Genetical and Biochemical Basis of Methane Monooxygenases of Methylosinus trichosporium OB3b in Response to Copper
Abstract
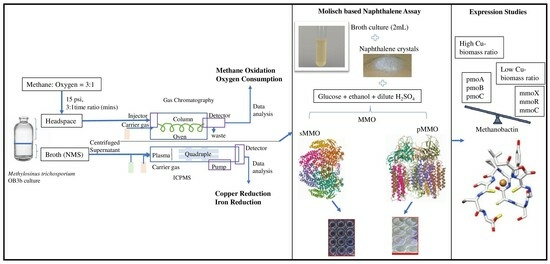
1. Introduction
2. Results and Discussion
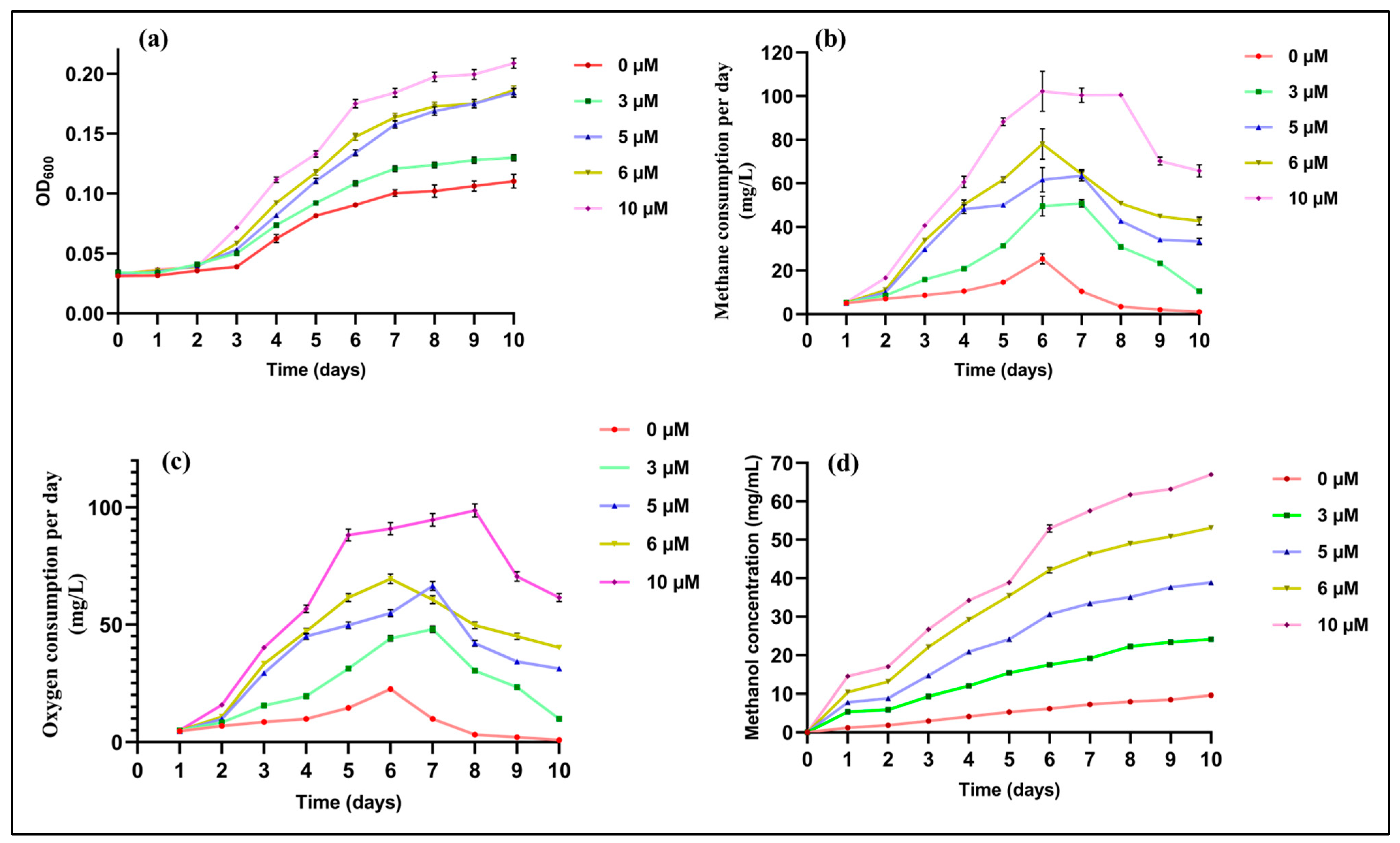
2.1. Effects of Cu on Growth and Methane Utilization
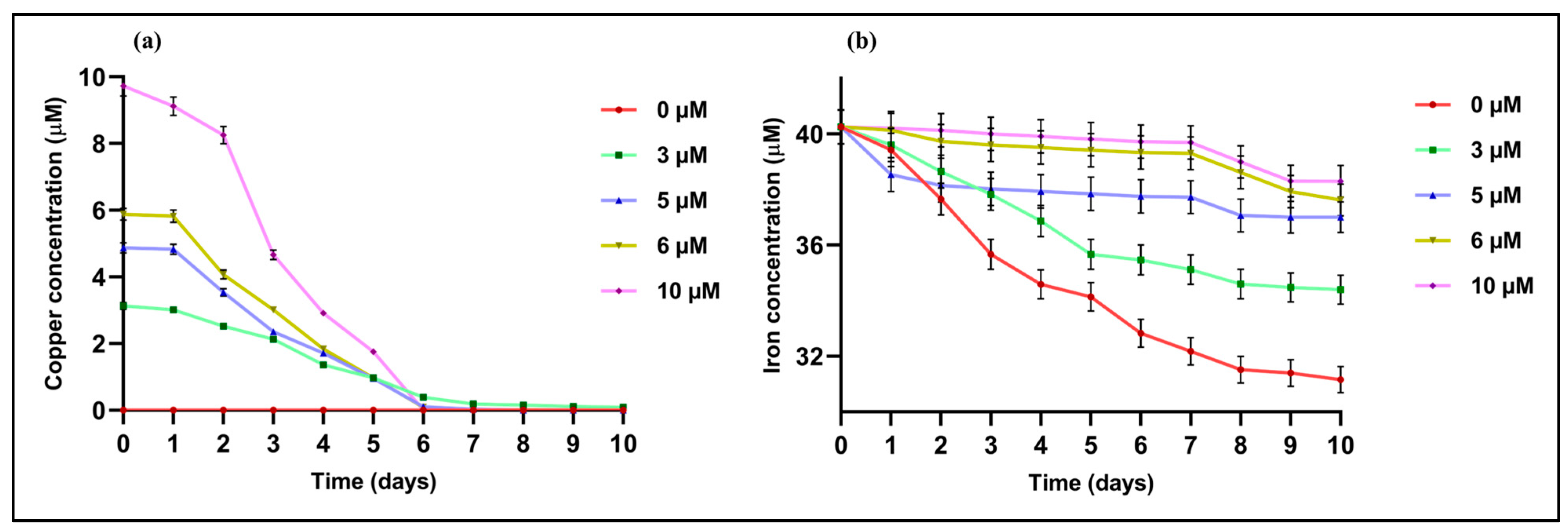
2.2. Effects of Cu Concentration on Metal (Cu and Fe) Uptake Rates
2.3. Effects of Metals on the Expression of pMMO and sMMO
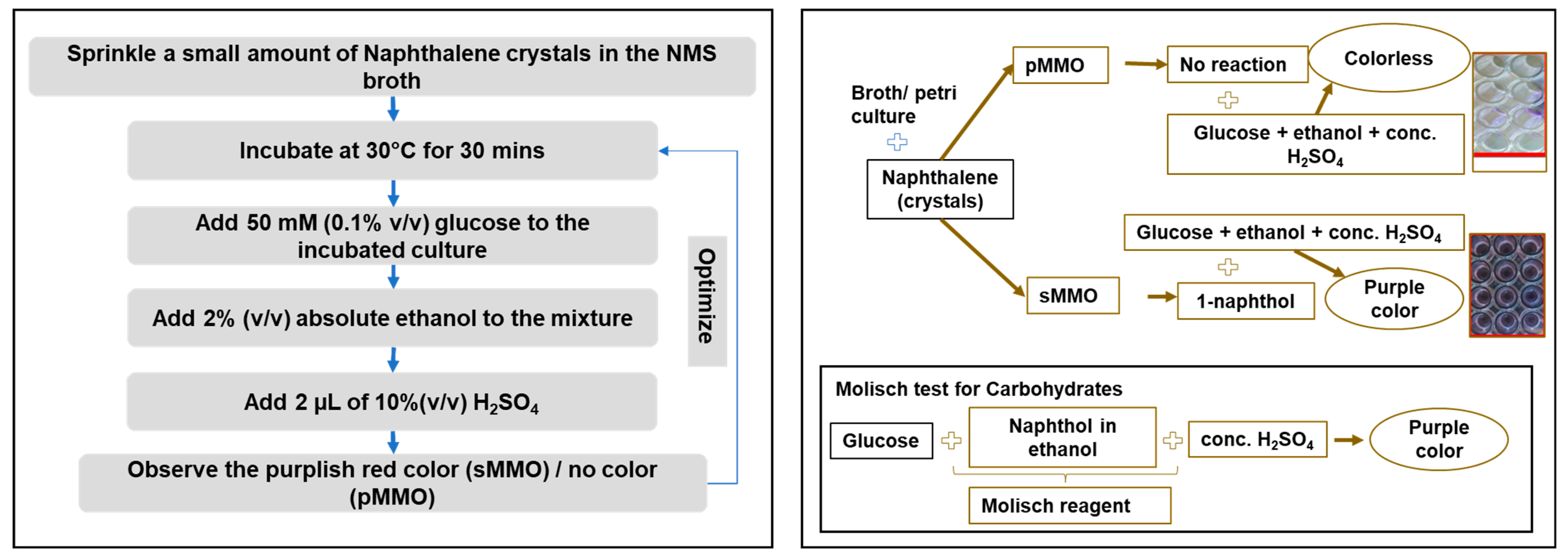
2.4. Molisch-Based Naphthalene Assay
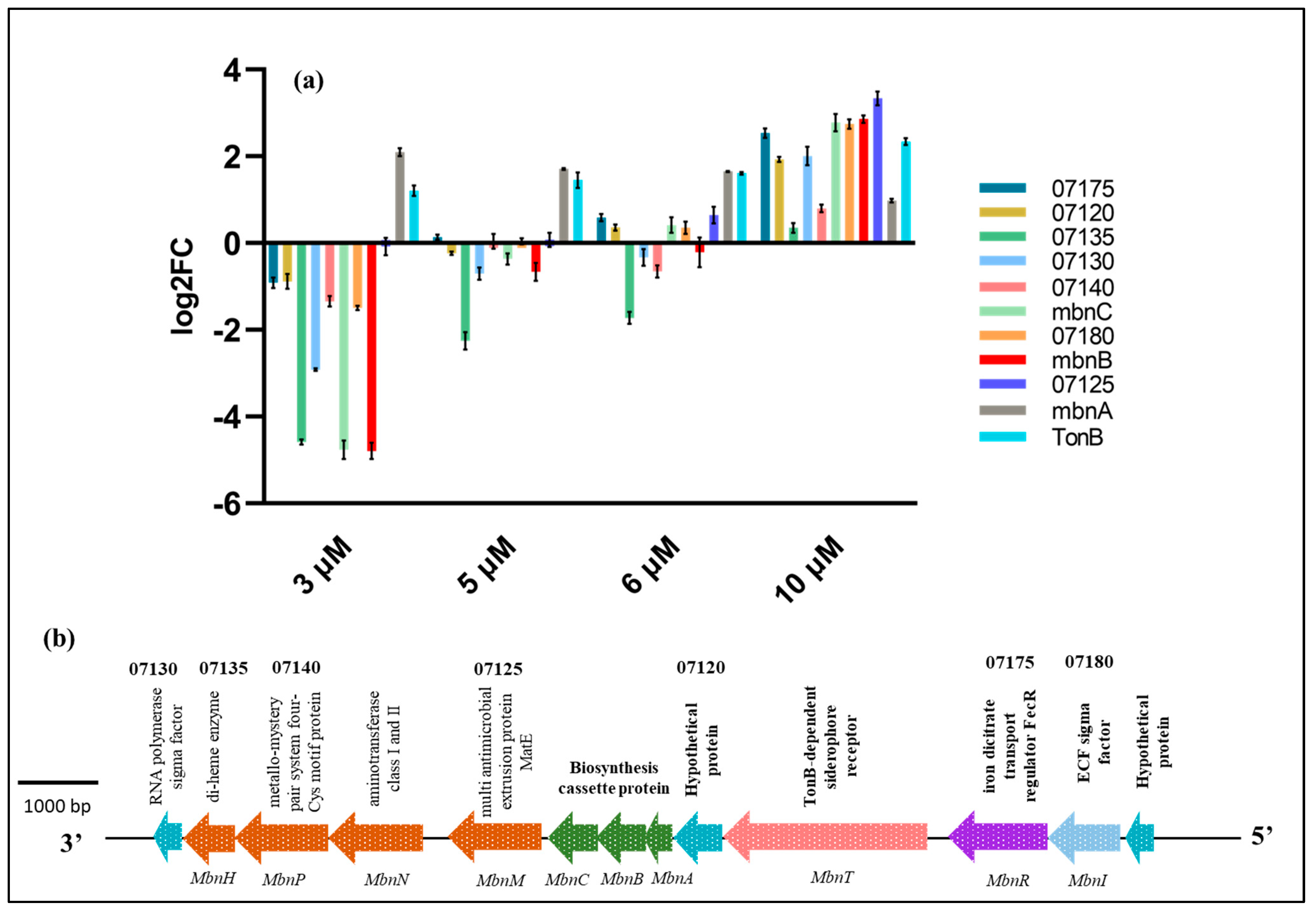
2.5. Effects of Cu on the Expression of Methanobactin Operon Genes
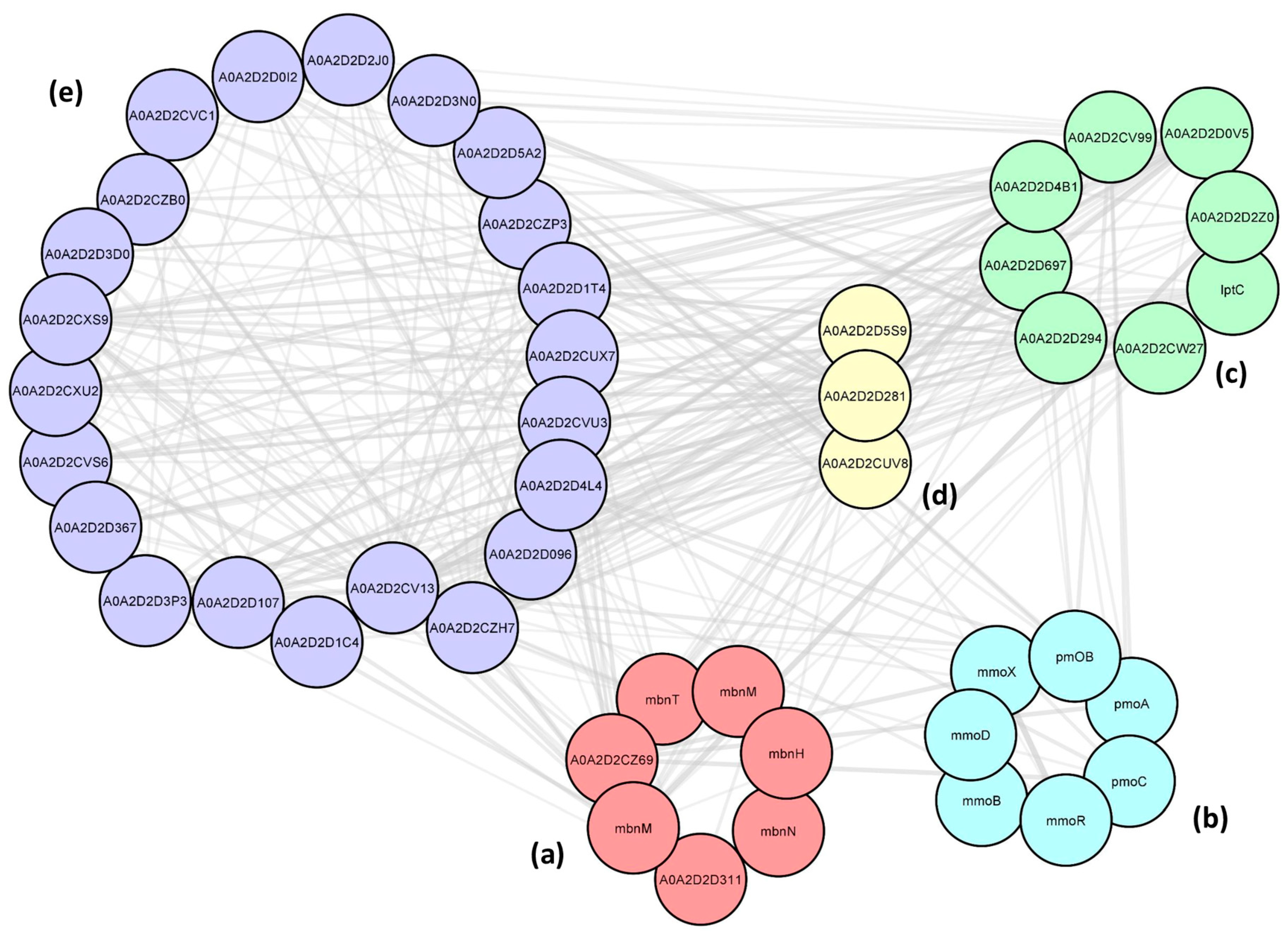
2.6. Protein–Protein Interactions (PPIs) between the Cu-Associated Genes
3. Materials and Methods
3.1. Growth Conditions
3.2. Measurement of Consumption Rates of the Gases
3.3. Measurement of Extracellular Methanol
3.4. Stoichiometry Calculations
3.5. Profiling of Copper and Iron Concentrations
3.6. Naphthalene–Ortho-Dianisidine Assay
3.7. Quantitative Molisch-Based Naphthalene Assay
3.8. Total RNA Isolation from Whole Cells
3.9. qPCR Gene Expression
3.10. Protein–Protein Interaction Using STRING Database and Cytoscape
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sahoo, K.K.; Goswami, G.; Das, D. Biotransformation of Methane and Carbon Dioxide into High-Value Products by Methanotrophs: Current State of Art and Future Prospects. Front. Microbiol. 2021, 12, 520. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Cruz, S.; Vaksmaa, A.; Horn, M.A.; Niemann, H.; Pijuan, M.; Ho, A. Methanotrophs: Discoveries, environmental relevance, and a perspective on current and future applications. Front. Microbiol. 2021, 12, 1057. [Google Scholar] [CrossRef] [PubMed]
- Lawton, T.J.; Rosenzweig, A.C. Methane-oxidizing enzymes: An upstream problem in biological gas-to-liquids conversion. J. Am. Chem. Soc. 2016, 138, 9327–9340. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.-W.; Kunz, R.C.; Boyd, E.S.; Semrau, J.D.; Antholine, W.E.; Han, J.-I.; Zahn, J.A.; Boyd, J.M.; de la Mora, A.M.; DiSpirito, A.A. The membrane-associated methane monooxygenase (pMMO) and pMMO-NADH: Quinone oxidoreductase complex from Methylococcus capsulatus Bath. J. Bacteriol. 2003, 185, 5755–5764. [Google Scholar] [CrossRef]
