite predation on toluene removal from off-gas. Appl. Microbiol. Biotechnol. 2002, 58, 690–694. [Google Scholar] [CrossRef]
- Verbeke, T.J.; Dedysh, S.N.; Dunfield, P.F. Methanotrophy in acidic environments, including northern wetlands. In Handbook of Hydrocarbon and Lipid Microbiology; Timmis, K.N., Ed.; Springer: Berlin/Heidelberg, Germany, 2018. [Google Scholar]
- Syed, R.; Saggar, S.; Tate, K.; Rehm, B.H.A.; Berben, P. Assessing the performance of floating biofilters for oxidation of methane from dairy effluent ponds. J. Environ. Qual. 2017, 46, 272–280. [Google Scholar] [CrossRef]
- Nikiema, J.; Bibeau, L.; Lavoie, J.; Brzezinski, R.; Vigneux, J.; Heitz, M. Biofiltration of methane: An experimental study. Chem. Eng. J. 2005, 113, 111–117. [Google Scholar] [CrossRef]
- Melse, R.W.; van der Werf, A.W. Biofiltration for mitigation of methane emission from animal husbandry. Environ. Sci. Technol. 2005, 39, 5460–5468. [Google Scholar] [CrossRef]
- Berger, J.; Fornes, L.V.; Ott, C.; Jager, J.; Wawra, B.; Zanke, U. Methane oxidation in a landfill cover with capillary barrier. Waste Manag. 2005, 25, 369–373. [Google Scholar] [CrossRef]
- du Plessis, C.A.; Strauss, J.M.; Sebapalo, E.M.T.; Riedel, K.-H.J. Empirical model for methane oxidation using a composted pine bark biofilter. Fuel 2003, 82, 1359–1365. [Google Scholar] [CrossRef]
- Hilger, H.A.; Wollum, A.G.; Barlaz, M.A. Landfill methane oxidation response to vegetation, fertilization, and liming. J. Environ. Qual. 2000, 29, 324–334. [Google Scholar] [CrossRef]
- De Visscher, A.; Schippers, M.; Van Cleemput, O. Short-term kinetic response of enhanced methane oxidation in landfill cover soils to environmental factors. Biol. Fertil. Soils 2001, 33, 231–237. [Google Scholar] [CrossRef]
- Zarei, M.; Bayati, M.R.; Ebrahimi-Nik, M.; Rohani, A.; Hejazi, B. Modelling the removal efficiency of hydrogen sulfide from biogas in a biofilter using multiple linear regression and support vector machines. J. Clean. Prod. 2023, 404, 136965. [Google Scholar] [CrossRef]
- Yao, X.Z.; Chu, Y.X.; Wang, C.; Li, H.J.; Kang, Y.R.; He, R. Enhanced removal of methanethiol and its conversion products in the presence of methane in biofilters. J. Clean. Prod. 2019, 215, 75–83. [Google Scholar] [CrossRef]
- Wu, X.; Lin, Y.; Wang, Y.; Wu, S.; Yang, C. Volatile organic compound removal via biofiltration: Influences, challenges, and strategies. Chem. Eng. J. 2023, 471, 144420. [Google Scholar] [CrossRef]
- Vikrant, K.; Kailasa, S.K.; Tsang, D.C.; Lee, S.S.; Kumar, P.; Giri, B.S.; Kim, K.H. Biofiltration of hydrogen sulfide: Trends and challenges. J. Clean. Prod. 2018, 187, 131–147. [Google Scholar] [CrossRef]
- Valenzuela-Heredia, D.; Aroca, G. Methane biofiltration for the treatment of a simulated diluted biogas emission containing ammonia and hydrogen sulfide. Chem. Eng. J. 2023, 469, 143704. [Google Scholar] [CrossRef]
- Choi, H.; Ryu, H.W.; Cho, K.S. Biocomplex textile as an alternative daily cover for the simultaneous mitigation of methane and malodorous compounds. Waste Manag. 2018, 72, 339–348. [Google Scholar] [CrossRef]
- Sun, M.T.; Zhao, Y.Z.; Yang, Z.M.; Shi, X.S.; Wang, L.; Dai, M.; Guo, R.B. Methane elimination using biofiltration packed with fly ash ceramsite as support material. Front. Bioeng. Biotechnol. 2020, 8, 351. [Google Scholar] [CrossRef]
- Sheoran, K.; Siwal, S.S.; Kapoor, D.; Singh, N.; Saini, A.K.; Alsanie, W.F.; Thakur, V.K. Air pollutants removal using biofiltration technique: A challenge at the frontiers of sustainable environment. ACS Eng. Au 2022, 2, 378–396. [Google Scholar] [CrossRef]
- Shareefdeen, Z. High-performance biofilters for air treatment applications. In From Biofiltration to Promising Options in Gaseous Fluxes Biotreatment; Elsevier: Amsterdam, The Netherlands, 2020; pp. 111–127. [Google Scholar]
- Schmitz, R.A.; Peeters, S.H.; Mohammadi, S.S.; Berben, T.; van Erven, T.; Iosif, C.A.; Pol, A. Simultaneous sulfide and methane oxidation by an extremophile. Nat. Commun. 2023, 14, 2974. [Google Scholar] [CrossRef]
