Abstract
The use of antibiotics in the veterinary and zootechnic sectors poses a challenge to the reduction in antibiotic resistance rates. We evaluated the presence of antibiotic-resistant bacteria in the wastewater of dairy farms in the Sicily Region, Italy. The samples were examined by isolating and identifying the bacterial strains, which were then tested for the main classes of antibiotics. Aeromonas spp. and Vibrio spp. were the more commonly isolated strains (18.2%), followed by Pseudomonas spp. (15.9%), Enterobacter spp. and Citrobacter spp. (13.6%). Macrolides were the drugs against which the highest resistance was detected, followed by tetracyclines, penicillins, and cephalosporins. The agri-food chain and zootechnic areas embody an important source of bacteria resistant to antibiotics, and their presence in wastewater from processing factories could play a pivotal role in spreading these microorganisms and in environmental contamination.
1. Introduction
Antimicrobial resistance (AMR) represents one of the major public health threats, contributing to remarkable increases in terms of morbidity, mortality, and sanitary costs [1,2].
In veterinary medicine, zootechnics, agriculture, and aquaculture, antibiotics are widely used not only to maintain animal welfare and food safety but also to prevent infections [3,4]. Bacterial strains resistant to antibiotics are associated with the use of antibiotics in animals raised for food [5]. Animals have the ability to propagate these strains in the environment and introduce them into slaughterhouses. The high number of slaughterhouses present in Italy makes slaughterhouse wastewater a potential route for antibiotic-resistant bacteria and environmental contamination [6].
Milk is a complex product that can be contaminated by various microorganisms and can be a source of human diseases such as brucellosis, which is still endemic in southern Italy [7,8]. However, its role in the transmission of antibiotic-resistant bacteria is still unclear. The impact of antibiotics on the proliferation of AR in dairy industries has not been fully documented [9], but a correlation has been established between AR in cattle and human infections [10,11,12]. The dairy production chain could represent one potential AR transmission route [13]. Among the approved antibiotics for dairy operations, penicillins, cephalosporins, and tetracyclines are the most commonly used, followed by macrolides and sulfonamides, and tetracyclines and sulfonamides are the most frequently used for prophylactic purposes [14].
The aim of this study is to assess the presence of antibiotic-resistant bacteria in the wastewater collected by some dairies located in Sicily, Italy, in order to determine the potential role played regarding these contexts in the spread of antibiotic resistance.
2. Materials and Methods
2.1. Samples and Typology of Milk Processing Plants
We focused on 31 facilities located in all nine provincial territories of Sicily. The processing facilities are distinguished by the type of milk processed: bovine, sheep, goat, and buffalo milk. The majority of the facilities we studied are related to the PP (Processing Plant) type. In only two cases, the plants perform the functions of both CC (Collecting Center) and PP as described in accordance with Regulation (EC) no. 853/2004 [15].
The nature of the wastewater was sampled before it was introduced into the purification plants. The sample we took, at the very least, was a mixture of milk, milk-based products, or derivatives with water. Before cleaning and disinfection, they were collected when the equipment and tools for dairy products, including containers used for dairy products, were rinsed.
2.2. Microbiological Analyses
2.2.1. Treatment of the Samples and Phenotypic Identification of Bacteria
To evaluate the Enterobacteriaceae load, 20 µL of the samples were inoculated directly on Hektoen Enteric Agar (H.E.A.) and 200 µL were inoculated in a test tube containing an enrichment liquid medium (EE Broth-Mossel). The selection of colonies occurred based on their phenotypic characteristics after incubating at 37 °C for 24 h. To obtain a general representation of the Enterobacteriaceae strains present in each sample, some of these were collected. API 20 NE and API 20 E profiles (bioMerieux, Marcy l’Etoile, France) were used to identify the isolates at the species or genus level, as per the manufacturer’s instructions.
2.2.2. Antibiotic Susceptibility
Among the isolates, some were chosen based on their greater frequency of isolation. Selected strains underwent the Kirby–Bauer test. Different antibiotics grouped into the main classes according to their mechanisms of action were used (M100. Performance Standards for Antimicrobial Susceptibility Testing. 31st Edition. 2021).
2.2.3. Statistical Analyses
Prism 4.0 software was used to collect and analyze all the obtained data. The percentages and the 95% confidence interval (CI) were determined using descriptive statistics. Using chi-squared tests, we evaluated the Odds Ratio (OR) and the 95% CI to assess the association between antibiotic resistance and different origins of strains. Significance was assessed at the p < 0.05 level.