- Haque, M.F.U.; Xu, H.-J.; Murrell, J.C.; Crombie, A. Facultative methanotrophs—Diversity, genetics, molecular ecology and biotechnological potential: A mini-review. Microbiology 2020, 166, 894. [Google Scholar]
- Smith, T.; Dalton, H. Biocatalysis by methane monooxygenase and its implications for the petroleum industry. Stud. Surf. Sci. Catal. 2004, 151, 177–192. [Google Scholar]
- Semrau, J.D.; DiSpirito, A.A.; Yoon, S. Methanotrophs and copper. FEMS Microbiol. Rev. 2010, 34, 496–531. [Google Scholar] [CrossRef]
- Chidambarampadmavathy, K.; Obulisamy, P.; Heimann, K. Role of copper and iron in methane oxidation and bacterial biopolymer accumulation. Eng. Life Sci. 2015, 15, 387–399. [Google Scholar] [CrossRef]
- Semrau, J.D.; DiSpirito, A.A.; Gu, W.; Yoon, S. Metals and methanotrophy. Appl. Environ. Microbiol. 2018, 84, e02289-17. [Google Scholar] [CrossRef]
- Fru, E.C. Copper biogeochemistry: A cornerstone in aerobic methanotrophic bacterial ecology and activity? Geomicrobiol. J. 2011, 28, 601–614. [Google Scholar] [CrossRef]
- Lontoh, S.T. Substrate Oxidation by Methanotrophs Expressing Particulate Methane Monooxygenase (pMMO): A Study of Whole-Cell Oxidation of Trichloroethylene and Its Potential Use for Environmental Remediation; University of Michigan: Ann Arbor, MI, USA, 2000. [Google Scholar]
- Lontoh, S.; Semrau, J.D. Methane and trichloroethylene degradation by Methylosinus trichosporium OB3b expressing particulate methane monooxygenase. Appl. Environ. Microbiol. 1998, 64, 1106–1114. [Google Scholar] [CrossRef] [PubMed]
- Morton, J.D.; Hayes, K.F.; Semrau, J.D. Effect of copper speciation on whole-cell soluble methane monooxygenase activity in Methylosinus trichosporium OB3b. Appl. Environ. Microbiol. 2000, 66, 1730–1733. [Google Scholar] [CrossRef]
- Gilbert, B.; McDonald, I.R.; Finch, R.; Stafford, G.P.; Nielsen, A.K.; Murrell, J.C. Molecular analysis of the pmo (particulate methane monooxygenase) operons from two type II methanotrophs. Appl. Environ. Microbiol. 2000, 66, 966–975. [Google Scholar] [CrossRef] [PubMed]