- Schirmer, W.N.; Stroparo, E.C.; Capanema, M.A.; Mazur, D.L.; Jucá, J.F.T.; Martins, K.G. Biofiltration of Methane Fugitive Emissions from Landfills Using Scum from Municipal Wastewater Treatment Plants (Wwtp) as Alternative Substrate. J. Mater. Cycles Waste Manag. 2021, 24, 2041–2053. [Google Scholar] [CrossRef]
- Haj-Amor, Z.; Araya, T.; Kim, D.G.; Bouri, S.; Lee, J.; Ghiloufi, W.; Lal, R. Soil salinity and its associated effects on soil microorganisms, greenhouse gas emissions, crop yield, biodiversity and desertification: A review. Sci. Total Environ. 2022, 843, 156946. [Google Scholar] [CrossRef]
- Samanta, D.; Sani, R.K. Methane Oxidation via Chemical and Biological Methods: Challenges and Solutions. Methane 2023, 2, 279–303. [Google Scholar] [CrossRef]
- Chan, R.; Chiemchaisri, W.; Chiemchaisri, C. Exploring Effective Bio-Cover Materials for Mitigating Methane Emission at a Tropical Landfill. Appl. Sci. 2023, 13, 1990. [Google Scholar] [CrossRef]
- Rossi, E.; Pecorini, I.; Iannelli, R. Methane oxidation of residual landfill gas in a full-scale biofilter: Human health risk assessment of volatile and malodours compound emissions. Environ. Sci. Pollut. Res. 2021, 28, 24419–24431. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, A.A.; García-Aguilar, B.P.; Jones, J.P.; Heitz, M. Improvement of methane biofiltration by the addition of non-ionic surfactants to biofilters packed with inert materials. Process Biochem. 2012, 47, 76–82. [Google Scholar] [CrossRef]
- Pecorini, I.; Rossi, E.; Iannelli, R. Mitigation of methane, NMVOCs and odor emissions in active and passive biofiltration systems at municipal solid waste landfills. Sustainability 2020, 12, 3203. [Google Scholar] [CrossRef]
- Nikiema, J.; Brzezinski, R.; Heitz, M. Elimination of methane generated from landfills by biofiltration: A review. Rev. Environ. Sci. Bio Technol. 2007, 6, 261–284. [Google Scholar] [CrossRef]
- Muliyadi, M.; Purwanto, P.; Sumiyati, S.; Soeprobowati, T.R. Removal of Pollutants in Wastewater using Plastic-Based Media Biofiltration: A Meta-Analysis. Pollution 2023, 9, 421–432. [Google Scholar]
- Casas, M.E.; Guivernau, M.; Viñas, M.; Fernández, B.; Cáceres, R.; Biel, C.; Matamoros, V. Use of wood and cork in biofilters for the simultaneous removal of nitrates and pesticides from groundwater. Chemosphere 2023, 313, 137502. [Google Scholar] [CrossRef]
- Ménard, C.; Ramirez, A.A.; Nikiema, J.; Heitz, M. Biofiltration of methane and trace gases from landfills: A review. Environ. Rev. 2012, 20, 40–53. [Google Scholar] [CrossRef]
- Menard, C.; Ramirez, A.A.; Nikiema, J.; Heitz, M. Analysis of the effects of temperature, the amount of nutrient solution and the carbon dioxide concentration on methane biofiltration. Int. J. Sustain. Dev. Plan. 2011, 6, 312–324. [Google Scholar] [CrossRef]
- Limbri, H.; Gunawan, C.; Rosche, B.; Scott, J. Challenges to developing methane biofiltration for coal mine ventilation air: A review. Water Air Soil Pollut. 2013, 224, 1566. [Google Scholar] [CrossRef]
- Li, Y.; Tabassum, S.; Chu, C.; Zhang, Z. Inhibitory effect of high phenol concentration in treating coal gasification wastewater in anaerobic biofilter. J. Environ. Sci. 2018, 64, 207–215. [Google Scholar] [CrossRef]
- Li, D.Z.; Chen, L.; Liu, G.; Yuan, Z.Y.; Li, B.F.; Zhang, X.; Wei, J.Q. Porous metal–organic frameworks for methane storage and capture: Status and challenges. New Carbon Mater. 2021, 36, 468–496. [Google Scholar] [CrossRef]
- Larmola, T.; Rissanen, A.J.; Ojanen, P.; Stenberg, L.; Kohl, L.; Mäkipää, R. Mosses as biofilters for ditch methane emissions from forestry drained peatlands (No. EGU23-1382). In Proceedings of the Copernicus Meetings, EGU General Assembly, Vienna, Austria, 24–28 April 2023. [Google Scholar]
- Lai, D.; Huang, Z.; Xie, J.; Ai, X.; Xin, X.; Hong, J. Effects of filling methods on the degradation of ethyl acetate and the microbial community in biofilters. Process Saf. Environ. Prot. 2023, 174, 188–199. [Google Scholar] [CrossRef]
