Abstract
Accurate cancer prognosis remains a major challenge, as single gene expression biomarkers often lack clinical reliability, and most ML approaches fail even when considering large gene panels. In this study, we used a novel AutoML framework (O2Pmgen) benchmarked with a well-established framework (TPOT) on TCGA transcriptomic data for breast, lung, and renal cancers to identify small gene panels predictive of patient survival. From 58 EMT-related genes, we found models based on panels of 6–10 genes that outperformed single-marker models and ML models that considered the 58 EMT genes, with performance gains up to 21%. Further, the generated models achieved good predictive power with AUCs of 71–83%. Our results demonstrated that affordable and efficient prognostic tools using small, biologically relevant gene sets can provide better risk stratification in clinical oncology.
1. Introduction
Cancer remains one of the leading causes of death globally, with over 10 million deaths recorded annually. While advances in diagnostics and treatment have improved outcomes for many patients, accurately predicting cancer progression and survival remains a significant challenge [1,2]. Current prognostic tools often rely on clinical staging and single-gene biomarkers, frequently lacking the sensitivity, specificity, and predictive power required to guide personalised treatment decisions [3,4]. Prognostic failure can lead to inappropriate therapeutic strategies, delayed interventions, and overall poorer clinical outcomes. In particular, the molecular heterogeneity of tumours is often underrepresented in standard models, limiting their ability to stratify patients effectively by risk [1]. As the field of precision oncology evolves, there is a growing need for robust, cost-effective prognostic tools that can integrate molecular data to enhance clinical decision-making. Recent developments in transcriptomic profiling and artificial intelligence (AI) have opened new avenues for improving cancer prognosis [5,6,7]. Machine learning and AutoML technologies, when applied to gene expression data, offer a scalable means of identifying complex molecular signatures that are predictive of patient outcomes [8,9]. However, models resulting from such ML frameworks are often considered as “black boxes” and therefore applied to large gene panels to generate highly accurate models [1,10]. This constrains their clinical application, making them expensive and less practical for routine use. In this study, we aim to address these challenges by applying a new AutoML algorithm, O2Pmgen, through the Digital Phenomics platform in an attempt to identify small, biologically meaningful gene panels capable of predicting cancer survival. Using transcriptomic data from The Cancer Genome Atlas (TCGA), we focused on breast, lung, and renal cancers, selecting gene candidates involved in epithelial-to-mesenchymal transition (EMT), a key process in metastasis [11,12].
2. Materials and Methods
Transcriptomic datasets were derived from The Cancer Genome Atlas (TCGA), focusing on primary tumour biopsies from patients with breast (BRCA), lung (LUAD, LUSC), and renal (KICH, KIRC, KIRP) cancers. Transcriptomic data were pre-normalised and obtained as FPKM (fragments per kilobase of transcripts per million mapped reads) expression values from the 2021 Human Protein Atlas transcriptomic profiles [4,13]. On these datasets, a subset of 58 genes was selected based on their roles in epithelial-to-mesenchymal transition (EMT), a hallmark of cancer progression as previously described in [5]. Publicly available data associated with transcriptomics datasets were downloaded from https://www.proteinatlas.org/ (accessed on 10 May 2025). Binarised prognostic outcomes (good/poor) were under the assumption that patients surviving over five years post-diagnosis were labelled as having a good prognosis, while those deceased within two years were classified as poor prognosis, as previously described in [14]. Final datasets consisted of 239 BRCA (40 poor, 199 good), 325 lung (231 poor, 94 good), and 318 renal cancer cases (108 poor, 210 good). All curated datasets were uploaded and made available via the Digital Phenomics platform UIv0.24 (https://digitalphenomics.com, accessed on 10 May 2025).
Three AutoML tools were employed in the same 58-selected genes cancer transcriptomics datasets to develop binary classifiers for prognosis inference: BMfinder, TPOT, and O2Pmgen. BMfinder software (v1.0) was applied for the generation of classifiers based on single-gene abundance thresholds. For each gene, optimal cutoff values were computed to maximise classifier sensitivity across the prognosis classes. The tool applies directionality of the decision rule (above or below the cutoff) depending on whether gene expression was elevated or reduced in poor-prognosis cases. TPOT (Tree-Based Pipeline Optimisation Tool, v0.12.0) was applied using a genetic programming framework to evolve optimal ML pipelines [8] For each dataset, TPOT performed 100 evolutionary generations with a population of 50 models, using 5-fold cross-validation and ROC-AUC as the optimisation criterion. Model generation using TPOT and BMfinder was executed on a 16-core VPS environment running custom Python 3.8 scripts. O2Pmgen (v1.1) was executed via the Digital Phenomics platform [14]. This proprietary AI-driven AutoML tool uses an evolution-inspired multi-objective optimisation algorithm to identify optimal combinations of gene expression patterns [14]. The algorithm was constrained to train on <50% of the dataset, retaining the remainder for testing. Models incorporated both directionality (up/down regulation) and binary presence logic of multiple biomarkers on a scoring function, as previously described in [14].