- Ross, M.O.; MacMillan, F.; Wang, J.; Nisthal, A.; Lawton, T.J.; Olafson, B.D.; Mayo, S.L.; Rosenzweig, A.C.; Hoffman, B.M. Particulate methane monooxygenase contains only mononuclear copper centers. Science 2019, 364, 566–570. [Google Scholar] [CrossRef] [PubMed]
- Hakemian, A.S.; Kondapalli, K.C.; Telser, J.; Hoffman, B.M.; Stemmler, T.L.; Rosenzweig, A.C. The metal centers of particulate methane monooxygenase from Methylosinus trichosporium OB3b. Biochemistry 2008, 47, 6793–6801. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Shen, X.; Yao, L.; Bentalib, A.; Peng, Z. Active sites in heterogeneous catalytic reaction on metal and metal oxide: Theory and practice. Catalysts 2018, 8, 478. [Google Scholar] [CrossRef]
- Samanta, D.; Govil, T.; Salem, D.R.; Krumholz, L.R.; Gerlach, R.; Gadhamshetty, V.; Sani, R.K. Methane Monooxygenases: Their Regulations and Applications in Biofuel Production. In Microbes for Sustainable Development and Bioremediation; CRC Press: Boca Raton, FL, USA, 2019; pp. 187–206. [Google Scholar]
- Samanta, D.; Govil, T.; Saxena, P.; Gadhamshetty, V.; Krumholz, L.R.; Salem, D.R.; Sani, R.K. Enhancement of Methane Catalysis Rates in Methylosinus trichosporium OB3b. Biomolecules 2022, 12, 560. [Google Scholar] [CrossRef]
- Peng, P.; Yang, J.; DiSpirito, A.A.; Semrau, J.D. MmoD regulates soluble methane monooxygenase and methanobactin production in Methylosinus trichosporium OB3b. Appl. Environ. Microbiol. 2023, 89, e01601-23. [Google Scholar] [CrossRef]
- Gu, W.; Farhan Ul Haque, M.; Semrau, J.D. Characterization of the role of copCD in copper uptake and the ‘copper-switch’in Methylosinus trichosporium OB3b. FEMS Microbiol. Lett. 2017, 364, fnx094. [Google Scholar] [CrossRef]
- Kenney, G.E.; Sadek, M.; Rosenzweig, A.C. Copper-responsive gene expression in the methanotroph Methylosinus trichosporium OB3b. Metallomics 2016, 8, 931–940. [Google Scholar] [CrossRef]
- Semrau, J.D.; DiSpirito, A.A.; Obulisamy, P.K.; Kang-Yun, C.S. Methanobactin from methanotrophs: Genetics, structure, function and potential applications. FEMS Microbiol. Lett. 2020, 367, fnaa045. [Google Scholar] [CrossRef]
- Kenney, G.E.; Rosenzweig, A.C. Chalkophores. Annu. Rev. Biochem. 2018, 87, 645–676. [Google Scholar] [CrossRef]
