Shotgun Proteomics: A Powerful Tool for Investigating the Chemical Complexity of Biscuit Melanoidins †
Abstract
1. Introduction
2. Materials and Methods
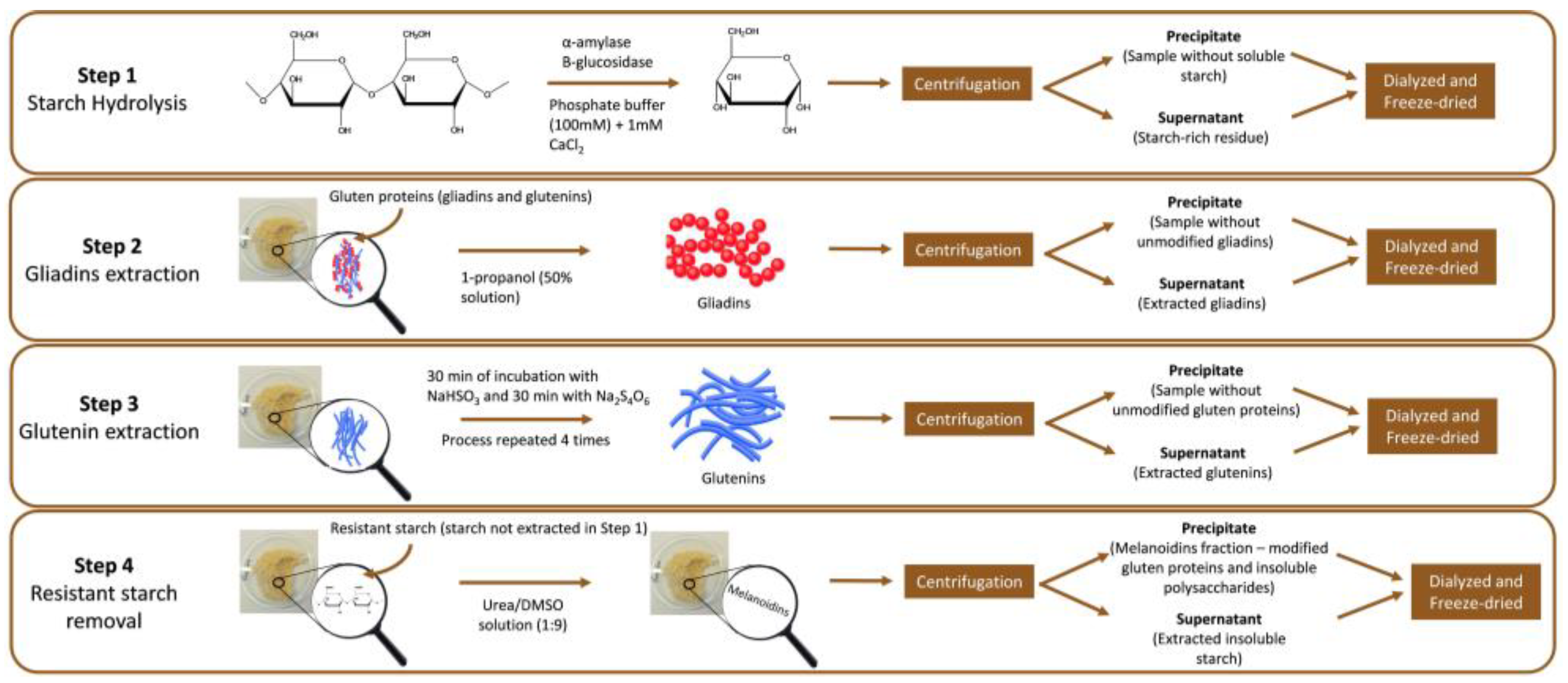
2.1. Sample Preparation
2.2. Sample Fractionation
2.3. Color Measurement
2.4. Peptide Analysis
2.5. Identification of Proteins and Possible Modifications Induced by MR
3. Results and Discussion
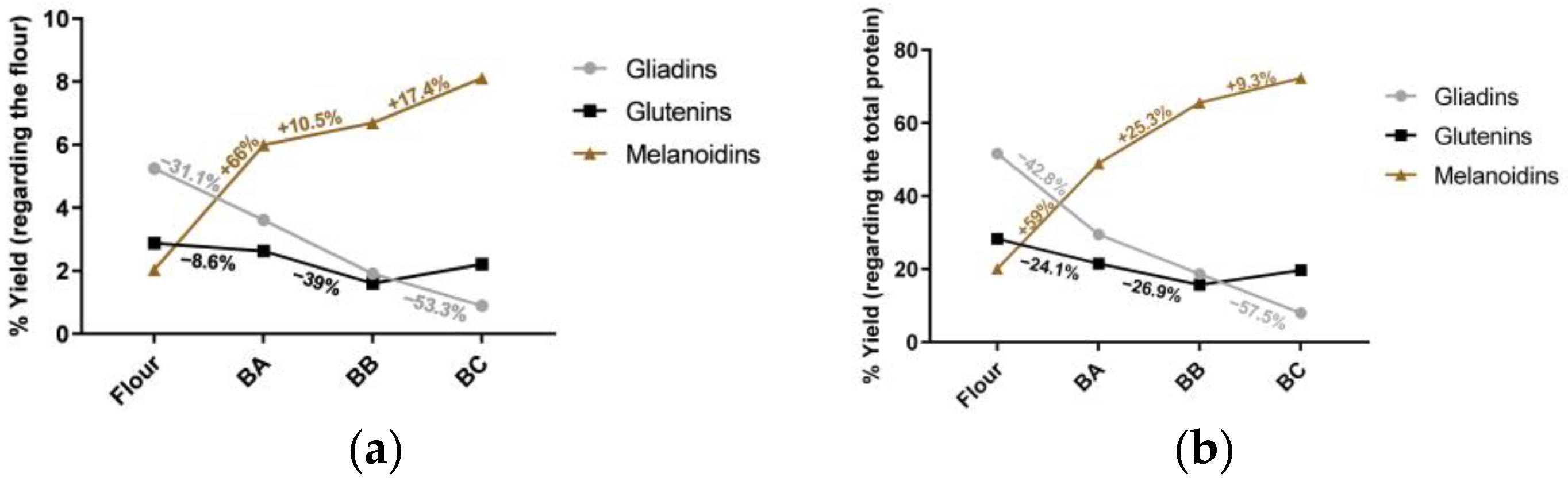
3.1. Fractionation Yields
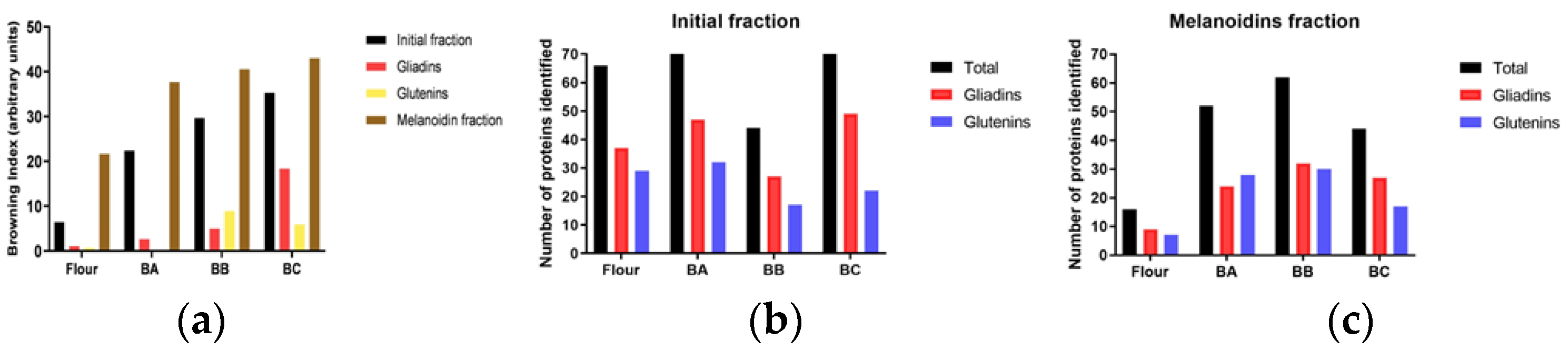
3.2. Color of the Samples
3.3. Identification of the Proteins of the Fractions
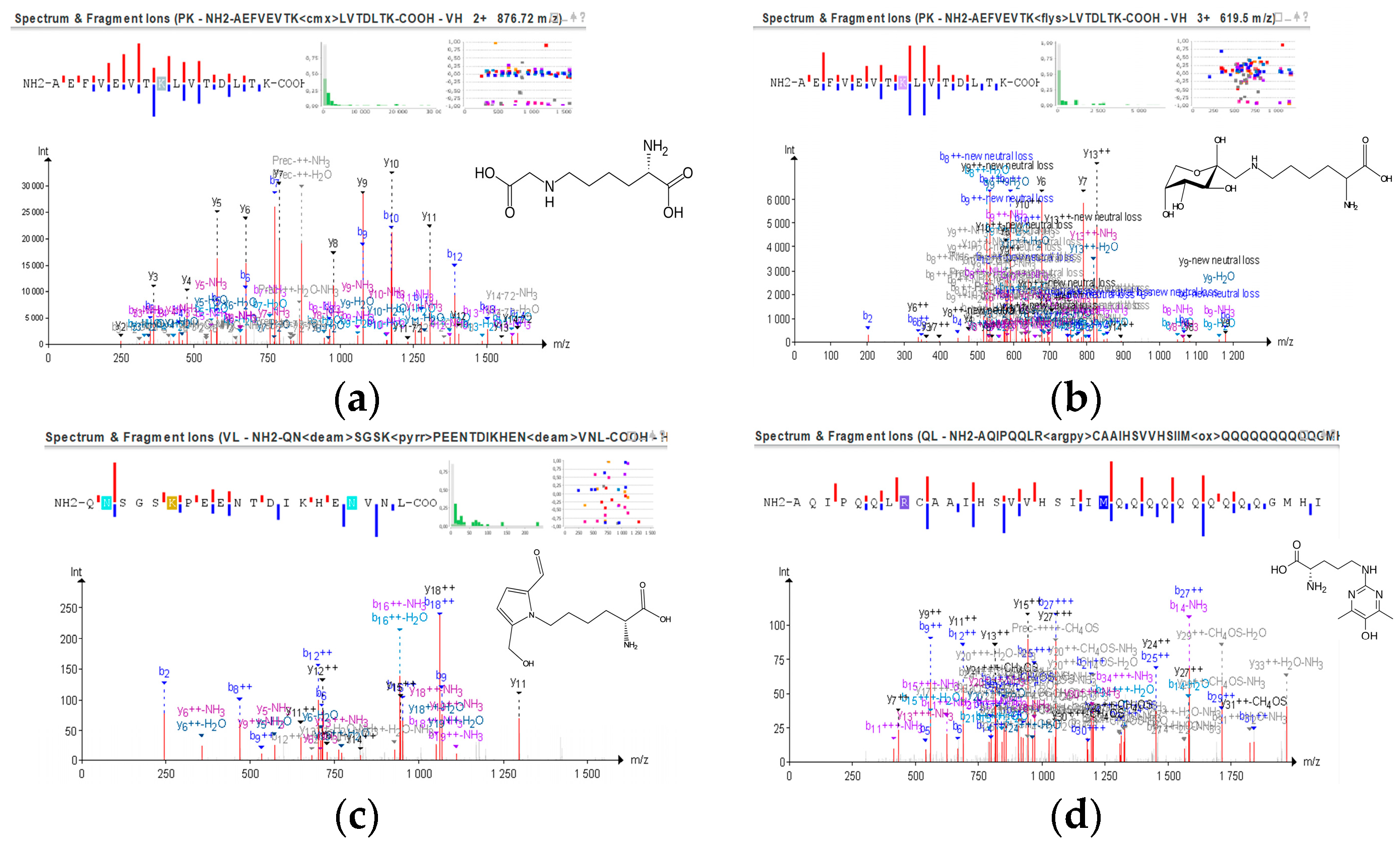
3.4. Assessment of Protein Modifications Induced by MR
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fogliano, V.; Morales, F.J. Estimation of dietary intake of melanoidins from coffee and bread. Food Funct. 2011, 2, 117–123. [Google Scholar] [CrossRef] [PubMed]
- AACC Approved Methods of Analysis, 11th ed.; Method 10-50.05. Baking Quality of Cookie Flour; Cereals & Grains Association: St. Paul, MN, USA, 1999.