3. Results
3.1. Bacterial Detection
Figure 1 displays the percentages of the bacterial genera that were found.
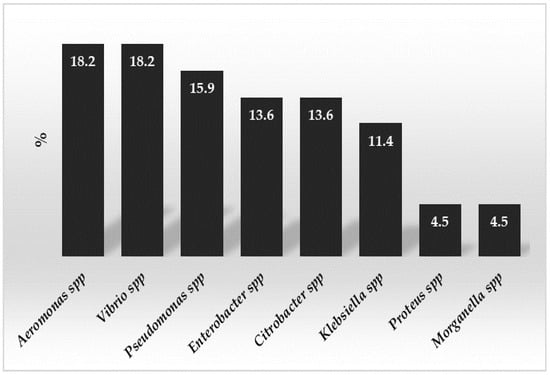
Figure 1.
Percentages of the bacterial genera detected in the analyzed wastewater samples.
The most frequently found genera were Aeromonas spp. and Vibrio spp., with equal percentages, comprising more than one third of all the bacteria found. Following Enterobacter spp. and Citrobacter spp. as the most often detected genera, Pseudomonas spp. came in second. Pseudomonas aeruginosa comprised 40% of all the strains in the genus Pseudomonas spp. Additionally, nearly half (48.6%) of the identified strains were found in wastewater from artisanal plants that did not have purification equipment and discharged their waste straight into the sewer or into an Imhoff tank through decanting and crop irrigation.
3.2. Evaluation of Antibiotic Resistance
Forty-four bacterial strains were picked from the analyzed samples and evaluated for antibiotic sensitivity. Figure 2 displays the overall mean percentages of antibiotic resistance of the selected bacterial strains.
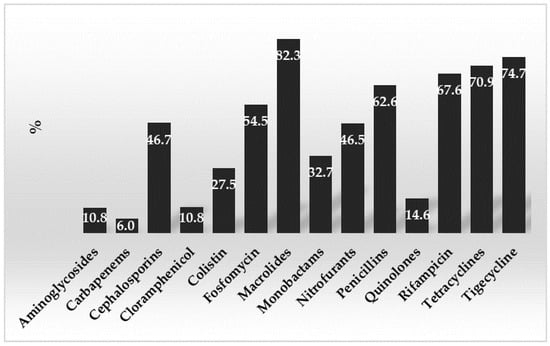
Figure 2.
Percentages of general resistance of the detected bacterial strains to the used antibiotics divided by family.
The macrolides, followed by tigecycline, tetracyclines, and rifampicin, were the medications toward which the highest mean percentage values of resistance were found. The penicillins and cephalosporins also had high percentage values. In contrast, low percentage values were found for the antibiotics that had the lowest mean % value, including carbapenems, quinolones, aminoglycosides, chloramphenicol, and others. For the cephalosporins, Fosfomycin, macrolides, monobactams (p = 0.000456), nitrofurans (p = 0.000456), and quinolones (p = 0.018422), decreases in the mean levels of antibiotic resistance were found. However, it should be noted that, for colistin and carbapenems, the results obtained highlighted greater resistance in Aeromonas compared to the other studied strains.
4. Discussion
Our study examined the wastewater gathered from dairy processing facilities situated in Sicily, a southern Italian region. The Gram-negative bacteria belonging to the genera Aeromonas, Vibrio, and Pseudomonas were the most frequently found among the bacterial species, likely as a result of their widespread environmental diffusion, particularly in aquatic environments. Enterobacteria from the families Enterobacter, Citrobacter, and Klebsiella were next in line. Additionally, Proteus spp. and Morganella morganii were found in smaller quantities. The previous evidence has focused attention on the role of MDR-Proteus spp. as an emergent cause of healthcare-associated infections and environmental contamination [16,17,18]. Due to their antibiotic resistance, Aeromonas spp. are becoming increasingly significant in human pathophysiology, as demonstrated by numerous earlier studies [19,20,21]. Aeromonas spp. showed less antibiotic resistance in our investigation than the other discovered genera. According to previously published research [6], these isolates exhibited moderate sensitivity to aminoglycosides, chloramphenicol, and quinolones and high resistance to penicillins, cephalosporins, and macrolides. However, we must draw attention to a few criticisms, such as the relatively high resistance to colistin and carbapenems in comparison to other strains.