- Kenney, G.E.; Rosenzweig, A.C. Genome mining for methanobactins. BMC Biol. 2013, 11, 17. [Google Scholar] [CrossRef]
- Sirajuddin, S.; Rosenzweig, A.C. Enzymatic oxidation of methane. Biochemistry 2015, 54, 2283–2294. [Google Scholar] [CrossRef]
- Aimen, H.; Khan, A.S.; Kanwal, N. Methanotrophs: The natural way to tackle greenhouse effect. J. Bioremediat. Biodegrad. 2018, 9, 432. [Google Scholar] [CrossRef]
- Park, S.; Hanna, L.; Taylor, R.T.; Droege, M.W. Batch cultivation of Methylosinus trichosporium OB3b. I: Production of soluble methane monooxygenase. Biotechnol. Bioeng. 1991, 38, 423–433. [Google Scholar] [CrossRef]
- Hwang, I.Y.; Hur, D.H.; Lee, J.H.; Park, C.-H.; Chang, I.S.; Lee, J.W.; Lee, E.Y. Batch conversion of methane to methanol using Methylosinus trichosporium OB3b as biocatalyst. J. Microbiol. Biotechnol. 2015, 25, 375–380. [Google Scholar] [CrossRef] [PubMed]
- DiSpirito, A.A.; Zahn, J.A.; Graham, D.W.; Kim, H.J.; Larive, C.K.; Derrick, T.S.; Cox, C.D.; Taylor, A. Copper-binding compounds from Methylosinus trichosporium OB3b. J. Bacteriol. 1998, 180, 3606–3613. [Google Scholar] [CrossRef] [PubMed]
- Van Hylckama, V.J.; de Koning, W.; Janssen, D.B. Transformation kinetics of chlorinated ethenes by Methylosinus trichosporium OB3b and detection of unstable epoxides by on-line gas chromatography. Appl. Environ. Microbiol. 1996, 62, 3304–3312. [Google Scholar] [CrossRef] [PubMed]
- Xing, Z.; Zhao, T.; Gao, Y.; He, Z.; Yang, X.; Peng, X. Depth profiles of methane oxidation kinetics and the related methanotrophic community in a simulated landfill cover. Huan Jing Ke Xue = Huanjing Kexue 2015, 36, 4302–4310. [Google Scholar] [PubMed]
- Cantera, S.; Lebrero, R.; García-Encina, P.A.; Muñoz, R. Evaluation of the influence of methane and copper concentration and methane mass transport on the community structure and biodegradation kinetics of methanotrophic cultures. J. Environ. Manag. 2016, 171, 11–20. [Google Scholar] [CrossRef]
- Guggenheim, C.; Brand, A.; Bürgmann, H.; Sigg, L.; Wehrli, B. Aerobic methane oxidation under copper scarcity in a stratified lake. Sci. Rep. 2019, 9, 4817. [Google Scholar] [CrossRef] [PubMed]