- La, H.; Hettiaratchi, J.P.A.; Achari, G.; Verbeke, T.J.; Dunfield, P.F. Biofiltration of methane using hybrid mixtures of biochar, lava rock and compost. Environ. Pollut. 2018, 241, 45–54. [Google Scholar] [CrossRef]
- La, H.; Hettiaratchi, J.P.A.; Achari, G.; Dunfield, P.F. Biofiltration of methane. Bioresour. Technol. 2018, 268, 759–772. [Google Scholar] [CrossRef]
- La, H.; Hettiaratchi, J.P.A.; Achari, G. The influence of biochar and compost mixtures, water content, and gas flow rate, on the continuous adsorption of methane in a fixed bed column. J. Environ. Manag. 2019, 233, 175–183. [Google Scholar] [CrossRef]
- Kriipsalu, M.; Somani, M.; Pehme, K.; Tamm, O.; Truu, J.; Truu, M.; Orupold, K. Performance of biocover in controlling methane emissions from landfill: A decade of full-scale investigation. Process Saf. Environ. Prot. 2023, 172, 486–495. [Google Scholar] [CrossRef]
- Kim, B.; Zhang, Y.G. Methane Index: Towards a quantitative archaeal lipid biomarker proxy for reconstructing marine sedimentary methane fluxes. Geochim. Cosmochim. Acta 2023, 354, 74–87. [Google Scholar] [CrossRef]
- Khabiri, B.; Ferdowsi, M.; Buelna, G.; Jones, J.P.; Heitz, M. Bioelimination of low methane concentrations emitted from wastewater treatment plants: A review. Crit. Rev. Biotechnol. 2022, 42, 450–467. [Google Scholar] [CrossRef] [PubMed]
- Kallistova, A.Y.; Koval, D.D.; Kadnikov, V.V.; Toshchakov, S.V.; Yusupov, S.K.; Izotova, A.O.; Pimenov, N.V. Methane Cycle in a Littoral Site of a Temperate Freshwater Lake. Microbiology 2023, 92, 153–170. [Google Scholar] [CrossRef]
- Josiane, N.; Michele, H. The influence of the gas flow rate during methane biofiltration on an inorganic packing material. Can. J. Chem. Eng. 2009, 87, 136–142. [Google Scholar] [CrossRef]
- Johannisson, J. Prospective Environmental Assessment of Technologies for Mitigating Methane Emissions. Doctoral Dissertation, Universität Ulm, Ulm, Germany, 2023. [Google Scholar]
- Jawad, J.; Khalil, M.J.; Sengar, A.K.; Zaidi, S.J. Experimental analysis and modeling of the methane degradation in a three stage biofilter using composted sawdust as packing media. J. Environ. Manag. 2021, 286, 112214. [Google Scholar] [CrossRef] [PubMed]
- Knief, C. Diversity and Habitat Preferences of Cultivated and Uncultivated Aerobic Methanotrophic Bacteria Evaluated Based on pmoA as Molecular Marker. Front. Microbiol. 2015, 6, 1346. [Google Scholar] [CrossRef]
- Gunasekera, S.S.; Hettiaratchi, J.P.; Bartholameuz, E.M.; Farrokhzadeh, H.; Irvine, E. A comparative evaluation of the performance of full-scale high-rate methane biofilter (HMBF) systems and flow-through laboratory columns. Environ. Sci. Pollut. Res. 2018, 25, 35845–35854. [Google Scholar] [CrossRef]
- Girard, M.; Ramirez, A.A.; Buelna, G.; Heitz, M. Biofiltration of methane at low concentrations representative of the piggery industry—Influence of the methane and nitrogen concentrations. Chem. Eng. J. 2011, 168, 151–158. [Google Scholar] [CrossRef]
- Frasi, N.; Rossi, E.; Pecorini, I.; Iannelli, R. Methane oxidation efficiency in biofiltration systems with different moisture content treating diluted landfill gas. Energies 2020, 13, 2872. [Google Scholar] [CrossRef]
- Fjelsted, L.; Scheutz, C.; Christensen, A.G.; Larsen, J.E.; Kjeldsen, P. Biofiltration of diluted landfill gas in an active loaded open-bed compost filter. Waste Manag. 2020, 103, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ferdowsi, M.; Khabiri, B.; Buelna, G.; Jones, J.P.; Heitz, M. Prolonged operation of a methane biofilter from acclimation to the failure stage. Environ. Technol. 2023, 105, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Hakobyan, A.; Liesack, W. Unexpected metabolic versatility among type II methanotrophs in the Alphaproteobacteria. Bio Chem. 2020, 401, 1469–1477. [Google Scholar] [CrossRef] [PubMed]
- Rani, V.; Kaushik, R.; Majumder, S.; Rani, A.T.; Devi, A.A.; Divekar, P.; Singh, J. Synergistic interaction of methanotrophs and methylotrophs in regulating methane emission. In Microbial Technology for Sustainable Environment; Springer: Singapore, 2021; pp. 419–437. [Google Scholar]