High resistance levels have been found in several important antibiotic groups that are widely used in human medicine, including cephalosporins, macrolides, penicillins, tetracyclines, and tigecycline. It was reported that macrolides had the highest resistance rate. Penicillins and cephalosporins were by far the compounds regarding which the highest levels of resistance were found among β-lactams. Fortunately, very low carbapenem resistance levels were reported. These results are most likely a result of the widespread use of these medications in veterinary medicine to treat a variety of clinical diseases, the subsequent selection of resistant clones that can contaminate milk, and, finally, the processing environment.
5. Conclusions
Antibiotic-resistant bacteria can be found in large quantities in the livestock and agri-food sectors, and their presence in wastewater from processing facilities may contribute to environmental pollution by these germs. Italy undoubtedly ranks among the nations with the largest milk production, and there are milk processing facilities located throughout many different nations and regions. According to our study, these plants’ wastewater collection systems are tainted with bacteria that are resistant to antibiotics, which can be a significant cause of environmental contamination. Due to the lack of purification plants, small facilities that process only small amounts of milk are especially harmful. Antibiotic resistance is a major worry, so it is critical to monitor the situation at all times to minimize danger and address this issue.
The authors believe that, although there are many publications on the topic in the literature, the data obtained from their study can enrich the information existing in the literature. Nowadays, from a One Health perspective, any scientific contribution can contribute to understanding the complex phenomenon of antibiotic resistance. Furthermore, in the literature, there are many generic studies on wastewater, but very few concern wastewater coming exclusively from specific sectors, such as dairies or other livestock establishments.
Author Contributions
Conceptualization, P.L. and A.V.; methodology, A.F., C.E.R., P.T., M.E.G. and I.L.S.; formal analysis, investigation, and data curation, A.F. and P.L.; resources, A.V.; writing—original draft preparation, A.F.; supervision, P.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are available from the corresponding author upon specific request of the interested party.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations the Review on Antimicrobial Resistance. 2016. Available online: https://www.biomerieuxconnection.com/wp-content/uploads/2018/04/Tackling-Drug-Resistant-Infections-Globally_-Final-Report-and-Recommendations.pdf (accessed on 12 March 2024).
- Mancuso, G.; De Gaetano, S.; Midiri, A.; Zummo, S.; Biondo, C. The Challenge of Overcoming Antibiotic Resistance in Carbapenem-Resistant Gram-Negative Bacteria: “Attack on Titan”. Microorganisms 2023, 11, 1912. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.L.; Caffrey, N.P.; Nóbrega, D.B.; Cork, S.C.; Ronksley, P.E.; Barkema, H.W.; Polachek, A.J.; Ganshorn, H.; Sharma, N.; Kellner, J.D.; et al. Restricting the use of antibiotics in food-producing animals and its associations with antibiotic resistance in food-producing animals and human beings: A systematic review and meta-analysis. Lancet Planet. Health 2017, 1, e316–e327. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.L.; Caffrey, N.P.; Nóbrega, D.B.; Cork, S.C.; Ronksley, P.E.; Barkema, H.W.; Polachek, A.J.; Ganshorn, H.; Sharma, N.; Kellner, J.D.; et al. Comparison of different approaches to antibiotic restriction in food-producing animals: Stratified results from a systematic review and meta-analysis. BMJ Glob. Health 2019, 4, e001710. [Google Scholar] [CrossRef] [PubMed]
- Marshall, B.M.; Levy, S.B. Food animals and antimicrobials: Impacts on human health. Clin. Microbiol. Rev. 2011, 24, 718–733. [Google Scholar] [CrossRef] [PubMed]
- Facciolà, A.; Virga, A.; Gioffrè, M.E.; Laganà, P. Evaluation of Antibiotic Resistance in Bacterial Strains Isolated from Sewage of Slaughterhouses Located in Sicily (Italy). Int. J. Environ. Res. Public Health 2021, 18, 9611. [Google Scholar] [CrossRef] [PubMed]
- Facciolà, A.; Palamara, M.A.R.; D’Andrea, G.; Marano, F.; Magliarditi, D.; Puglisi, G.; Picerno, I.; Di Pietro, A.; Visalli, G. Brucellosis is a public health problem in southern Italy: Burden and epidemiological trend of human and animal disease. J. Infect. Public Health 2018, 11, 861–866. [Google Scholar] [CrossRef] [PubMed]
- Facciolà, A.; Visalli, G.; D’Andrea, G.; Laganà, A.; Varvarà, M.; Spataro, P.; Di Pietro, A. The Italian Mandatory Notification System: An Important Public Health Tool for Continuous Monitoring of Infectious Diseases. New Microbiol. 2022, 45, 115–123. [Google Scholar] [PubMed]
- Oliver, S.P.; Murinda, S.E.; Jayarao, B.M. Impact of antibiotic use in adult dairy cows on antimicrobial resistance of veterinary and human pathogens: A comprehensive review. Foodborne Pathog. Dis. 2011, 8, 337–355. [Google Scholar] [CrossRef]
- Food and Drug Administration. The Judicious Use of Medically Important Antimicrobial Drugs in Food-Producing Animals. 2012. Available online: https://www.fda.gov/media/79140/download (accessed on 12 March 2024).