Classifier performance was assessed using Receiver Operating Characteristic Area Under the Curve (ROC-AUC), sensitivity, and specificity metrics computed using the software tool ROCplot version 1.0 [14]. Optimal sensitivity (True Positive Rate) and specificity (True Negative Rate) were computed using ROCplot on the entire dataset to ensure an accurate and deterministic calculation of model performance metrics without requiring confidence intervals.
3. Results
Using autoML approaches, we obtained a bespoke predictive model for the prognosis of each cancer type. NFKB2, PTK2 and TGFB1 genes were identified as single bespoke predictive biomarkers for breast, lung and renal cancers, respectively. Bespoke combinations of 10 biomarkers that maximise the predictive capacity were identified for breast cancer and combinations of 6 biomarkers for the remaining cancer types (Figure 1). Breast, lung, and renal cancer types exhibited distinct gene expression patterns and shared only two genes (ILK and EGF) between different cancer types. Interestingly, none of the identified single bespoke biomarkers were present in the bespoke combinations.
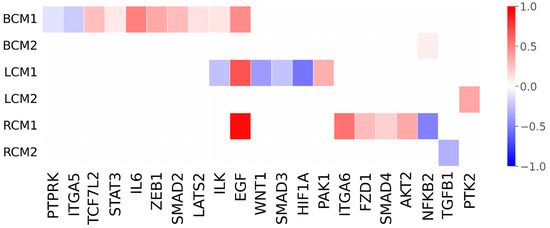
Figure 1.
Predicted single biomarkers and combinations of biomarkers identified for breast, lung and renal cancers. Gene names are indicated on the x-axis, and the normalised degree of variation between cancer survival medians and controls is represented on the y-axis. BCM1, LCM1 and RCM1 indicate the predictive combination of biomarkers for breast, lung and renal cancers, respectively. BCM2, LCM2 and RCM2 indicate the single predictive biomarkers for breast, lung and renal cancers, respectively. Gene downregulations in expression are depicted by negative numbers represented by a blue gradient scale, and up-regulations by positive numbers in a red gradient scale. Null values in white represent that the gene was not identified as a predictive biomarker.
Performance evaluation of the generated predictive models (Table 1) showed substantial improvement of performance metrics (AUC, sensitivity and specificity) of models composed of combinations of biomarkers (O2Pmgen-derived) in comparison with the single biomarker approach (BMfinder-derived). Models derived by autoML using all genes (TPOT-derived) showed equivalent improvements on sensitivity but only mild improvements on the AUC and specificity.

Table 1.
Performance of the models generated using different ML approaches.
4. Discussion
The results of this study underscore the potential of AI-driven approaches for identifying bespoke predictive models and clinically relevant biomarkers. Notably, we demonstrated that employing advanced modelling strategies such as O2Pmgen, which iteratively searches, evaluates, and selects optimal biomarker combinations, can lead to substantial gains in predictive performance. Across the three cancer types analysed, this approach yielded up to a 21% increase in AUC compared with both a widely used AutoML framework (TPOT) and the best-performing single-biomarker classifier.
The obtained sensitivities and specificities suggest that O2Pmgen tends to generate models with higher specificity than traditional approaches, explaining the observed gains in predictive power. These models consistently showed improved specificity relative to baseline models, often without a corresponding loss in sensitivity. The algorithm tends to prioritise specificity, potentially reducing false positive rates. In clinical contexts, this may be advantageous where the goal is to minimise overtreatment or unnecessary follow-up of patients incorrectly identified as high risk. However, such specificity could lead to under-identification of true positive cases, which may be unacceptable in settings where early detection is critical. The clinical appropriateness of this trade-off depends on the intended application and patient population.
Importantly, the predictive models generated by O2Pmgen were based on compact and biologically interpretable gene panels, offering a valuable opportunity for developing cost-effective and practical prognostic tools to support clinical decision-making in oncology. Furthermore, the selected gene combinations exhibited distinct patterns of upregulation and downregulation of signalling components specific to each cancer type, suggesting the presence of tumour-specific molecular signatures associated with favourable prognosis.
Interestingly, the expression patterns identified align with transcriptomic and signalling features characteristic of mesenchymal-like cancer cells, while being incompatible with highly invasive or metastable hybrid phenotypes [11,15]. These findings support the hypothesis that the gene panels discovered through our AI-guided approach reflect stable, less aggressive tumour states and correlate with improved clinical outcomes.
Although the models showed promising performance, limitations should be acknowledged. These include reliance on retrospective TCGA data without independent validation, potential bias from unbalanced datasets (notably in breast cancer), and restriction to 58 genes, which may limit broader model optimisation. Thus, results should be interpreted cautiously and viewed primarily as a qualitative comparison across AutoML approaches.
5. Conclusions
This work demonstrates that autoML strategies for developing models based on finding optimal combinations of small gene sets can outperform the predictive power of larger, less practical panels. The findings highlight the potential for affordable, scalable, and clinically applicable prognostic tools that support personalised oncology and improved patient management. As next steps, we aim to extend this approach by exploring the full transcriptomic space to identify improved gene set combinations. Additionally, external validation using independent datasets will be pursued to strengthen the clinical relevance and generalisability of the models.