- Xing, Z.; Zhao, T.; Zhang, L.; Gao, Y.; Liu, S.; Yang, X. Effects of copper on expression of methane monooxygenases, trichloroethylene degradation, and community structure in methanotrophic consortia. Eng. Life Sci. 2018, 18, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Koo, C.W.; Rosenzweig, A.C. Biochemistry of aerobic biological methane oxidation. Chem. Soc. Rev. 2021, 50, 3424–3436. [Google Scholar] [CrossRef] [PubMed]
- Tentori, E.F.; Richardson, R.E. Methane monooxygenase gene transcripts as quantitative biomarkers of methanotrophic activity in Methylosinus trichosporium OB3b. Appl. Environ. Microbiol. 2020, 86, e01048-20. [Google Scholar] [CrossRef] [PubMed]
- Scheutz, C.; Kjeldsen, P. Environmental factors influencing attenuation of methane and hydrochlorofluorocarbons in landfill cover soils. J. Environ. Qual. 2004, 33, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhou, J.; Wang, X.; Zhang, Y. Coupled effects of methane monooxygenase and nitrogen source on growth and poly-β-hydroxybutyrate (PHB) production of Methylosinus trichosporium OB3b. J. Environ. Sci. 2017, 52, 49–57. [Google Scholar] [CrossRef]
- Duan, C.; Luo, M.; Xing, X. High-rate conversion of methane to methanol by Methylosinus trichosporium OB3b. Bioresour. Technol. 2011, 102, 7349–7353. [Google Scholar] [CrossRef]
- Takeguchi, M.; Furuto, T.; Sugimori, D.; Okura, I. Optimization of methanol biosynthesis by Methylosinus trichosporium OB3b: An approach to improve methanol accumulation. Appl. Biochem. Biotechnol. 1997, 68, 143–152. [Google Scholar] [CrossRef]
- Begonja, A.; Hrsak, D. Effect of growth conditions on the expression of soluble methane monooxygenase. Food Technol. Biotechnol. 2001, 39, 29–36. [Google Scholar]
- Collins, J.F.; Prohaska, J.R.; Knutson, M.D. Metabolic crossroads of iron and copper. Nutr. Rev. 2010, 68, 133–147. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, M.; Retamal-Morales, G.; Tischler, D. Metal binding ability of microbial natural metal chelators and potential applications. Nat. Prod. Rep. 2020, 37, 1262–1283. [Google Scholar] [CrossRef] [PubMed]