- Casey, J.A.; Curriero, F.C.; Cosgrove, S.E.; Nachman, K.E.; Schwartz, B.S. High-density livestock operations, crop field application of manure, and risk of community-associated methicillin-resistant Staphylococcus aureus infection in Pennsylvania. JAMA Intern. Med. 2013, 173, 1980–1990. [Google Scholar] [CrossRef] [PubMed]
- Madec, J.Y.; Haenni, M.; Nordmann, P.; Poirel, L. Extended-spectrum β-lactamase/AmpC- and carbapenemase-producing Enterobacteriaceae in animals: A threat for humans? Clin. Microbiol. Infect. 2017, 23, 826–833. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulos, P.; Papadopoulos, T.; Angelidis, A.S.; Boukouvala, E.; Zdragas, A.; Papa, A.; Hadjichristodoulou, C.; Sergelidis, D. Prevalence of Staphylococcus aureus and of methicillin-resistant S. aureus (MRSA) along the production chain of dairy products in north-western Greece. Food Microbiol. 2018, 69, 43–50. [Google Scholar] [CrossRef] [PubMed]
- National Animal Health Monitoring System. Dairy 2014–Health and Management Practices on U.S. Dairy Operations, 2014. USDA, Animal and Plant Health Inspection Service, Veterinary Services, Center for Epidemiology and Animal Health, National Animal Health Monitoring System. 2018. Available online: https://www.aphis.usda.gov/animal_health/nahms/dairy/downloads/dairy14/Dairy14_dr_PartIII.pdf (accessed on 15 March 2024).
- The European Parliament and the Council of the European Union. Regulation (EC) No 853/2004 of the European Parliament and of the Council. Available online: https://www.legislation.gov.uk/eur/2004/853/contents (accessed on 11 March 2024).
- Laganà, P.; Votano, L.; Caruso, G.; Azzaro, M.; Lo Giudice, A.; Delia, S. Bacterial isolates from the Arctic region (Pasvik River, Norway): Assessment of biofilm production and antibiotic susceptibility profiles. Environ. Sci. Pollut. Res. Int. 2018, 25, 1089–1102. [Google Scholar] [CrossRef] [PubMed]
- Badger-Emeka, L.; Al Rashed, A.S.; Aljindan, R.Y.; Emeka, P.M.; Quadri, S.A.; Almutairi, H.H. Incidence of Drug-Resistant Hospital-Associated Gram-Negative Bacterial Infections, the Accompanying Risk Factors, and Clinical Outcomes with Treatment. Antibiotics 2023, 12, 1425. [Google Scholar] [CrossRef] [PubMed]
- Facciolà, A.; Gioffrè, M.E.; Chiera, D.; Ferlazzo, M.; Virga, A.; Laganà, P. Evaluation of antibiotic resistance in Proteus spp: A growing trend that worries Public Health. Results of 10 Years of Analysis. New Microbiol. 2022, 45, 269–277. [Google Scholar] [PubMed]
- Yuwono, C.; Wehrhahn, M.C.; Liu, F.; Riordan, S.M.; Zhang, L. The Isolation of Aeromonas Species and Other Common Enteric Bacterial Pathogens from Patients with Gastroenteritis in an Australian Population. Microorganisms 2021, 9, 1440. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Yuwono, C.; Tay, A.C.Y.; Wehrhahn, M.C.; Riordan, S.M.; Zhang, L. Analysis of global Aeromonas veronii genomes provides novel information on source of infection and virulence in human gastrointestinal diseases. BMC Genom. 2022, 23, 166. [Google Scholar] [CrossRef]
- Lo, C.C.; Liao, W.Y.; Chou, M.C.; Wu, Y.Y.; Yeh, T.H.; Lo, H.R. Overexpression of Resistance-Nodulation-Division Efflux Pump Genes Contributes to Multidrug Resistance in Aeromonas hydrophila Clinical Isolates. Microb. Drug Resist. 2022, 28, 153–160. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).