Space and Time in the Universe of the Cell Nucleus after Ionizing Radiation Attacks: A Comparison of Cancer and Non-Cancer Cell Response †
Abstract
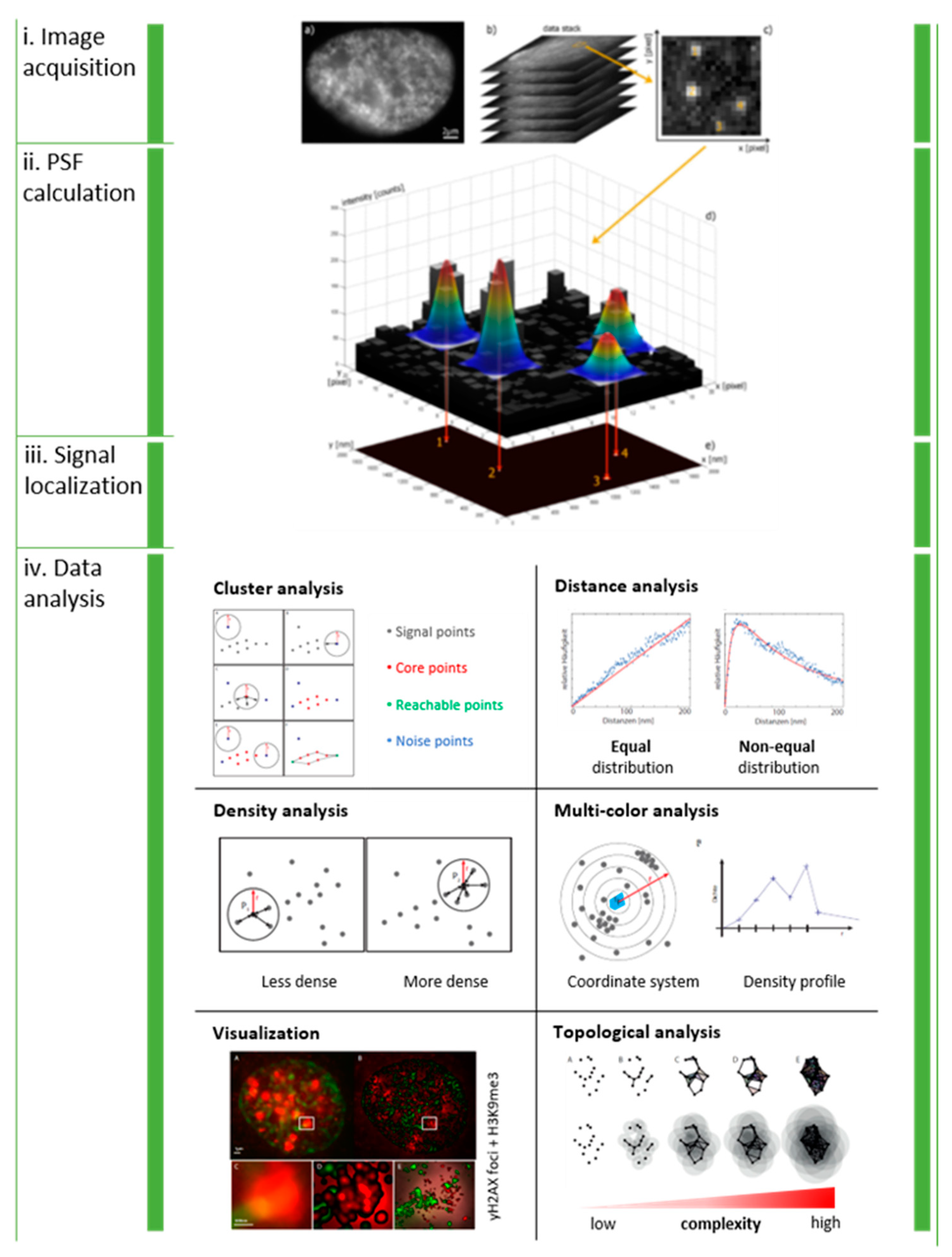
:1. Introduction
2. Spatial Analysis of Repair Cluster Formation after Low-LET Photon Irradiation
3. Spatial Analysis of Repair Cluster Formation within α-Particle Tracks and β-Particle Damage Sites
4. Spatial Analysis of Repair Cluster Formation within Damage Tracks of High-LET 15N Ions
5. Dose-Dependent ALU Signaling
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lee, J.-H.; Hausmann, M. Super-Resolution Radiation Biology: From Bio-Dosimetry towards Nano-Studies of DNA Repair Mechanisms; IntechOpen: London, UK, 2021. [Google Scholar] [CrossRef]
- Falk, M.; Hausmann, M. A Paradigm Revolution or Just Better Resolution—Will Newly Emerging Superresolution Techniques Identify Chromatin Architecture as a Key Factor in Radiation-Induced DNA Damage and Repair Regulation? Cancers 2020, 13, 18. [Google Scholar] [CrossRef] [PubMed]
- Eberle, J.P.; Rapp, A.; Krufczik, M.; Eryilmaz, M.; Gunkel, M.; Erfle, H.; Hausmann, M. Super-resolution microscopy tech-niques and their potential for applications in radiation biophysics. In Super-resolution Microscopy—Methods and Proto-cols. Meth. Molec. Biol. 2017, 1663, 1–13. [Google Scholar]
- Falk, M.; Hausmann, M.; Lukášová, E.; Biswas, A.; Hildenbrand, G.; Davídková, M.; Krasavin, E.; Kleibl, Z.; Falková, I.; Ježková, L.; et al. Determining OMICS spatiotemporal dimensions using exciting new nanoscopy techniques to asses complex cell responses to DNA damage—PART A (Radiomics). Crit. Rev. Eukaryot. Gene Express. 2014, 24, 205–223. [Google Scholar] [CrossRef] [PubMed]
- Falk, M.; Hausmann, M.; Lukášová, E.; Biswas, A.; Hildenbrand, G.; Davídková, M.; Krasavin, E.; Kleibl, Z.; Falková, I.; Ježková, L.; et al. Determining OMICS spatiotemporal dimensions using exciting new nanoscopy techniques to asses complex cell responses to DNA damage—PART B (Structuromics). Crit. Rev. Eukaryot. Gene Express. 2014, 24, 225–247. [Google Scholar] [CrossRef] [PubMed]