- Warren, M.J.; Lin, X.; Gaby, J.C.; Kretz, C.B.; Kolton, M.; Morton, P.L.; Pett-Ridge, J.; Weston, D.J.; Schadt, C.W.; Kostka, J.E. Molybdenum-based diazotrophy in a Sphagnum peatland in northern Minnesota. Appl. Environ. Microbiol. 2017, 83, e01174-17. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Su, T.; Li, X.; Xing, X. Research progresses of methanotrophs and methane monooxygenases. Sheng Wu Gong Cheng Xue Bao = Chin. J. Biotechnol. 2008, 24, 1511–1519. [Google Scholar]
- Brusseau, G.A.; Tsien, H.-C.; Hanson, R.S.; Wackett, L.P. Optimization of trichloroethylene oxidation by methanotrophs and the use of a colorimetric assay to detect soluble methane monooxygenase activity. Biodegradation 1990, 1, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Henry, S.M.; Grbic-Galic, D. Effect of mineral media on trichloroethylene oxidation by aquifer methanotrophs. Microb. Ecol. 1990, 20, 151–169. [Google Scholar] [CrossRef]
- Nguyen, H.-H.T.; Elliott, S.J.; Yip, J.H.-K.; Chan, S.I. The particulate methane monooxygenase from Methylococcus capsulatus (Bath) is a novel copper-containing three-subunit enzyme: Isolation and characterization. J. Biol. Chem. 1998, 273, 7957–7966. [Google Scholar] [CrossRef]
- DiSpirito, A.A.; Semrau, J.D.; Murrell, J.C.; Gallagher, W.H.; Dennison, C.; Vuilleumier, S. Methanobactin and the link between copper and bacterial methane oxidation. Microbiol. Mol. Biol. Rev. 2016, 80, 387–409. [Google Scholar] [CrossRef]
- Vita, N.; Platsaki, S.; Baslé, A.; Allen, S.J.; Paterson, N.G.; Crombie, A.T.; Murrell, J.C.; Waldron, K.J.; Dennison, C. A four-helix bundle stores copper for methane oxidation. Nature 2015, 525, 140–143. [Google Scholar] [CrossRef]
- Wymore, R.A.; Lee, M.H.; Keener, W.K.; Miller, A.R.; Colwell, F.S.; Watwood, M.E.; Sorenson, K.S., Jr. Field evidence for intrinsic aerobic chlorinated ethene cometabolism by methanotrophs expressing soluble methane monooxygenase. Bioremediat. J. 2007, 11, 125–139. [Google Scholar] [CrossRef][Green Version]
- Kenney, G.E.; Rosenzweig, A.C. Methanobactins: Maintaining copper homeostasis in methanotrophs and beyond. J. Biol. Chem. 2018, 293, 4606–4615. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Baral, B.S.; DiSpirito, A.A.; Semrau, J.D. An aminotransferase is responsible for the deamination of the N-terminal leucine and required for formation of oxazolone ring A in methanobactin of Methylosinus trichosporium OB3b. Appl. Environ. Microbiol. 2017, 83, e02619-16. [Google Scholar] [CrossRef] [PubMed]