- Im, J.; Lee, S.W.; Yoon, S.; DiSpirito, A.A.; Semrau, J.D. Characterization of a novel facultative Methylocystis species capable of growth on methane, acetate and ethanol. Environ. Microbiol. Rep. 2011, 3, 174–181. [Google Scholar] [CrossRef]
- González-Cortés, J.J.; Quijano, G.; Ramírez, M.; Cantero, D. Methane concentration and bacterial communities’ dynamics during the anoxic desulfurization of landfill biogas under diverse nitrate sources and hydraulic residence times. J. Environ. Chem. Eng. 2023, 11, 109285. [Google Scholar] [CrossRef]
- He, L.; Groom, J.D.; Wilson, E.H.; Fernandez, J.; Konopka, M.C.; Beck, D.A.; Lidstrom, M.E. A methanotrophic bacterium to enable methane removal for climate mitigation. Proc. Natl. Acad. Sci. USA 2023, 120, e2310046120. [Google Scholar] [CrossRef]
- Amodeo, C.; Sofo, A.; Tito, M.T.; Scopa, A.; Masi, S.; Pascale, R.; Caniani, D. Environmental factors influencing landfill gas biofiltration: Lab scale study on methanotrophic bacteria growth. J. Environ. Sci. Health Part A 2018, 53, 825–831. [Google Scholar] [CrossRef]
- Khabiri, B.; Ferdowsi, M.; Benyoussef, E.H.; Malhautier, L.; Buelna, G.; Jones, J.P.; Heitz, M. Biofiltration of methane in presence of ethylbenzene or xylene. Atmos. Pollut. Res. 2022, 13, 101271. [Google Scholar]
- Arslan, M.; Ganiyu, S.O.; Lillico, D.M.; Stafford, J.L.; El-Din, M.G. Fate of dissolved organics and generated sulfate ions during biofiltration of oil sands process water pretreated with sulfate radical advanced oxidation process. Chem. Eng. J. 2023, 458, 141390. [Google Scholar] [CrossRef]
- La, H.; Hettiaratchi, J.P.A.; Achari, G.; Kim, J.J.; Dunfield, P.F. Investigation of biologically stable biofilter medium for methane mitigation by methanotrophic bacteria. J. Hazard. Toxic Radioact. Waste 2018, 22, 04018013. [Google Scholar] [CrossRef]
- Ménard, C.; Ramirez, A.A.; Nikiema, J.; Heitz, M. Effect of trace gases, toluene and chlorobenzene, on methane biofiltration: An experimental study. Chem. Eng. J. 2012, 204, 8–15. [Google Scholar] [CrossRef]
- Pecorini, I.; Iannelli, R. Landfill GHG reduction through different microbial methane oxidation biocovers. Processes 2020, 8, 591. [Google Scholar] [CrossRef]
- Pearce, D. Ecology of Methanotrophs in a Landfill Methane Biofilter. Doctoral Dissertation, University of East Anglia, Norwich, UK, 2023. [Google Scholar]
- Pachaiappan, R.; Cornejo-Ponce, L.; Rajendran, R.; Manavalan, K.; Femilaa Rajan, V.; Awad, F. A review on biofiltration techniques: Recent advancements in the removal of volatile organic compounds and heavy metals in the treatment of polluted water. Bioengineered 2022, 13, 8432–8477. [Google Scholar] [CrossRef] [PubMed]
- Sana, N.; Arnepalli, D.N.; Krishnan, C. A bio-augmented system with Methylosarcina sp. LC-4 immobilized on bio-carriers: Towards an integrated approach to mitigate and valorize methane emissions from landfills to biodiesel. Chemosphere 2023, 341, 139992. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.T.; Yang, Z.M.; Lu, J.; Fan, X.L.; Guo, R.B.; Fu, S.F. Improvement of bacterial methane elimination using porous ceramsite as biocarrier. J. Chem. Technol. Biotechnol. 2018, 93, 2406–2414. [Google Scholar] [CrossRef]
- Sun, M.T.; Yang, Z.M.; Fu, S.F.; Fan, X.L.; Guo, R.B. Improved methane removal in exhaust gas from biogas upgrading process using immobilized methane-oxidizing bacteria. Bioresour. Technol. 2018, 256, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Thomasen, T.B.; Scheutz, C.; Kjeldsen, P. Treatment of landfill gas with low methane content by biocover systems. Waste Manag. 2019, 84, 29–37. [Google Scholar] [CrossRef]
- Vergara-Fernández, A.; Morales, P.; Scott, F.; Guerrero, S.; Yañez, L.; Mau, S.; Aroca, G. Methane biodegradation and enhanced methane solubilization by the filamentous fungi Fusarium solani. Chemosphere 2019, 226, 24–35. [Google Scholar] [CrossRef]
- Visvanathan, C.; Pokhrel, D.; Cheimchaisri, W.; Hettiaratchi, J.P.A.; Wu, J.S. Methanotrophic activities in tropical landfill cover soils: Effects of temperature, moisture content and methane concentration. Waste Manag. Res. 1999, 17, 313–323. [Google Scholar] [CrossRef]