- Hausmann, M.; Ilić, N.; Pilarczyk, G.; Lee, J.-H.; Logeswaran, A.; Borroni, A.P.; Krufczik, M.; Theda, F.; Waltrich, N.; Bestvater, F.; et al. Challenges for Super-Resolution Localization Microscopy and Biomolecular Fluorescent Nano-Probing in Cancer Research. Int. J. Mol. Sci. 2017, 18, 2066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.-H.; Tchetgna, F.L.D.; Krufczik, M.; Schmitt, E.; Cremer, C.; Estvater, F.B.; Hausmann, M. COMBO-FISH: A Versatile Tool Beyond Standard FISH to Study Chromatin Organization by Fluorescence Light Microscopy. OBM Genet. 2018, 3, 1. [Google Scholar] [CrossRef]
- Zhang, Y.; Máté, G.; Müller, P.; Hillebrandt, S.; Krufczik, M.; Bach, M.; Kaufmann, R.; Hausmann, M.; Heermann, D.W. Radiation Induced Chromatin Conformation Changes Analysed by Fluorescent Localization Microscopy, Statistical Physics, and Graph Theory. PLoS ONE 2015, 10, e0128555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hausmann, M.; Wagner, E.; Lee, J.-H.; Schrock, G.; Schaufler, W.; Krufczik, M.; Papenfuss, F.; Port, M.; Bestvater, F.; Scherthan, H. Super-resolution localization microscopy of radiation-induced histone H2AX-phosphorylation in relation to H3K9-trimethylation in HeLa cells. Nanoscale 2018, 10, 4320–4331. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, A.; Krufczik, M.; Heermann, D.W.; Hausmann, M. Using Persistent Homology as a New Approach for Super-Resolution Localization Microscopy Data Analysis and Classification of γH2AX Foci/Clusters. Int. J. Mol. Sci. 2018, 19, 2263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eryilmaz, M.; Schmitt, E.; Krufczik, M.; Theda, F.; Lee, J.-H.; Cremer, C.; Bestvater, F.; Schaufler, W.; Hausmann, M.; Hildenbrand, G. Localization Microscopy Analyses of MRE11 Clusters in 3D-Conserved Cell Nuclei of Different Cell Lines. Cancers 2018, 10, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scherthan, H.; Lee, J.-H.; Maus, E.; Schumann, S.; Muhtadi, R.; Chojowski, R.; Port, M.; Lassmann, M.; Bestvater, F.; Hausmann, M. Nanostructure of Clustered DNA Damage in Leukocytes after In-Solution Irradiation with the Alpha Emitter Ra. Cancers 2019, 11, 1877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Depes, D.; Lee, J.-H.; Bobkova, E.; Jezkova, L.; Falkova, I.; Bestvater, F.; Pagacova, E.; Kopecna, O.; Zadneprianetc, M.; Bacikova, A.; et al. Single-molecule localization microscopy as a promising tool for γH2AX/53BP1 foci exploration. Eur. Phys. J. 2018, 72, 158. [Google Scholar] [CrossRef]
