RhsP2 Protein as a New Antibacterial Toxin Targeting RNA †
Abstract
1. Introduction
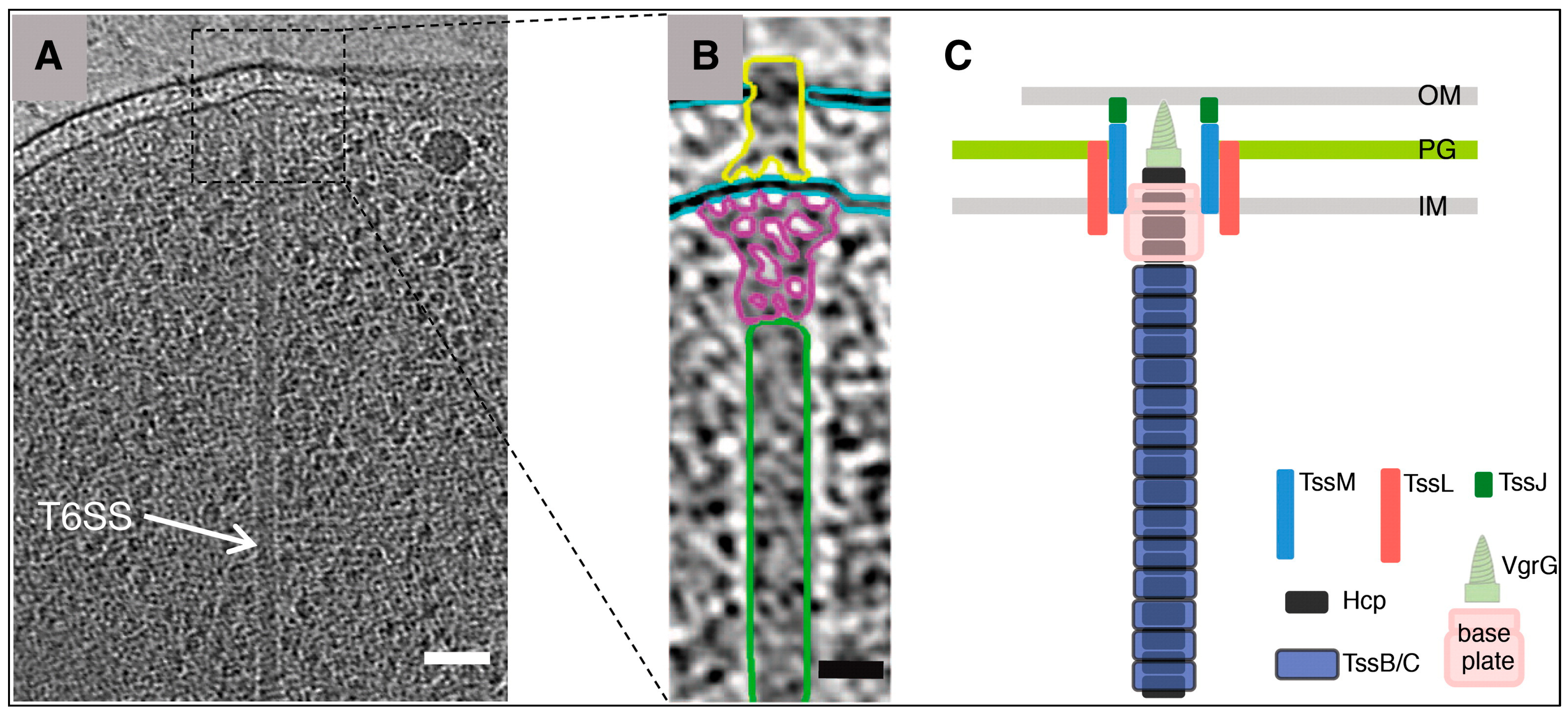
1.1. H2T6SS System (Type Six Secretion System Encodes loci Named H2)
1.2. Mechanism of Action of RhsP2
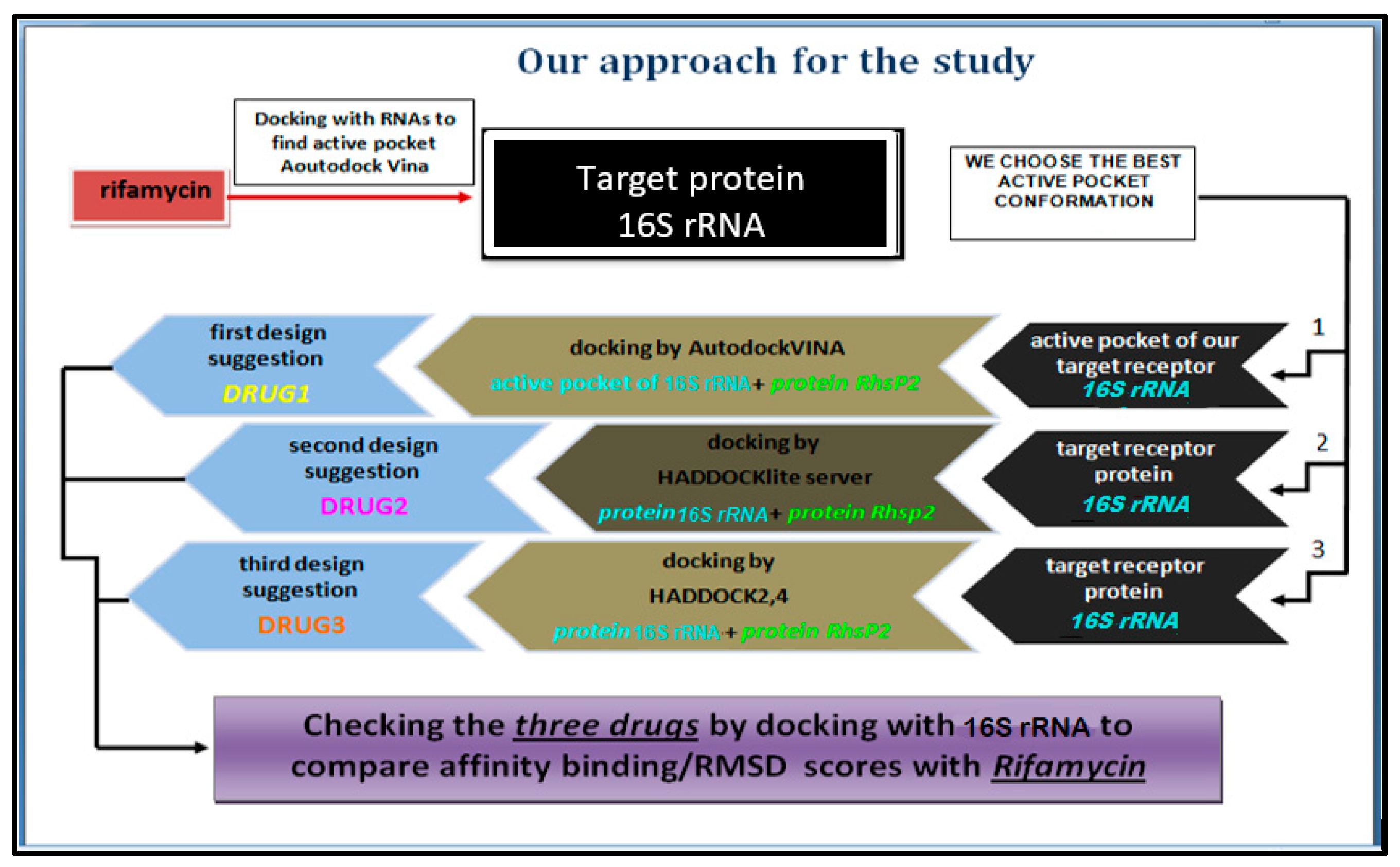
2. Methodology (Rigid Receptor + Flexible Ligand)
2.1. Part I:Preparing Target Protein, RhsP2toxin, and Bioreference in Order to Generate Protein–Protein Interaction (PPI)
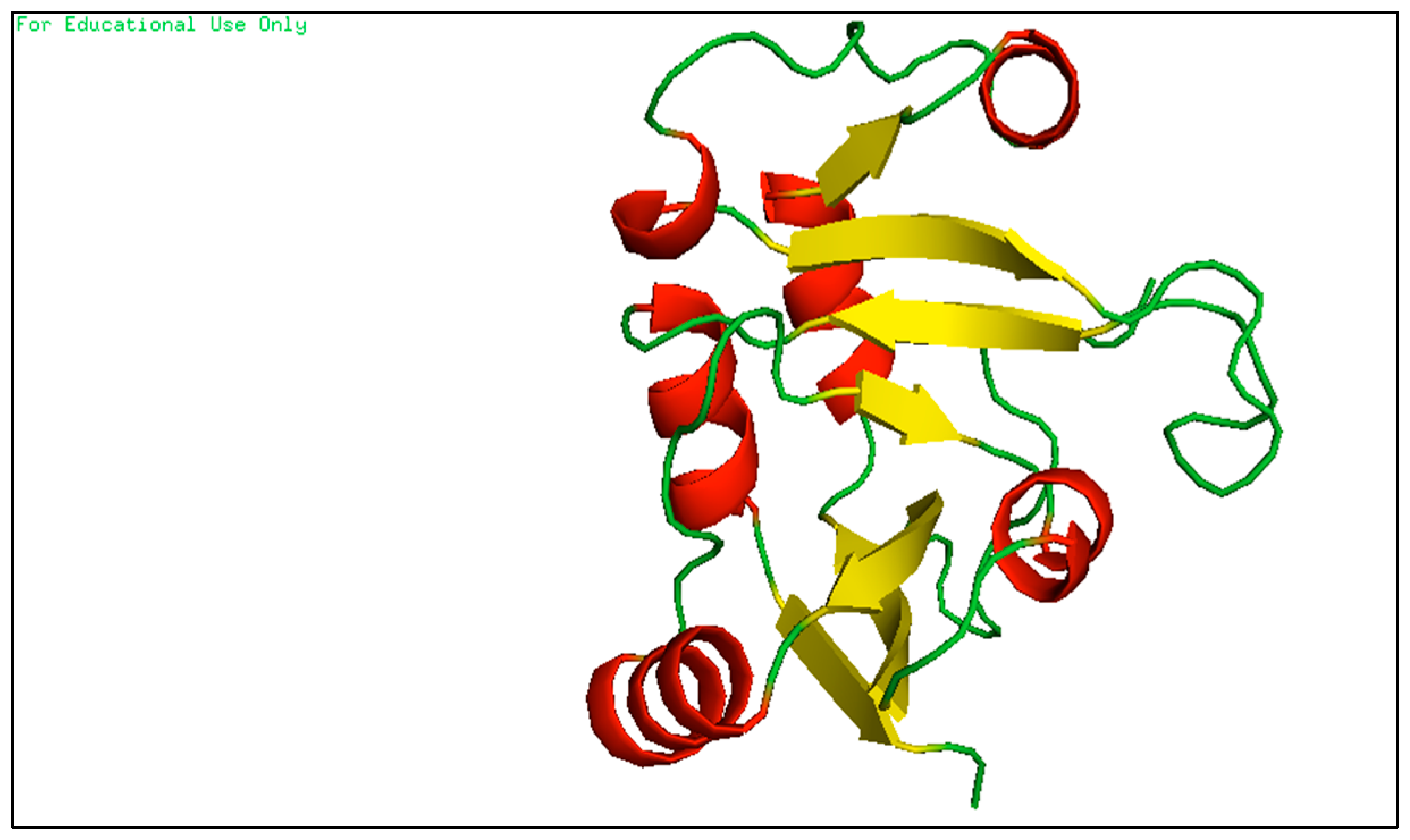
2.1.1. Prepare Target Protein
2.1.2. Prepare RhsP2 Toxin
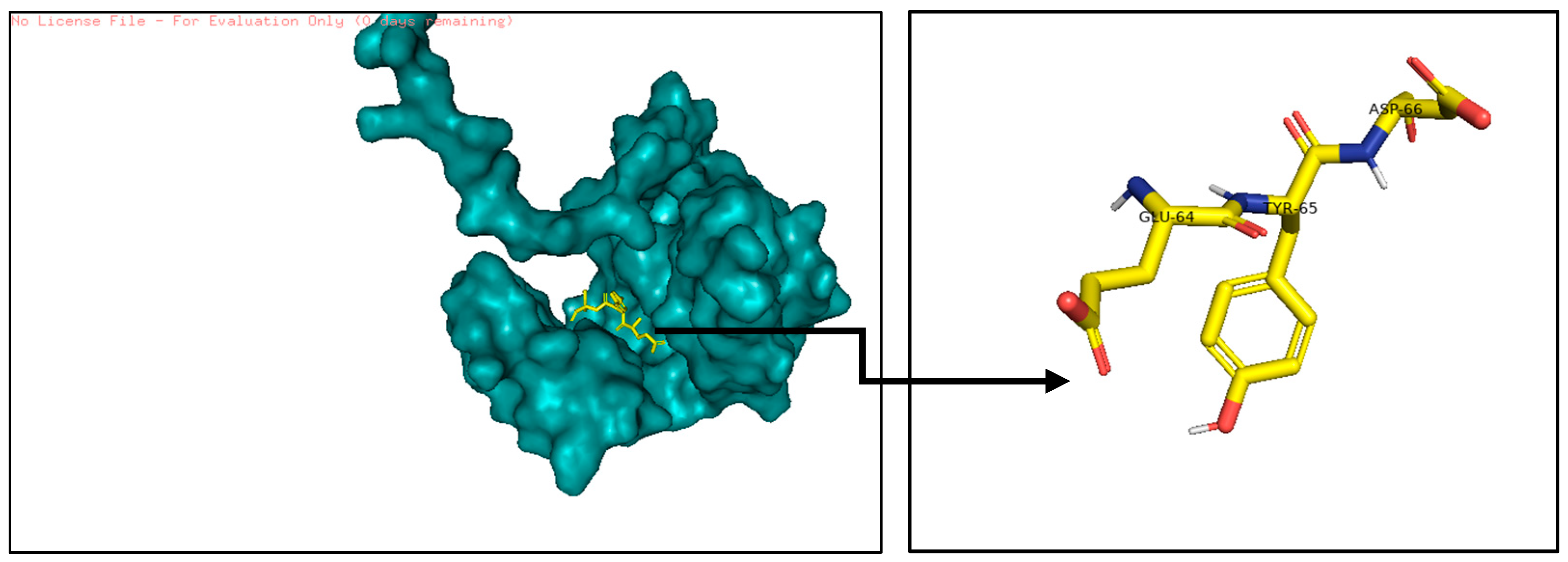
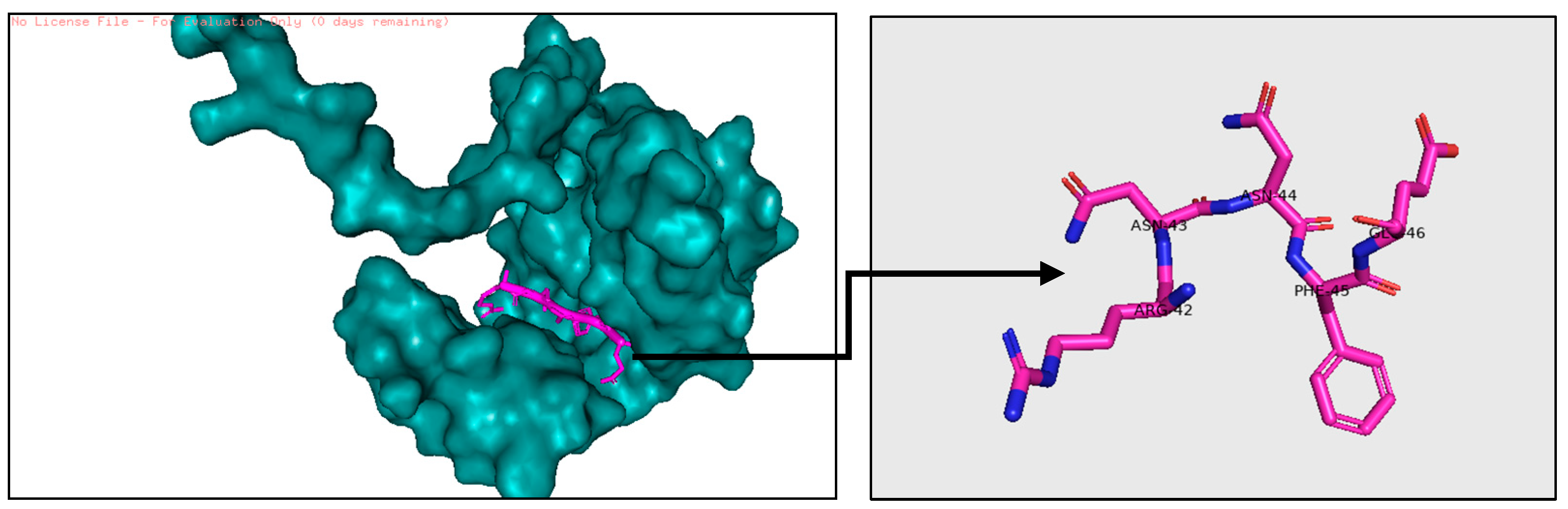
2.1.3. Determine Bioreference and Target Protein Active Site
2.2. Part II: Protein–Protein Interaction (PPI) Docking