- Stein, V.B.; Hettiaratchi, J.P.A. Methane oxidation in three alberta soils: Influence of soil parameters and methane flux rates. Environ. Technol. 2001, 22, 101–111. [Google Scholar] [CrossRef]
- Dalcin Martins, P.; de Jong, A.; Lenstra, W.K.; van Helmond, N.A.; Slomp, C.P.; Jetten, M.S.; Rasigraf, O. Enrichment of novel Verrucomicrobia, Bacteroidetes, and Krumholzibacteria in an oxygen-limited methane-and iron-fed bioreactor inoculated with Bothnian Sea sediments. MicrobiologyOpen 2021, 10, e1175. [Google Scholar] [CrossRef]
- Scheutz, C.; Kjeldsen, P. Capacity for biodegradation of CFCs and HCFCs in a methane oxidative counter-gradient laboratory system simulating landfill soil covers. Environ. Sci. Technol. 2003, 37, 5143–5149. [Google Scholar] [CrossRef]
- Streese, J.; Stegmann, R. Microbial oxidation of methane from old landfills in biofilters. Waste Manag. 2003, 23, 573–580. [Google Scholar] [CrossRef]
- Buckley, D.H.; Baumgartner, L.K.; Visscher, P.T. Vertical distribution of methane metabolism in microbial mats of the Great Sippewissett salt marsh. Environ. Microbiol. 2008, 10, 967–977. [Google Scholar] [CrossRef]
- Chen, F.; Zheng, Y.; Hou, L.; Zhou, J.; Yin, G.; Liu, M. Denitrifying anaerobic methane oxidation in marsh sediments of Chongming eastern intertidal flat. Mar. Pollut Bullet. 2020, 150, 110681. [Google Scholar] [CrossRef]
- El Mashad, H.M.; Barzee, T.J.; Franco, R.B.; Zhang, R.; Kaffka, S.; Mitloehner, F. Anaerobic Digestion and Alternative Manure Management Technologies for Methane Emissions Mitigation on Californian Dairies. Atmosphere 2023, 14, 120. [Google Scholar] [CrossRef]
- Hettiarachchi, H.; Irandoost, E.; Hettiaratchi, J.P.; Pokhrel, D. A Field-Verified Model to Estimate Landfill Methane Flux Using Surface Methane Concentration Measurements under Calm Wind Conditions. J. Hazard. Toxic Radioact. Waste 2023, 27, 04023019. [Google Scholar] [CrossRef]
- Fedrizzi, F.; Cabana, H.; Ndanga, É.M.; Cabral, A.R. Biofiltration of methane from cow barns: Effects of climatic conditions and packing bed media acclimatization. Waste Manag. 2018, 78, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Al-Heetimi, O.T.; Van De Ven, C.J.; Van Geel, P.J.; Rayhani, M.T. Performance of Biocover Materials in Mitigating Methane Emissions from Landfills under Different Loading Rates. J. Environ. Eng. 2023, 149, 04023035. [Google Scholar] [CrossRef]
- Derviş, H. Bibliometric analysis using bibliometrix an R package. J. Scientometr. Res. 2019, 8, 156–160. [Google Scholar] [CrossRef]

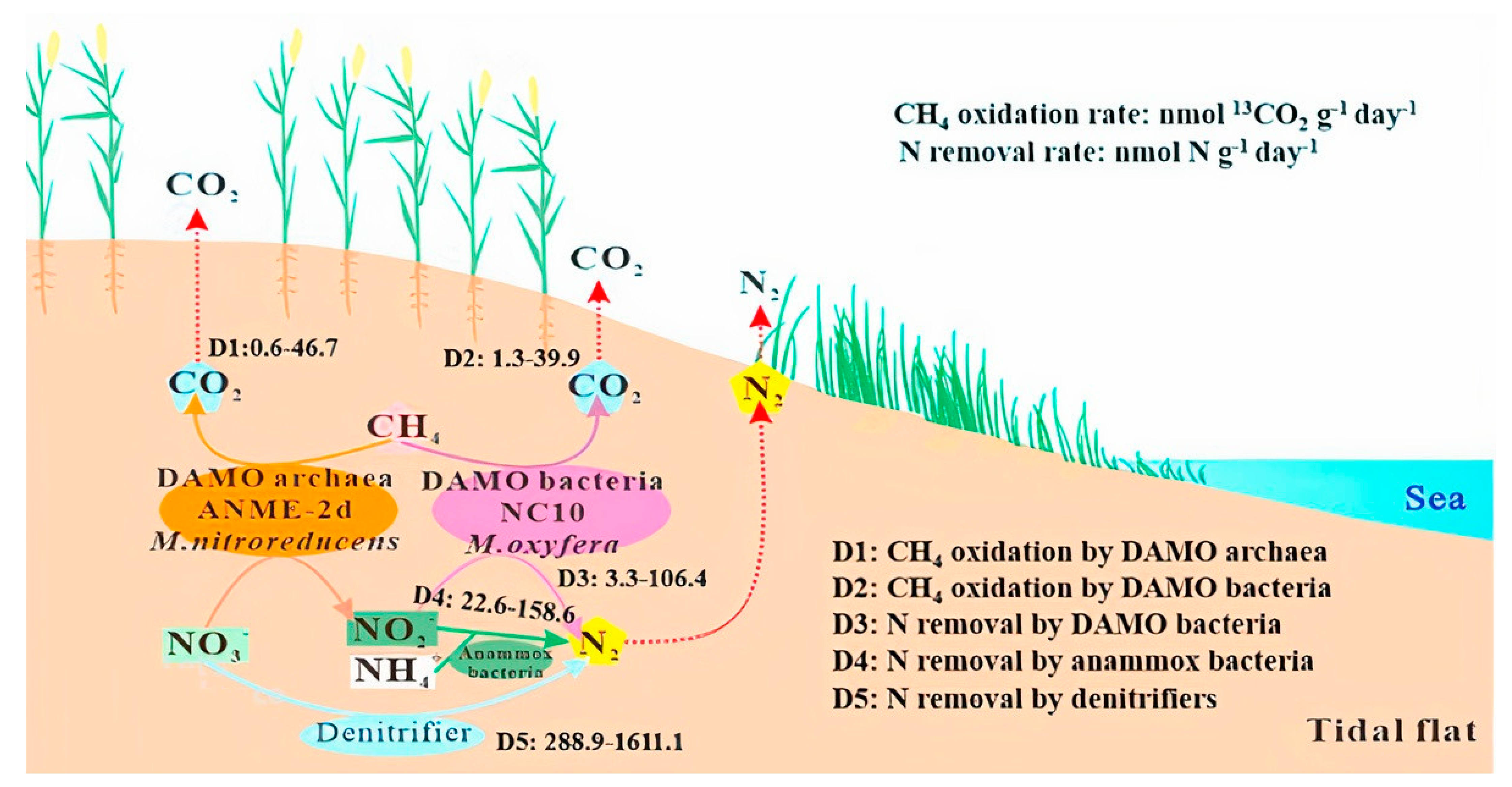
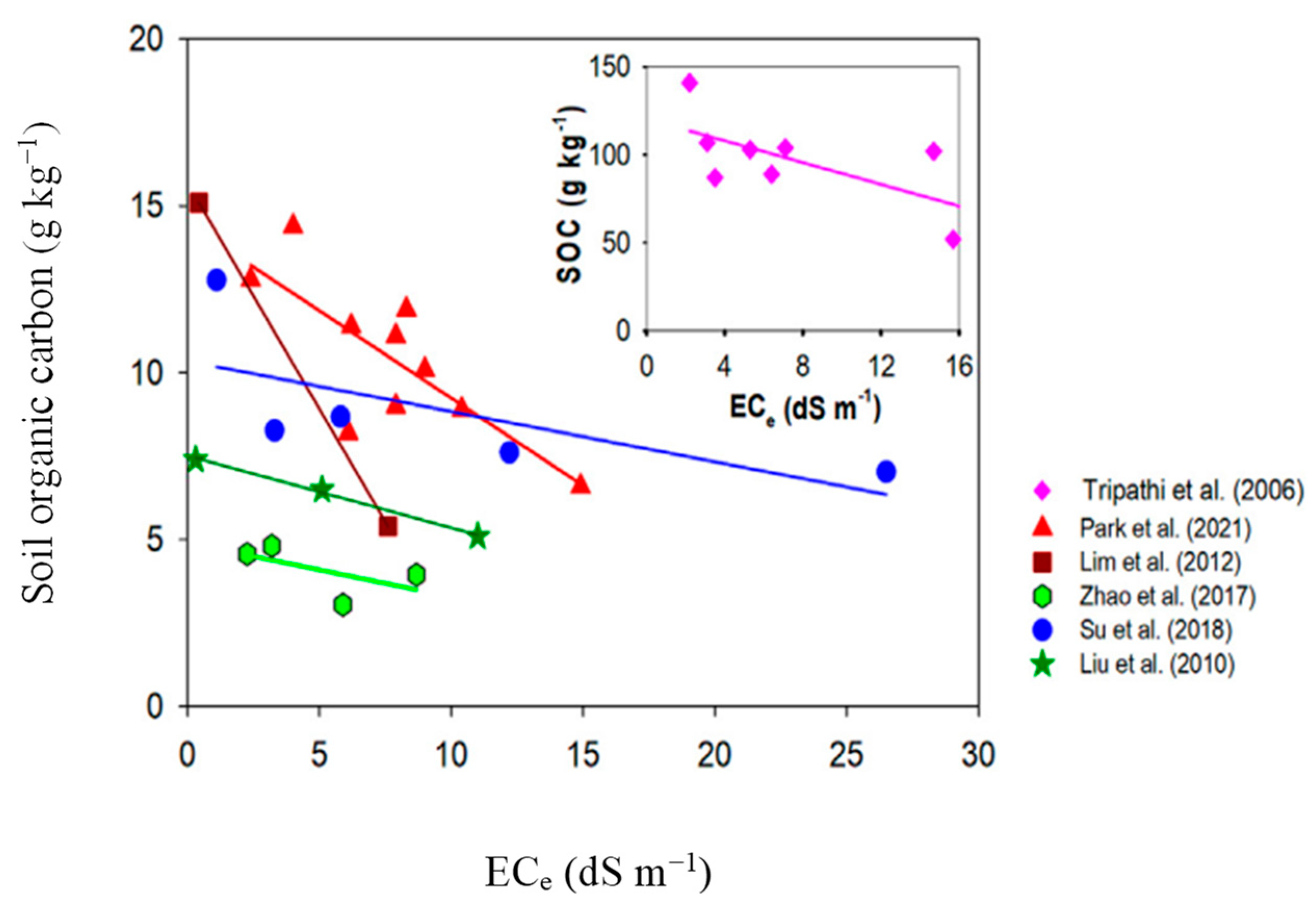

| Media | Nutrient Source | O2:CH4 | EBRT (min) | Maximum CH4 Conversion (%) | References |
|---|---|---|---|---|---|
| Landfill cover soil: coarse sand; clay top soil | NH4NO3, K2HPO4, and sewage sludge | 13:1 | 52.1 | 61% | [67] |
| Agricultural soil and landfill cover soil | Organic amendments: wheat straw and beet leaves | 30:1 | 7.8 | 78% | [68] |
| Soil (70% sand) | None | 21:1 | 92 to 93 | 83% | [69] |
| Landfill cover soil (closed landfill) | NH4Cl, KNO3, KCl | 11.7:1 | 103 | 36% | [70] |
| Alberta, Canada, soils | None | 2:1 | 43 | 50% | [71] |
| 2:1 compost and perlite | Nutrient solution | 12.5:1 | 57 to 1136 | >70% | [72] |
| Landfill cover soil | None | 25:1 | 40 to 45 | 40% | [73] |
| Compost and sand | None | 8:1 and 210:1 | 447 to 1162 | 88% | [74] |
| 40:60 (by volume) perlite and compost | None | 2.6:1 to 52:1 | 7 to 80 | 24% | [75] |
| Inorganic material and compost | Nutrient solution | 2:1 to 7:1 | 4.3 | 98% | [76] |
| Compost, wood fibres, peat, and mixture | None | 28:1 to 46:1 | 5.2 (bench) | 98% | [77] |
| Compost; recycling paper pellets | None | 28:1 to 30:1 | 2.4 (pilot) | 90% | [78] |
| 4:2:4 (ww) of compost, de-inking waste, and sand | None | 6:1 to 70:1 | 98 to 558 | 80% | [79] |
| Gravel | Nutrient solution | 10:1 | 495 to 515 | 85% | [80] |