Author Contributions
Conceptualisation, R.J.P. and U.L.F.; methodology, R.J.P. and U.L.F.; software, R.J.P. and U.L.F.; validation, T.A.P.; formal analysis, T.A.P.; data curation, T.A.P.; writing—original draft preparation, R.J.P.; writing—review and editing, R.J.P.; supervision, R.J.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the UK Government through the Innovation Navigator—Flexible Fund grant GMBS-FF-180057, which was awarded for the development of the Digital Phenomics platform.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data in this research is available at https://digitalphenomics.com, accessed on 10 May 2025.
Acknowledgments
The authors acknowledge Bioenhancer Systems Ltd. for supporting the resources necessary to conduct the analysis and maintain the tools online. The authors also acknowledge the Manchester Growth Business Innovation Hub for helping with grant acquisition and management, which gave rise to this work.
Conflicts of Interest
R. Pais declares a potential conflict of interest as he is the director of Bioenhancer Systems. U. Filho and T. Pais declare no conflict of interest.
References
- Kourou, K.; Exarchos, K.P.; Papaloukas, C.; Sakaloglou, P.; Exarchos, T.; Fotiadis, D.I. Applied Machine Learning in Cancer Research: A Systematic Review for Patient Diagnosis, Classification and Prognosis. Comput. Struct. Biotechnol. J. 2021, 19, 5546–5555. [Google Scholar] [CrossRef]
- Boeri, C.; Chiappa, C.; Galli, F.; De Berardinis, V.; Bardelli, L.; Carcano, G.; Rovera, F. Machine Learning Techniques in Breast Cancer Prognosis Prediction: A Primary Evaluation. Cancer Med. 2020, 9, 3234–3243. [Google Scholar] [CrossRef]
- Šutić, M.; Vukić, A.; Baranašić, J.; Försti, A.; Džubur, F.; Samaržija, M.; Jakopović, M.; Brčić, L.; Knežević, J. Diagnostic, Predictive, and Prognostic Biomarkers in Non-Small Cell Lung Cancer (NSCLC) Management. J. Pers. Med. 2021, 11, 1102. [Google Scholar] [CrossRef] [PubMed]
- Pontén, F.; Jirström, K.; Uhlen, M. The Human Protein Atlas—A Tool for Pathology. J. Pathol. 2008, 216, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Pais, R.J.; Lopes, F.; Parreira, I.; Silva, M.; Silva, M.; Moutinho, M.G. Predicting Cancer Prognostics from Tumour Transcriptomics Using an Auto Machine Learning Approach. Med. Sci. Forum 2023, 22, 6. [Google Scholar]
- Yang, D.; Ma, X.; Song, P. A Prognostic Model of Non Small Cell Lung Cancer Based on TCGA and ImmPort Databases. Sci. Rep. 2022, 12, 437. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Xu, Y.; Han, C.; Wang, Z.; Li, J.; Wang, Q.; Che, X. Identification of a Prognostic Risk Signature of Kidney Renal Clear Cell Carcinoma Based on Regulating the Immune Response Pathway Exploration. J. Oncol. 2020, 2020, 6657013. [Google Scholar] [CrossRef] [PubMed]
- Le, T.T.; Fu, W.; Moore, J.H. Scaling Tree-Based Automated Machine Learning to Biomedical Big Data with a Feature Set Selector. Bioinformatics 2020, 36, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Telikani, A.; Gandomi, A.H.; Tahmassebi, A.; Banzhaf, W. Evolutionary Machine Learning: A Survey. ACM Comput. Surv. 2021, 54, 161. [Google Scholar] [CrossRef]
- Roman-Naranjo, P.; Parra-Perez, A.M.; Lopez-Escamez, J.A. A Systematic Review on Machine Learning Approaches in the Diagnosis and Prognosis of Rare Genetic Diseases. J. Biomed. Inform. 2023, 143, 104429. [Google Scholar] [CrossRef] [PubMed]
- Liao, T.-T.; Yang, M.-H. Hybrid Epithelial/Mesenchymal State in Cancer Metastasis: Clinical Significance and Regulatory Mechanisms. Cells 2020, 9, 623. [Google Scholar] [CrossRef] [PubMed]
- Garg, M. Epithelial, Mesenchymal and Hybrid Epithelial/Mesenchymal Phenotypes and Their Clinical Relevance in Cancer Metastasis. Expert Rev. Mol. Med. 2017, 19, e3. [Google Scholar] [CrossRef]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A Pathology Atlas of the Human Cancer Transcriptome. Science 2017, 357. [Google Scholar] [CrossRef] [PubMed]
- Filho, U.L.; Pais, T.A.; Pais, R.J. Facilitating “Omics” for Phenotype Classification Using a User-Friendly AI-Driven Platform: Application in Cancer Prognostics. BioMedInformatics 2023, 3, 1071–1082. [Google Scholar] [CrossRef]
- Pais, R.J. Simulation of Multiple Microenvironments Shows a Pivot Role of RPTPs on the Control of Epithelial-to-Mesenchymal Transition. Biosystems 2020, 198, 104268. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).