- Prabakaran, S.; Lippens, G.; Steen, H.; Gunawardena, J. Post-translational modification: Nature’s escape from genetic imprisonment and the basis for dynamic information encoding. Wiley Interdiscip. Rev. Syst. Biol. Med. 2012, 4, 565–583. [Google Scholar] [CrossRef] [PubMed]
- Dershwitz, P.; Gu, W.; Roche, J.; Kang-Yun, C.S.; Semrau, J.D.; Bobik, T.A.; Fulton, B.; Zischka, H.; DiSpirito, A.A. MbnC Is Not Required for the Formation of the N-Terminal Oxazolone in the Methanobactin from Methylosinus trichosporium OB3b. Appl. Environ. Microbiol. 2022, 88, e01841-21. [Google Scholar] [CrossRef] [PubMed]
- Semrau, J.D.; Jagadevan, S.; DiSpirito, A.A.; Khalifa, A.; Scanlan, J.; Bergman, B.H.; Freemeier, B.C.; Baral, B.S.; Bandow, N.L.; Vorobev, A. Methanobactin and MmoD work in concert to act as the ‘copper-switch’ in methanotrophs. Environ. Microbiol. 2013, 15, 3077–3086. [Google Scholar] [CrossRef]
- Peng, P.; Kang-Yun, C.S.; Chang, J.; Gu, W.; DiSpirito, A.A.; Semrau, J.D. Two TonB-Dependent Transporters in Methylosinus trichosporium OB3b Are Responsible for Uptake of Different Forms of Methanobactin and Are Involved in the Canonical “Copper Switch”. Appl. Environ. Microbiol. 2022, 88, e01793-21. [Google Scholar] [CrossRef]
- Peng, P.; Gu, W.; DiSpirito, A.A.; Semrau, J.D. Multiple Mechanisms for Copper Uptake by Methylosinus trichosporium OB3b in the Presence of Heterologous Methanobactin. mBio 2022, 13, e02239-22. [Google Scholar] [CrossRef]
- Samanta, D.; Singh, R.N.; Goh, K.M.; Sani, R.K. Integrating metabolomics and whole genome sequencing to elucidate the metabolic pathways in Methylosinus trichosporium OB3b. Syst. Microbiol. Biomanuf. 2023, 1–14. [Google Scholar] [CrossRef]
- Paul, B.G.; Ding, H.; Bagby, S.C.; Kellermann, M.Y.; Redmond, M.C.; Andersen, G.L.; Valentine, D.L. Methane-oxidizing bacteria shunt carbon to microbial mats at a marine hydrocarbon seep. Front. Microbiol. 2017, 8, 186. [Google Scholar] [CrossRef]