- The active pocket of 16S rRNA was displayed by the pyMOL TOOL version 2.5.8, which was used in the second docking with RhsP2; because VINA is not programmed for protein–protein docking, RhsP2 is considered as a ligand instead of a protein when using AutoDock VINA. After the docking results are obtained, extract the active sites of RhsP2 via pyMOL. This resulted in the first suggested drug design (DRUG1) (stochastic shape-based method).
- Using Online Docking Programs, HADDOCKlite and HADDOCK 2,4 Server, for Protein-Protein Docking to Suggest New Pharmacophore Models (DRUG2 and DRUG3).
2.3. Part III: Evaluate Pharmacophors (DRUG1,2,3)
3. Results and Discussion
4. Conclusions
- The first part provides information on the studied protein secreted by the bacterium P. aeruginosa RhsP2 as a newly discovered protein of the Rhs family. Rifamycin was used in this study to compare the accuracy of docking programs. The second part describes in detail the docking procedures, which were used in order to dock the protein–protein complexes, and the software and criteria used in the assessment. The third part presents the docking results obtained from the individual programs, AutoDock VINA, HADDOCKlite, and HADDOCK 2.4, and compares them in terms of the binding energy and RMSD.
- These results show that the recently developed HADDOCKlite server obtained similar drugs that were the closest to the Rifamycin reference in terms of their interaction with the active pocket of the 16S rRNA target protein, followed by HADDOCK 2.4 server and the stochastic method using AutoDock VINA, which showed a deviation in predicting the binding energies of the docking complex.
- The results were reasonable as a preliminary prediction with one target 16S rRNA, which was selected as a special bacterial ribosome that is different from the human 18S rRNA ribosome; thus, the toxin RhsP2 has a good interaction with 16S rRNA, suggesting that it could be an inhibitor of 16S rRNA by binding to the active pocket (similar to the action of Rifamycin) and inhibiting its function in translating amino acids into proteins.