- Bobkova, E.; Depes, D.; Lee, J.-H.; Jezkova, L.; Falkova, I.; Pagacova, E.; Kopecna, O.; Zadneprianetc, M.; Bacikova, A.; Kulikova, E.; et al. Recruitment of 53BP1 Proteins for DNA Repair and Persistence of Repair Clusters Differ for Cell Types as Detected by Single Molecule Localization Microscopy. Int. J. Mol. Sci. 2018, 19, 3713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hausmann, M.; Neitzel, C.; Bobkova, E.; Nagel, D.; Hofmann, A.; Chramko, T.; Smirnova, E.; Kopečná, O.; Pagáčová, E.; Boreyko, A.; et al. Single Molecule Localization Microscopy Analyses of DNA-Repair Foci and Clusters Detected Along Particle Damage Tracks. Front. Phys. 2020, 8, 473. [Google Scholar] [CrossRef]
- Krufczik, M.; Sievers, A.; Hausmann, A.; Lee, J.-H.; Hildenbrand, G.; Schaufler, W.; Hausmann, M. Combining Low Temperature Fluorescence DNA-Hybridization, Immunostaining, and Super-Resolution Localization Microscopy for Nano-Structure Analysis of ALU Elements and Their Influence on Chromatin Structure. Int. J. Mol. Sci. 2017, 18, 1005. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hausmann, M.; Neitzel, C.; Hahn, H.; Winter, R.; Falkova, I.; Heermann, D.W.; Pilarczyk, G.; Hildenbrand, G.; Scherthan, H.; Falk, M. Space and Time in the Universe of the Cell Nucleus after Ionizing Radiation Attacks: A Comparison of Cancer and Non-Cancer Cell Response. Med. Sci. Forum 2021, 3, 15. https://doi.org/10.3390/IECC2021-09219
Hausmann M, Neitzel C, Hahn H, Winter R, Falkova I, Heermann DW, Pilarczyk G, Hildenbrand G, Scherthan H, Falk M. Space and Time in the Universe of the Cell Nucleus after Ionizing Radiation Attacks: A Comparison of Cancer and Non-Cancer Cell Response. Medical Sciences Forum. 2021; 3(1):15. https://doi.org/10.3390/IECC2021-09219
Chicago/Turabian StyleHausmann, Michael, Charlotte Neitzel, Hannes Hahn, Ruth Winter, Iva Falkova, Dieter W. Heermann, Götz Pilarczyk, Georg Hildenbrand, Harry Scherthan, and Martin Falk. 2021. "Space and Time in the Universe of the Cell Nucleus after Ionizing Radiation Attacks: A Comparison of Cancer and Non-Cancer Cell Response" Medical Sciences Forum 3, no. 1: 15. https://doi.org/10.3390/IECC2021-09219
APA StyleHausmann, M., Neitzel, C., Hahn, H., Winter, R., Falkova, I., Heermann, D. W., Pilarczyk, G., Hildenbrand, G., Scherthan, H., & Falk, M. (2021). Space and Time in the Universe of the Cell Nucleus after Ionizing Radiation Attacks: A Comparison of Cancer and Non-Cancer Cell Response. Medical Sciences Forum, 3(1), 15. https://doi.org/10.3390/IECC2021-09219








