In Silico Approach to Assessing the Polyphenols from Krishna Tulsi (Ocimum tenuiflorum L.) as a Keap1/Nrf2 Receptor for the Treatment of Inflammatory Bowel Disease †
Abstract
1. Introduction
2. Materials and Methods
- Protein preparation: the Keap1 receptor was obtained from a protein data bank (4ZY3). The protein was then modeled using AutoDock Tools and Biovia discovery studio.
- Selection of ligand: three-dimensional structures of syringic acid, caffeic acid, ferulic acid, catechin and epicatechin were obtained from PubChem database.
- Active site of target protein: PyMOL software was used to analyze the target binding site of the receptor protein.
- Molecular docking: the target protein was prepared using AutoDock Tools and Biovia discovery studio. The grid box on the active binding site was generated for docking. The ligands were docked to the Keap1 receptor using the default settings in Autodock Vina. The program was run for a specified number of generations, and the binding affinity and interactions of the compounds with the receptor were analyzed.
- Data analysis: The results of the docking simulations were analyzed using the Biovia discovery studio, which generates a summary of the best-docked complexes and their binding energies. The binding modes and binding affinities of the Ocimum tenuiflorum compounds to the Keap1/Nrf2 receptor were analyzed and compared to identify the most promising compounds for further study.
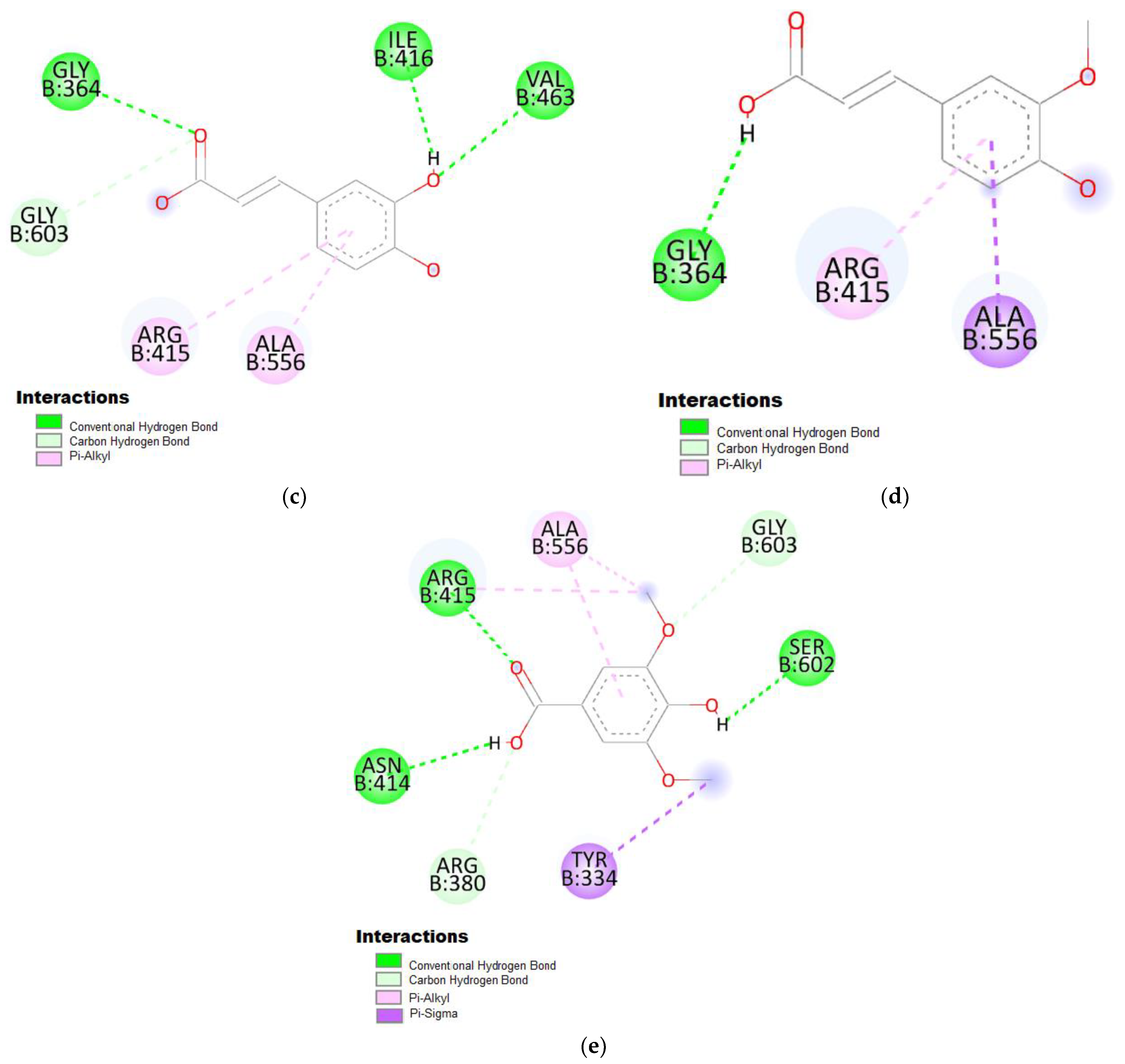
3. Results
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tsopmejio, I.S.N.; Ding, M.; Wei, J.; Zhao, C.; Jiang, Y.; Li, Y.; Song, H. Auricularia polytricha and Flammulina velutipes ameliorate inflammation and modulate the gut microbiota via regulation of NF-κB and Keap1/Nrf2 signaling pathways on DSS-induced inflammatory bowel disease. Food Biosci. 2022, 47, 101426. [Google Scholar] [CrossRef]
- Jakubczyk, D.; Leszczyńska, K.; Górska, S. The Effectiveness of Probiotics in the Treatment of Inflammatory Bowel Disease (IBD)—A Critical Review. Nutrients 2020, 12, 1973. [Google Scholar] [CrossRef] [PubMed]
- Ha, C.; McGovern, D.; Melmed, G.Y. Crohn’s disease: Clinical manifestations and management. In Yamada’s Textbook of Gastroenterology; Wiley Online Library: Hoboken, NJ, USA, 2022; pp. 1294–1323. [Google Scholar] [CrossRef]