| Stone | Nutrient solution | 2.5:1 | 4.2 | [81] | |
| Landfill cover soil and earth worm cast | None | 2.7:1 to 3.7:1 | 3.2 to 7.5 | 41% | [82] |
| Compost, 2:1 ceramsite and compost | None | 21:1 to 161:1 | 3.8 to 280 | 23% | [54] |
| gravel | Nutrient solution | 21:1 to 161:1 | 884 | 11% | [63] |
| Various: compost, sewage sludge, sand, soil, and Mixtures | None | 0.4:1, 1.7:1, 3.8:1 | 4.2 | 96% | [29] |
| Stones | Nutrient solution | Atmospheric diffusion | 310 | 97% | [50] |
| BTF: Clay spheres, Polypropylene sphere, Stones | Nutrient solution | 840:1 | 4.2 | 38% | [36] |
| 1:1 Perlite and volcanic pumice soils from landfill | Nutrient solution | 49:1 | 4.25 | >90% | [51] |
| cap | None | 2.5:1 | 4.2 | 100% | [42] |
| Pumice soils (top soil and subsoil) | None | 28.5:1 | >120 | 65% | [64] |
| Soil, compost and mixtures (1:1, 3:1 w/w) | None | 29:1 | 90 | 36% | [23] |
| 52.8% plastic waste and 47.2% stabilized organic Waste | Nutrient solution | 40:1 to 393:1 | 15.7 | Up to 100% | [17] |
| GAC and pumice | Mineral Salt Medium | 2:1 | 9425 to 28,274 | 65% | [29] |
| BTF: polyurethane foam | Nutrient solution | 3:1 to 206:1 | 20.1 | 50% | [27] |
| Perlite | Nutrient solution | 6:1 | 4 | 43% | [34] |
| Tobermolite | Nutrient solution | Atmospheric diffusion | 20 | Up to 100% | [18] |
| Mixture: wood pine bark chips, perlite, compost | None | 3.4:1 to 52.3:1 | 20 | 100% | [34] |
| Mixtures: compost (60%, v/v), burst furnace slag | Nutrient solution | 9:1 | 4.4 | 70–100% | [28] |
| Stone material | Mineral Salt Medium | 4:1 | 7.4 to 42.8 | Up to 97% | [18] |
| Compost and fungal strains | Nitrate Mineral Salts | 4:1 | 1 to 6 | 91–99% | [30] |
| Activated carbon, plastic bio-balls, gravel | None | 85:1 to 161:1 | 20 to 40 | 82.8% | [25] |
| Fungal strains and spores; bacterial consortium | None | 6.7:1 to 59.8:1 | 200 to 998 | 12% | [10] |
| 0:50 (v/v) volcanic pumic soil and perlite | Nitrate Mineral Salts | 210:1 | 0.42 (or 25 s) | 51.3% | [39] |
| BTF: Polyethylene rings | None | 161:1 | 50 | 33% | [47] |
| Compost | None | 123:1 | 10 to 100 | 62% | [14] |
| Compost | Nitrate Mineral Salts | 20:1 | 54 to 163 | 90% to 95% | [28] |
| Site Name | Sample Frequency a | Salinity (mS m−1) | Soil Surface Flooding b | Daily Flux Reported | Annual Flux Reported c | References |
|---|---|---|---|---|---|---|
| (mg CH4 m−2d−1) | (g CH4 m−2 yr−1) | |||||
| Fresh | 13 (17 mo) | 0.4 | not systematic | 213.3 | [74] | |
| Brackish | 13 (17 mo) | 1.8 | not systematic | 97.3 | [62] | |
| Salt | 13 (17 mo) | 18.1 | not systematic | 5.7 | [54] | |
| Creek bank | 16 (20 mo) | 18.7 | exposed | 1.2 | [33] | |
| High marsh | 13 (13 mo) | 22.6 | exposed | 0.4 | [20] | |
| Short Spartina | 21 (24 mo) | 26.3 | exposed | 1.3 | [72] | |
| Site 1 | 11 (12 mo) | 5.1 | not reported | 18.2 | [60] | |
| Site 2 | 11 (12 mo) | 12.8 | not reported | 22.4 | [30] | |
| Site 3 | 11 (12 mo) | 16.6 | not reported | 5.6 | [29] | |
| GI Near Bank | 8 (13 mo) | 0.25 | exposed | 8.2 | [52] | |
| GI Far Bank | 8 (13 mo) | 0.25 | exposed | 5.7 | [38] | |
| UF Near Bank | 8 (13 mo) | 0.25 | exposed | 5.1 | [40] | |
| UF Far Bank | 8 (13 mo) | 0.25 | exposed | 3.5 | [22] | |
| Upland edge | 6 (1.5 mo) | 23.5 | not systematic | 3.7 | [12] | |
| High marsh | 6 (1.5 mo) | 31.6 | not systematic | 0.5 | [26] | |
| Middle marsh | 6 (1.5 mo) | 33.7 | not systematic | 0.6 | [82] | |
| Low marsh | 6 (1.5 mo) | 35.1 | not systematic | 0.6 | [30] | |
| Scirpus Close | 68 (24 mo) | 2.5 | exposed | 4.5 | [11] | |
| Phragmites Far | 68 (24 mo) | 2.5 | exposed | 75.4 | [29] | |
| Sweet Hall | 8 (15 mo) | 0.25 | exposed | 96.0 | [32] | |
| Lower site | 17 (20 mo) | 0.25 | exposed | 1.3 | [81] | |
| Upper site | 16 (20 mo) | 0.25 | exposed | 1.8 | [55] | |
| Alresford Creek | 12 (12 mo) | 0.25 | not reported | 0.3 | [38] | |
| Colne Point | 12 (12 mo) | 33.0 | not reported | 0.4 | [62] | |
| C3 Ambient CO2 | 14 (24 mo) | 6.8 | exposed | 13.9 | [57] | |
| C4 Ambient CO2 | 7 (24 mo) | 6.8 | exposed | 9.6 | [36] | |
| Salt marsh | - | - | flooded | 600.0 | [84] | |