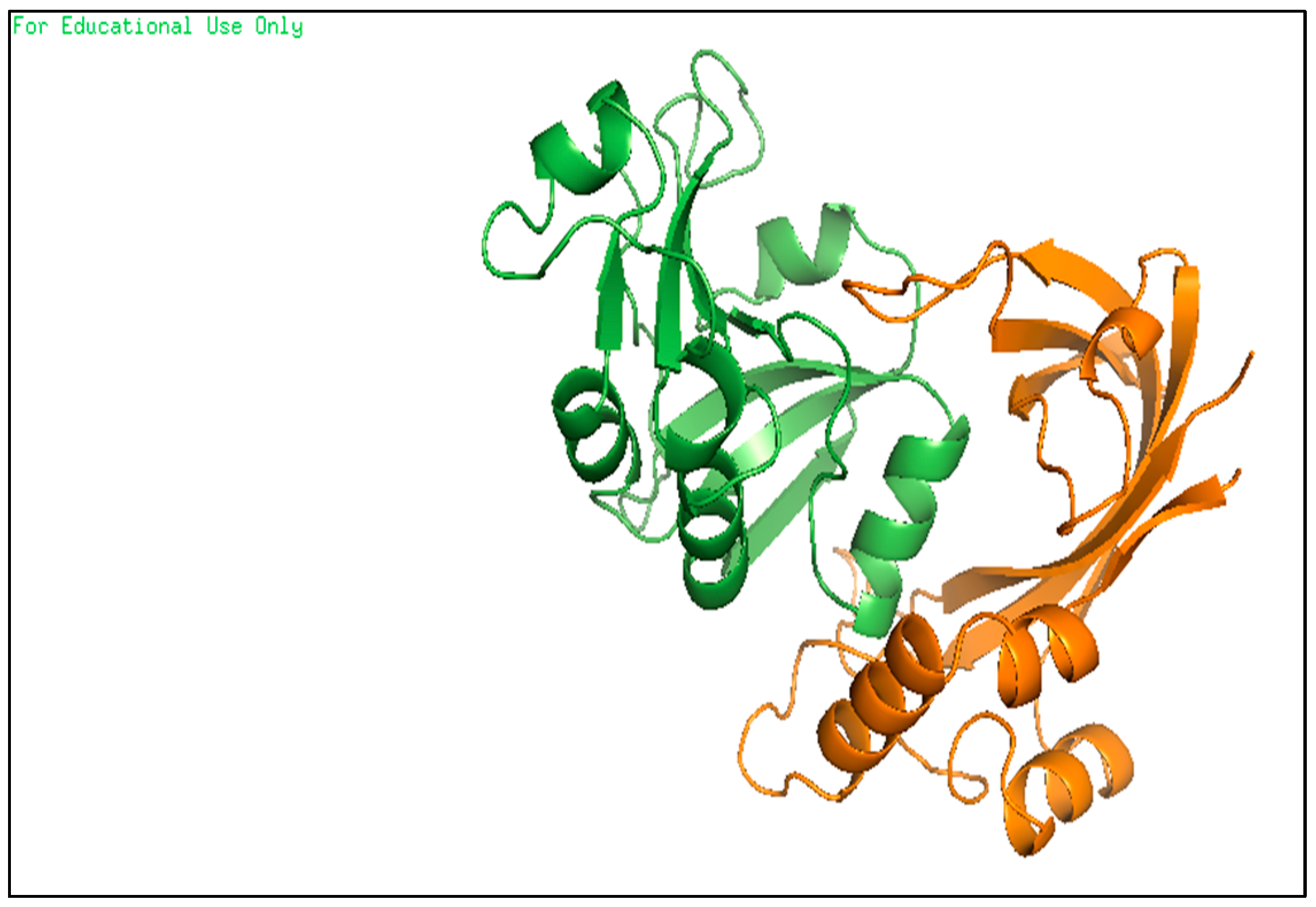
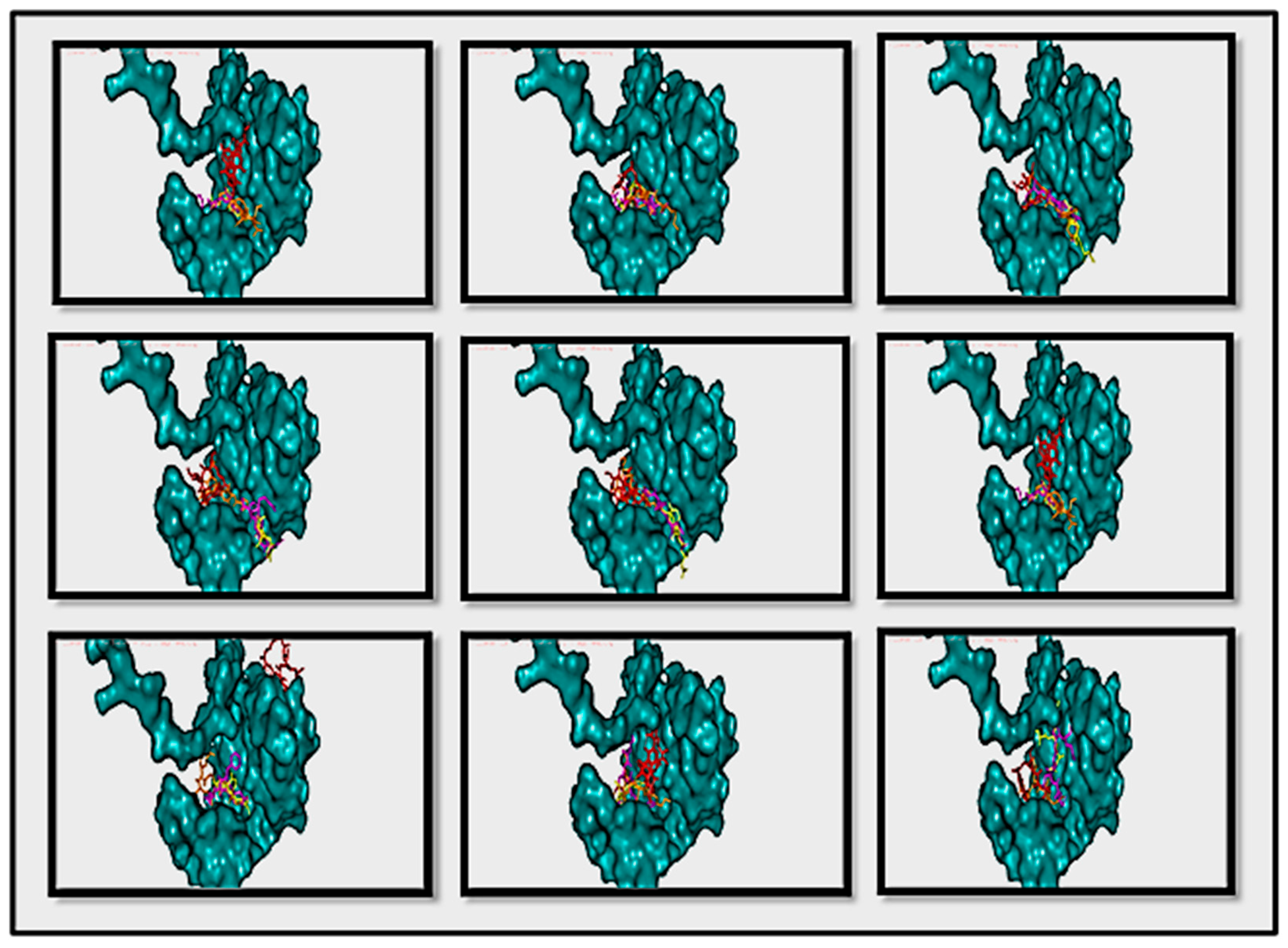
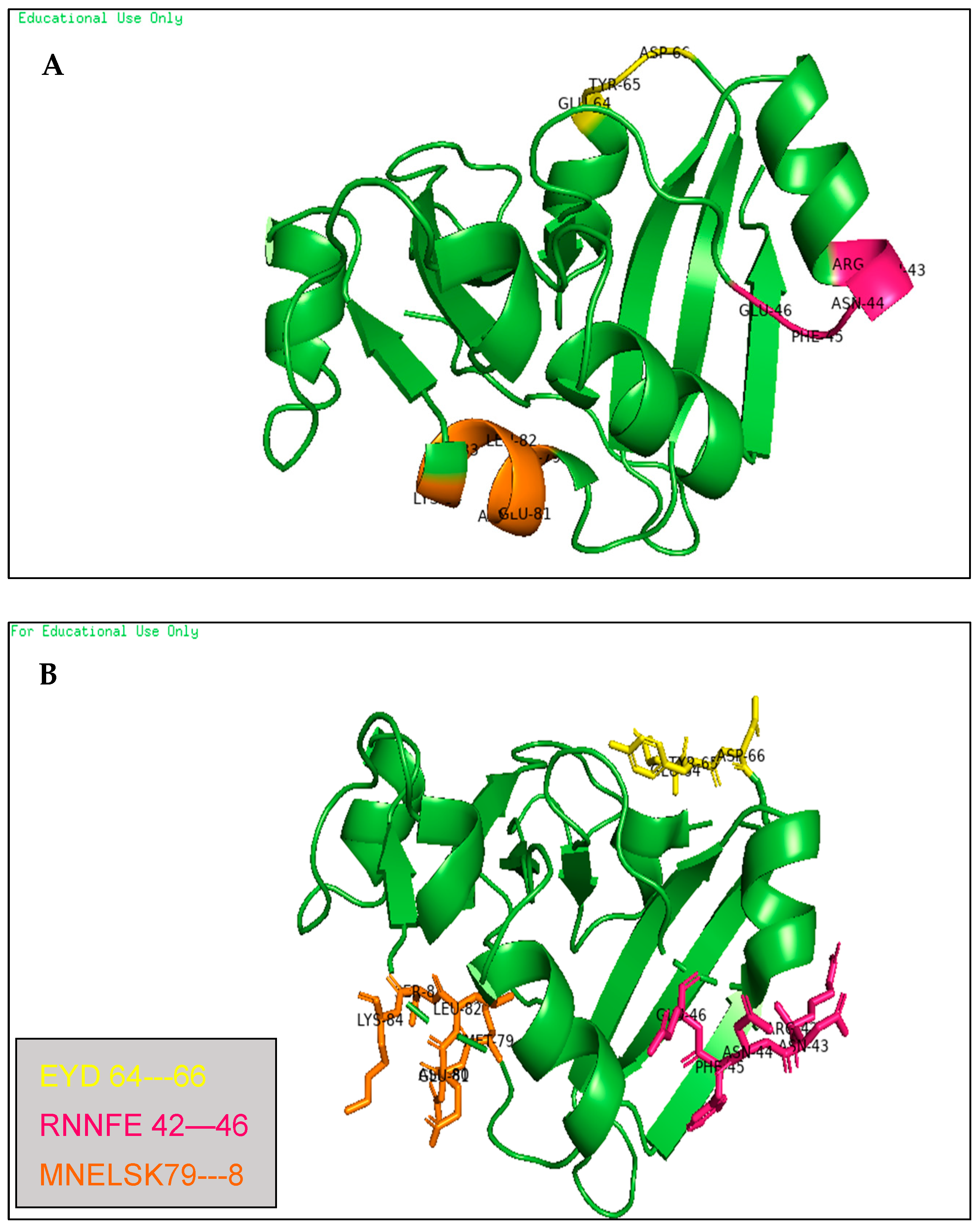
- These binding sites were predicted by molecular docking methods, with at least three sites, as shown in Figure 13 in two ways of pyMOL displaying; carton with a colored parts which refer to the three predicted drugs (A) and, carton with a sticky colored parts which refer to the three predicted drugs (B).
5. Future Work
- Finish calculation for 18S rRNA: Due to the lack of confirmed studies on the selectivity of the RhsP2 complex with human RNA proteins, it is necessary to re-conduct the experimental studies, which requires repeating the docking processes with other human protein targets, comparing the new binding scores with the previous RNA bacterial target ones, and determining the effectiveness and toxicity of the studied toxin.