- Williams, I.; Pandey, S.; Haller, W.; Huynh, H.Q.; Chan, A.; Düeker, G.; Bettels, R.; Peyrin-Biroulet, L.; Dike, C.R.; DeGeeter, C.; et al. Anti-TNF therapy for inflammatory bowel disease in patients with neurodegenerative Niemann-Pick disease Type C. Wellcome Open Res. 2022, 7, 11. [Google Scholar] [CrossRef] [PubMed]
- Alemany-Cosme, E.; Sáez-González, E.; Moret, I.; Mateos, B.; Iborra, M.; Nos, P.; Sandoval, J.; Beltrán, B. Oxidative Stress in the Pathogenesis of Crohn’s Disease and the Interconnection with Immunological Response, Microbiota, External Environmental Factors, and Epigenetics. Antioxidants 2021, 10, 64. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Chang, E.B. Inflammatory Bowel Diseases (IBD) and the Microbiome—Searching the Crime Scene for Clues. Gastroenterology 2021, 160, 524–537. [Google Scholar] [CrossRef] [PubMed]
- Pinzi, L.; Rastelli, G. Molecular Docking: Shifting Paradigms in Drug Discovery. Int. J. Mol. Sci. 2019, 20, 4331. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Chao, X.; Wu, J.; Ma, X.; Yang, Y.; Wu, Y.; Lin, J. Exploring the Potential Mechanism of Guchang Zhixie Wan for Treating Ulcerative Colitis by Comprehensive Network Pharmacological Approaches and Molecular Docking Validation as Well as Cell Experiments. Chem. Biodivers. 2021, 18, e2000810. [Google Scholar] [CrossRef] [PubMed]
- Dakshayani, L.; Merchant, N.; Smitha, S.; Surendra, G.; Reddy, T.G.; Mamatha, D.; Deepthi, G.; Ramana, D.V.; Chandrasekhar, T.; Reddy, M.C. Holy Basil: A Potential Herbal Source for Therapeutic Applications. Curr. Trends Biotechnol. Pharm. 2021, 15, 87–100. [Google Scholar] [CrossRef]
- Antonescu, A.-I.; Miere, F.; Fritea, L.; Ganea, M.; Zdrinca, M.; Dobjanschi, L.; Antonescu, A.; Vicas, S.I.; Bodog, F.; Sindhu, R.K.; et al. Perspectives on the Combined Effects of Ocimum basilicum and Trifolium pratense Extracts in Terms of Phytochemical Profile and Pharmacological Effects. Plants 2021, 10, 1390. [Google Scholar] [CrossRef] [PubMed]
- Hussain, A.I.; Chatha, S.A.S.; Kamal, G.M.; Ali, M.A.; Hanif, M.A.; Lazhari, M.I. Chemical composition and biological activities of essential oil and extracts from Ocimum sanctum. Int. J. Food Prop. 2017, 20, 1569–1581. [Google Scholar] [CrossRef]
- Joshi, R.K.; Hoti, S.L. Chemical composition of the essential oil of Ocimum tenuiflorum L. (Krishna Tulsi) from North West Karnataka, India. Plant Sci. Today 2014, 1, 99–102. [Google Scholar] [CrossRef]