| Salt marsh 24-h Day | - | 2.1 | flooded | 2365.7 | [34] | |
| CD Marsh | - | - | exposed | 1585.8 | 14.4 | [21] |
| CD Marsh 24-h Day | 9 (12 mo) | 5.5 | exposed | 13.8 | [30] | |
| Wildlife | 6 (6 mo) | 11.6 | exposed | 90.0 | 14.1 | [17] |
| Barbados | 6 (6 mo) | 12.9 | exposed | 94.0 | [54] | |
| Shanyutan wetland | - | 2.3 | exposed | 122.4 | [41] | |
| Shanyutan wetland | - | 4.2 | flooded | 48.0 | [68] | |
| Shanyutan wetland | - | 2.3 | exposed | 112.8 | [71] | |
| All flood stages | 10 (12 mo) | 2.3 | flooded + exposed | - | 32.6 | [64] |
| Experiment Type | Treatment Detail | Observation (GHG Emissions) | Key Findings and Reasoning | References |
|---|---|---|---|---|
| Pot experiment Initial soil pH = 7.8, EC = 5.6 dS m−1, OC = 1.48% | 25 nM salinity. 25 nM + phosphogypsum | Biochar amendment to saline soil reduced CH4 emission by 16.4% (25 mM) to 19.6% (at 75 mM) | Phospho-gypsum and biochar mitigate CH4 emission due to improved soil redox potential (Eh), increased SO42− and decreased soil EC. | [62] |
| Field experiment growing rice conducted in Jiangsu Province, China | N1 (300 kg N ha−1). N1 + humic acid. N1 + gypsum. N1 + humic acid + gypsum | CH4 emissions increased with Humic acid (6.2%), gypsum (19.4%), decreased with gypsum + humic acid (27.3%). Humic acid and gypsum application increase N2O emission | Humic acid and gypsum application with N300 kg ha−1 is the better management for coastal saline soils of China to mitigate CH4 emission. | [50] |
| Field experiment with rice. | No by-product gypsum fertiliser (BGF); BGF (2 Mg ha−1); BGF (4 Mg ha−1); BGF (8 Mg ha−1) | CH4 flux decreased with increasing level of BGF, and BGF (8 Mg ha−1) reduced it by 60.6% compared to control. | BGF application could be a better management practice for CH4 mitigation from paddy soils. | [42] |
| Field experiment with rice in upland soil. | Urea (250 kg ha−1). Urea + Phosphogypsum (90 kg ha−1). Urea + silicate slag (150 kg ha−1) | Silicate slag and phosphogypsum reduced CH4 emission by 18.0–23.5% and 14.7–18.6%, respectively. | Silicate slag and phosphogypsum decreased CH4 due to high free iron oxide and SO42− content which acted as electron acceptors | [30] |
| Field experiment with rice. | Urea (165 kg N ha−1); Urea + gypsum (6.60 t ha−1) | The CH4 emissions from gypsum amended plots were reduced by 55–70% compared to non-amended plots. | Inhibition of methanogenesis by sulfate-reducing bacteria caused a reduction in CH4 emission. | [64] |
| Field experiment with rice | N1 (300 kg N ha−1). N1 + 20 t biochar ha−1. N1 + 40 t biochar ha−1 | Biochar amendment increased N2O emissions b 13.7–38.1% and had no significant effects on CH4 emissions | Thus, long-term observations are needed to evaluate the environmental impacts of biochar and N fertilisers | [51] |
| 30 days incubation experiment | Control; Biochar | Biochar amendment to saline soil decrease CH4 uptake (8.8%), CO2 (11.9%), and N2O (9.8%) emissions | Biochar amendment to soils mitigates GHG emissions where CO2 and N2O are driven by soil rewetting events. | [63] |
| Rice experiment in irrigated saline soils of Gadakujang (a fishing hamlet) of coastal Odisha, India | Prilled urea (40 kg N ha−1); Sesbania green manure (5 Mg ha−1) + Prilled urea (20 kg N ha−1). Ipomoea lacunose (5 Mg ha−1) + Prilled urea (28 kg N ha−1) | Sesbania and Ipomoea lacunose green manure reduced CH4 emission by 23.2 and 29.9%. | Locally available Ipomoea lacunose green manure ca use CH4 mitigation and yield enhancement from the coastal saline rice ecosystems | [71] |
| Field experiment with rice | GM (S. Rostrata: 20 t ha−1) + urea (30 kg urea ha−1); GM + urea + gypsum (6.60 t ha−1) | Green manure addition enhances CH4 emissions by 10 times than that of urea application alone, further gypsum addition reduced CH4 emission by about 71.1% | Database for CH4 emissions mitigation from rice grow on high-sulfate containing soils | [36] |