- Buera, M.P.; Petriella, C.; Lozano, R.D. Definition of Colour in the Non-Enzymatic Browning. Die Farbe 1985, 33, 316–326. [Google Scholar]
- Wang, F.; Chen, R.; Zhu, J.; Sun, D.; Song, C.; Wu, Y.; Ye, M.; Wang, L.; Zou, H. A fully automated system with online sample loading, isotope dimethyl labeling and multidimensional separation for high-throughput quantitative proteome analysis. Anal. Chem. 2010, 82, 3007–3015. [Google Scholar] [CrossRef] [PubMed]
- Barsnes, H.; Huber, S.; Sickmann, A.; Eidhammer, I.; Martens, L. OMSSA Parser: An open-source library to parse and extract data from OMSSA MS/MS search results. Proteomics 2009, 9, 3772–3774. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Wang, Q.; Chen, Y.; Lei, L.; Lei, X.; Zhao, J.; Zhang, Y.; Ming, J. Changes in the structural and physicochemical properties of wheat gliadin and maize amylopectin conjugates induced by dry-heating. J. Food Sci. 2022, 8, 3459–3471. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Siopa, J.; Ribeiro, M.; Cosme, F.; Nunes, F.M. Shotgun Proteomics: A Powerful Tool for Investigating the Chemical Complexity of Biscuit Melanoidins. Med. Sci. Forum 2023, 23, 7. https://doi.org/10.3390/msf2023023007
Siopa J, Ribeiro M, Cosme F, Nunes FM. Shotgun Proteomics: A Powerful Tool for Investigating the Chemical Complexity of Biscuit Melanoidins. Medical Sciences Forum. 2023; 23(1):7. https://doi.org/10.3390/msf2023023007
Chicago/Turabian StyleSiopa, João, Miguel Ribeiro, Fernanda Cosme, and Fernando M. Nunes. 2023. "Shotgun Proteomics: A Powerful Tool for Investigating the Chemical Complexity of Biscuit Melanoidins" Medical Sciences Forum 23, no. 1: 7. https://doi.org/10.3390/msf2023023007
APA StyleSiopa, J., Ribeiro, M., Cosme, F., & Nunes, F. M. (2023). Shotgun Proteomics: A Powerful Tool for Investigating the Chemical Complexity of Biscuit Melanoidins. Medical Sciences Forum, 23(1), 7. https://doi.org/10.3390/msf2023023007