- After first docking with RNA as a rigid receptor target, it has become increasingly clear that side-chain flexibility plays a crucial role in ligand–protein complexes. These changes allow the receptor to alter its binding site according to the orientation of the ligand; therefore, it is necessary to re-conduct flexible docking using Monte Carlo or molecular dynamics docking methods.
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| RhsP2 | Toxin secreted by Pseudomonas aeruginosa organisms belonging to the group of cytoplasmic enzymes affecting RNA through the mechanism of ADP-ribosyltransferase (from the group of ARTs) |
| ARTs | ADP-RibosylTransferases |
| ADPr | Adenosine DiPhosphate ribosyl |
| ncRNAs | Non-coding RNAs |
| H2T6SS | Type Six Secretion System Encodes Loci Named H2 |
| AMR | Antimicrobial Resistance |
| RNAP | RNA Polymerase |
| PPI | Protein–Protein Interaction |
| RMSD | Root Mean Square Deviation of Particle Coordinates |
| 16S rRNA | 16 Subunit Ribosomal Ribonucleic Acid |
References
- New Data Shows 148 Severe Antibiotic-Resistant Infections a Day in 2021. Available online: https://www.gov.uk/government/news/new-data-shows-148-severe-antibiotic-resistant-infections-a-day-in-2021 (accessed on 21 November 2022).
- ESPAUR Report 2022—The Latest Findings on Antimicrobial Resistance; UK Health Security Agency: London, UK, 21 November 2022. Available online: https://ukhsa.blog.gov.uk/2022/11/21/espaur-report-2022/ (accessed on 21 November 2022).