| Field experiment was conducted in saline sodic soils in the upper Yellow River basin, Northwest China | Organic fertiliser (CK), sheep manure (FYM), lignite bioorganic fertiliser (LBF1) (1.5 t ha−1) LBF2 (3 t ha−1), LBF3 (4.5 t ha−1), and LBF4 (7.5 t ha−1) | LBF treatments decreased CH4 and CO2 and increasing N2O emissions beyond 3 t ha−1 application rate. FYM acted as a CH4 source, and LBF2 and LBF3 treatments acted as CH4 sinks | The application of lignite bioorganic fertiliser at 3.0–4.5 t ha−1 is appropriate for GHG mitigation in saline-sodic farmlands | [27] |
| Microcosm experiments of 80 days incubation | Interaction of salinity (0 and 1.2% salt) with biochar | 5–10 times higher N2O emissions occurred from saline soils than that from non-saline soils. Aged biochar decreased N2O emissions and increased CO2 emissions in saline soils. | Aged biochar could be a better option for mitigation of N2O emissions from saline soils | [73] |
| Field experiment with rice crop | Nonsaline (NS) soi; NS soil + DMPP (0.8% w/w of N); low saline (LS) soil; LS soil + DMPP; high saline (HS) soil; HS soil + DMPP | The nitrification inhibitor DMPP (3,4-dimethyl pyrazole phosphate) reduced cumulative N2O emissions by 61% in non-saline soil and by 75% in low saline soil. | DMPP offsets low salinity-induced high N2O emissions by inhibiting ammonia oxidation. | [19] |
| Temperature | Specific temperature | CH4 Concentration | Ecosystem | Molecular Biomarker | Genus/Species/Type of Methanotroph | References |
|---|---|---|---|---|---|---|
| (°C) | (°C) | (%) | ||||
| 6–70 | 6 | 5 | Landfill cover soil | Shotgun sequencing, 16S rRNA | Methylobacter luteus, Methylobacter tundripaludum, Methylotenera | [38] |
| 6–70 | 23 | Methylobacter luteus, Methylocystis | ||||
| 6–70 | 30 | Methylobacter luteus, Methylovorus glucosetrophus | ||||
| 6–70 | 40 | Methylocaldum sp. SAD2, Methylocaldum sp.14B | ||||
| 6–70 | 50 | Methylocaldum Szegediense | ||||
| 5–45 | 5, 10, 15, 25, 35 | 4 | Rice field and forest soil | TRFLP-pmoA gene | Methylobacter Methylococcus/ Methylocaldum Methylocystis/ Methylosinus Methylomonas Methanica | [41] |
| 3–20 | 5, 10 | 5 | Landfill cover soil | PLFA | Type-I methanotrophs | [75] |
| 3–20 | 20 | Type-II methanotrophs | ||||
| 4–21 | 4 | 10 | Arctic lake | DNA-SIP | Methylophilus, Methylobacter | [32] |
| 4–21 | 10 | Methylobacter, Methylomonas, Methylosoma | ||||
| 4–21 | 21 | Methylocystis, Methylophilus, Methylobacter, Methylomonas | ||||
| 5–40 | 5 | 5–50 | Landfill cover soil | 16S rRNA gene analysis (DGGE) | Methylotenera versatilis | [63] |
| 5–40 | 10 | Methylobacter tundripaludum, Methylovorus glucosetrophus Methylocella tundrae, Methylobacter marinus, Methylosinus Sporium | ||||
| 5–40 | 20 | Methylobacter marinus, Methylobacter luteus, Methylobacter tundripaludum, Methylosinus trichosporium, Methylosinus Sporium | ||||
| 5–40 | 40 | Methylocaldum Gracile | ||||
| 4–20 | 4 | - | Hydrocarbon contaminated aquifer | FISH, TRFLP-pmoA gene | Methylococcaceae, Methylobacteriaceae sp., Methylomonas sp. | [51] |
| 4–20 | 12 | Methylococcaceae, Methylobacteraceae sp. | ||||
| 4–20 | 20 | Methylocystis sp., Methylococcaceae, Methylobacteraceae sp. | ||||
| 7.5–9.5 | - | - | Tundra bog soil | Immunofuorescence | Methylomonas, Methylobacter, Methylococcus, Methylocystis, Methylosinus | [70] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmadi, F.; Bodraya, T.; Lackner, M. Methane Biofiltration Processes: A Summary of Biotic and Abiotic Factors. Methane 2024, 3, 122-148. https://doi.org/10.3390/methane3010008
Ahmadi F, Bodraya T, Lackner M. Methane Biofiltration Processes: A Summary of Biotic and Abiotic Factors. Methane. 2024; 3(1):122-148. https://doi.org/10.3390/methane3010008
Chicago/Turabian StyleAhmadi, Fatemeh, Tatiana Bodraya, and Maximilian Lackner. 2024. "Methane Biofiltration Processes: A Summary of Biotic and Abiotic Factors" Methane 3, no. 1: 122-148. https://doi.org/10.3390/methane3010008
APA StyleAhmadi, F., Bodraya, T., & Lackner, M. (2024). Methane Biofiltration Processes: A Summary of Biotic and Abiotic Factors. Methane, 3(1), 122-148. https://doi.org/10.3390/methane3010008