- Palla, R.; Elumalai, A.; Eswaraiah, M.C.; Raju, K. A review on krishna tulsi, Ocimum tenuiflorum Linn. Int. J. Res. Ayurveda Pharm. 2012, 3, 291–293. [Google Scholar]
- Sharma, Y.; Bharadwaj, M.; Srivastava, N.; Kaur, A.; Kumar, M.; Agarwal, M.; Bahl, Y.; Bala, K. In vitro antioxidant activity of defatted seed extracts of Ocimum sanctum on rat PC-12 cells and its inhibitory efficacy with receptors of oral squamous cell carcinoma. Ind. Crops Prod. 2020, 154, 112668. [Google Scholar] [CrossRef]
- Elzoheiry, A.; Ayad, E.; Omar, N.; Elbakry, K.; Hyder, A. Anti-liver fibrosis activity of curcumin/chitosan-coated green silver nanoparticles. Sci. Rep. 2022, 12, 18403. [Google Scholar] [CrossRef] [PubMed]
- Dodson, M.; Redmann, M.; Rajasekaran, N.S.; Darley-Usmar, V.; Zhang, J. Correction: KEAP1–NRF2 signalling and autophagy in protection against oxidative and reductive proteotoxicity. Biochem. J. 2015, 471, 431. [Google Scholar] [CrossRef] [PubMed]




| Ligands | Estimated Free Binding Energy (kcal/mol) | Estimated Inhibition Constant (Ki) (µM) |
|---|---|---|
| Catechin | −8.2 | 0.960 |
| Epicatechin | −7.9 | 1.595 |
| Caffeic acid | −6.4 | 20.116 |
| Ferulic acid | −5.9 | 46.823 |
| Syringic acid | −5.6 | 77.734 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, S.; Sarkar, B. In Silico Approach to Assessing the Polyphenols from Krishna Tulsi (Ocimum tenuiflorum L.) as a Keap1/Nrf2 Receptor for the Treatment of Inflammatory Bowel Disease. Med. Sci. Forum 2023, 21, 13. https://doi.org/10.3390/ECB2023-14207
Kumar S, Sarkar B. In Silico Approach to Assessing the Polyphenols from Krishna Tulsi (Ocimum tenuiflorum L.) as a Keap1/Nrf2 Receptor for the Treatment of Inflammatory Bowel Disease. Medical Sciences Forum. 2023; 21(1):13. https://doi.org/10.3390/ECB2023-14207
Chicago/Turabian StyleKumar, Satish, and Biswatrish Sarkar. 2023. "In Silico Approach to Assessing the Polyphenols from Krishna Tulsi (Ocimum tenuiflorum L.) as a Keap1/Nrf2 Receptor for the Treatment of Inflammatory Bowel Disease" Medical Sciences Forum 21, no. 1: 13. https://doi.org/10.3390/ECB2023-14207
APA StyleKumar, S., & Sarkar, B. (2023). In Silico Approach to Assessing the Polyphenols from Krishna Tulsi (Ocimum tenuiflorum L.) as a Keap1/Nrf2 Receptor for the Treatment of Inflammatory Bowel Disease. Medical Sciences Forum, 21(1), 13. https://doi.org/10.3390/ECB2023-14207