- Silver, L.L. Challenges of Antibacterial Discovery. Clin. Microbiol. Rev. 2011, 24, 71–109. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Yang, X.; Lewis, P.J. Bacterial transcription as a target for antibacterial drug development. Microbiol. Mol. Biol. Rev. 2016, 80, 139–160. [Google Scholar] [CrossRef] [PubMed]
- Wright, G.D. Q&A: Antibiotic resistance: Where does it come from and what can we do about it? BMC Biol. 2010, 8, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, R.N.; Pattanaik, S.; Pattnaik, G.; Mallick, S.; Mohapatra, R. Review on the use of Molecular Docking as the First Line Tool in Drug Discovery and Development. Indian J. Pharm. Sci. 2022, 84, 1334–1337. [Google Scholar] [CrossRef]
- Chen, G.; Seukep, A.J.; Guo, M. Recent advances in molecular docking for the research and discovery of potential marine drugs. Mar. Drugs 2020, 18, 545. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.; Hachani, A.; Manoli, E.; Filloux, A. An rhs gene linked to the second type VI secretion cluster is a feature of the Pseudomonas aeruginosa strain PA14. J. Bacteriol. 2014, 196, 800–810. [Google Scholar] [CrossRef] [PubMed]
- Zoued, A.; Brunet, Y.R.; Durand, E.; Aschtgen, M.S.; Logger, L.; Douzi, B.; Journet, L.; Cambillau, C.; Cascales, E. Architecture and assembly of the Type VI secretion system. Biochim. Et Biophys. Acta Mol. Cell Res. 2014, 1843, 1664–1673. [Google Scholar] [CrossRef] [PubMed]
- Bullen, N.P.; Prehna, G.; Whitney, J.C. Crystal Structure of the RhsP2 C-Terminal Toxin Domain in Complex with Its Immunity Protein, RhsI2; Canadian Institutes of Health Research (CIHR); Natural Sciences and Engineering Research Council (NSERC): Ottawa, ON, Canada, 2022. [Google Scholar] [CrossRef]
- Popova, K.B.; Otcheva, L.A.; Traykovska, M.; Penchovsky, R. RNA as a potent target for antibacterial drug discovery. Biomed. J. Sci. Tech. Res. 2018, 10, 7752–7754. [Google Scholar] [CrossRef]
- Bullen, N.P.; Sychantha, D.; Thang, S.S.; Culviner, P.H.; Rudzite, M.; Ahmad, S.; Shah, V.S.; Filloux, A.; Prehna, G.; Whitney, J.C. An ADP-ribosyltransferase toxin kills bacterial cells by modifying structured non-coding RNAs. Mol. Cell 2022, 82, 3484–3498.e11. [Google Scholar] [CrossRef] [PubMed]




| Rifamycin | DRUG1 | DRUG2 | DRUG3 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mode | Affinity (kcal/mol) | rmsd l.b | rmsd u.b | Mode | (Kcal/mol) Affinity | rmsd l.b | rmsd u.b | Mode | (Kcal/mol) Affinity | rmsd l.b | rmsd u.b | Mode | (Kcal/mol) Affinity | rmsd l.b | rmsd u.b |
| 1 | −7.6 | 0 | 0 | 1 | −6.4 | 0 | 0 | 1 | −6.6 | 0 | 0 | 1 | −6 | 0 | 0 |
| 2 | −7.4 | 2.801 | 8.55 | 2 | −5.8 | 13.498 | 15.878 | 2 | −6 | 3.333 | 6.851 | 2 | −5.7 | 3.945 | 10.73 |
| 3 | −7.4 | 13.288 | 16.755 | 3 | −5.7 | 9.984 | 14.566 | 3 | −5.9 | 3.778 | 7.635 | 3 | −5.4 | 3.949 | 10.854 |
| 4 | −7.3 | 2.389 | 7.856 | 4 | −5.4 | 1.62 | 2.636 | 4 | −5.9 | 3.957 | 11.092 | 4 | −5.3 | 4.835 | 8.465 |
| 5 | −7.3 | 5.49 | 8.71 | 5 | −5.4 | 2.446 | 7.418 | 5 | −5.7 | 7.367 | 11.731 | 5 | −5.2 | 4.44 | 7.674 |
| 6 | −6.9 | 2.735 | 7.854 | 6 | −5.3 | 8.995 | 13.875 | 6 | −5.7 | 5.314 | 11.974 | 6 | −5.1 | 4.367 | 7.817 |
| 7 | −6.8 | 3.184 | 6.622 | 7 | −5.3 | 12.989 | 16.88 | 7 | −5.7 | 7.455 | 11.238 | 7 | −5.1 | 5.397 | 9.726 |
| 8 | −6.8 | 11.22 | 16.225 | 8 | −5.2 | 13.791 | 17.285 | 8 | −5.7 | 6.59 | 11.053 | 8 | −5 | 4.511 | 11.925 |
| 9 | −6.8 | 27.567 | 30.534 | 9 | −5.1 | 7.275 | 10.846 | 9 | −5.6 | 3.666 | 8.22 | 9 | −5 | 5.503 | 9.228 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haj Marza, T.N. RhsP2 Protein as a New Antibacterial Toxin Targeting RNA. Med. Sci. Forum 2025, 35, 3. https://doi.org/10.3390/msf2025035003
Haj Marza TN. RhsP2 Protein as a New Antibacterial Toxin Targeting RNA. Medical Sciences Forum. 2025; 35(1):3. https://doi.org/10.3390/msf2025035003
Chicago/Turabian StyleHaj Marza, Tamara Nami. 2025. "RhsP2 Protein as a New Antibacterial Toxin Targeting RNA" Medical Sciences Forum 35, no. 1: 3. https://doi.org/10.3390/msf2025035003
APA StyleHaj Marza, T. N. (2025). RhsP2 Protein as a New Antibacterial Toxin Targeting RNA. Medical Sciences Forum, 35(1), 3. https://doi.org/10.3390/msf2025035003