- Graham, D.W.; Korich, D.G.; LeBlanc, R.P.; Sinclair, N.A.; Arnold, R.G. Applications of a colorimetric plate assay for soluble methane monooxygenase activity. Appl. Environ. Microbiol. 1992, 58, 2231–2236. [Google Scholar] [CrossRef]
- Foulger, J.H. The use of the Molisch (α-naphthol) reactions in the study of sugars in biological fluids. J. Biol. Chem. 1931, 92, 345–353. [Google Scholar] [CrossRef]
- Gafar, S.; El-Kelany, M.; El-Shawadfy, S. Spectrophotometric properties of azo dye metal complex and its possible use as radiation dosimeter. J. Radiat. Res. Appl. Sci. 2018, 11, 190–194. [Google Scholar] [CrossRef]
- Jarad, A.J.; Kadhim, Z.S.; Mahmood, M.A.; Obaid, S.M. Synthesis, Spectral Studies and Biological Activity of Azo dye Complexes with Some MetalIons. J. Glob. Pharma Technol. 2009, 10, 97–106. [Google Scholar]
- Tripathi, A.K.; Saxena, P.; Thakur, P.; Rauniyar, S.; Samanta, D.; Gopalakrishnan, V.; Singh, R.N.; Sani, R.K. Transcriptomics and functional analysis of Copper stress response in the sulfate-reducing bacterium Desulfovibrio alaskensis G20. Int. J. Mol. Sci. 2022, 23, 1396. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Samanta, D.; Govil, T.; Saxena, P.; Krumholz, L.; Gadhamshetty, V.; Goh, K.M.; Sani, R.K. Genetical and Biochemical Basis of Methane Monooxygenases of Methylosinus trichosporium OB3b in Response to Copper. Methane 2024, 3, 103-121. https://doi.org/10.3390/methane3010007
Samanta D, Govil T, Saxena P, Krumholz L, Gadhamshetty V, Goh KM, Sani RK. Genetical and Biochemical Basis of Methane Monooxygenases of Methylosinus trichosporium OB3b in Response to Copper. Methane. 2024; 3(1):103-121. https://doi.org/10.3390/methane3010007
Chicago/Turabian StyleSamanta, Dipayan, Tanvi Govil, Priya Saxena, Lee Krumholz, Venkataramana Gadhamshetty, Kian Mau Goh, and Rajesh K. Sani. 2024. "Genetical and Biochemical Basis of Methane Monooxygenases of Methylosinus trichosporium OB3b in Response to Copper" Methane 3, no. 1: 103-121. https://doi.org/10.3390/methane3010007
APA StyleSamanta, D., Govil, T., Saxena, P., Krumholz, L., Gadhamshetty, V., Goh, K. M., & Sani, R. K. (2024). Genetical and Biochemical Basis of Methane Monooxygenases of Methylosinus trichosporium OB3b in Response to Copper. Methane, 3(1), 103-121. https://doi.org/10.3390/methane3010007