Cost-Effective Markers and Candidate Genes Analysis at Wheat MQTL Loci †
Abstract
1. Introduction
2. Experiments
2.1. Insertion Site-Based Polymorphism Markers
2.2. Candidate Genes
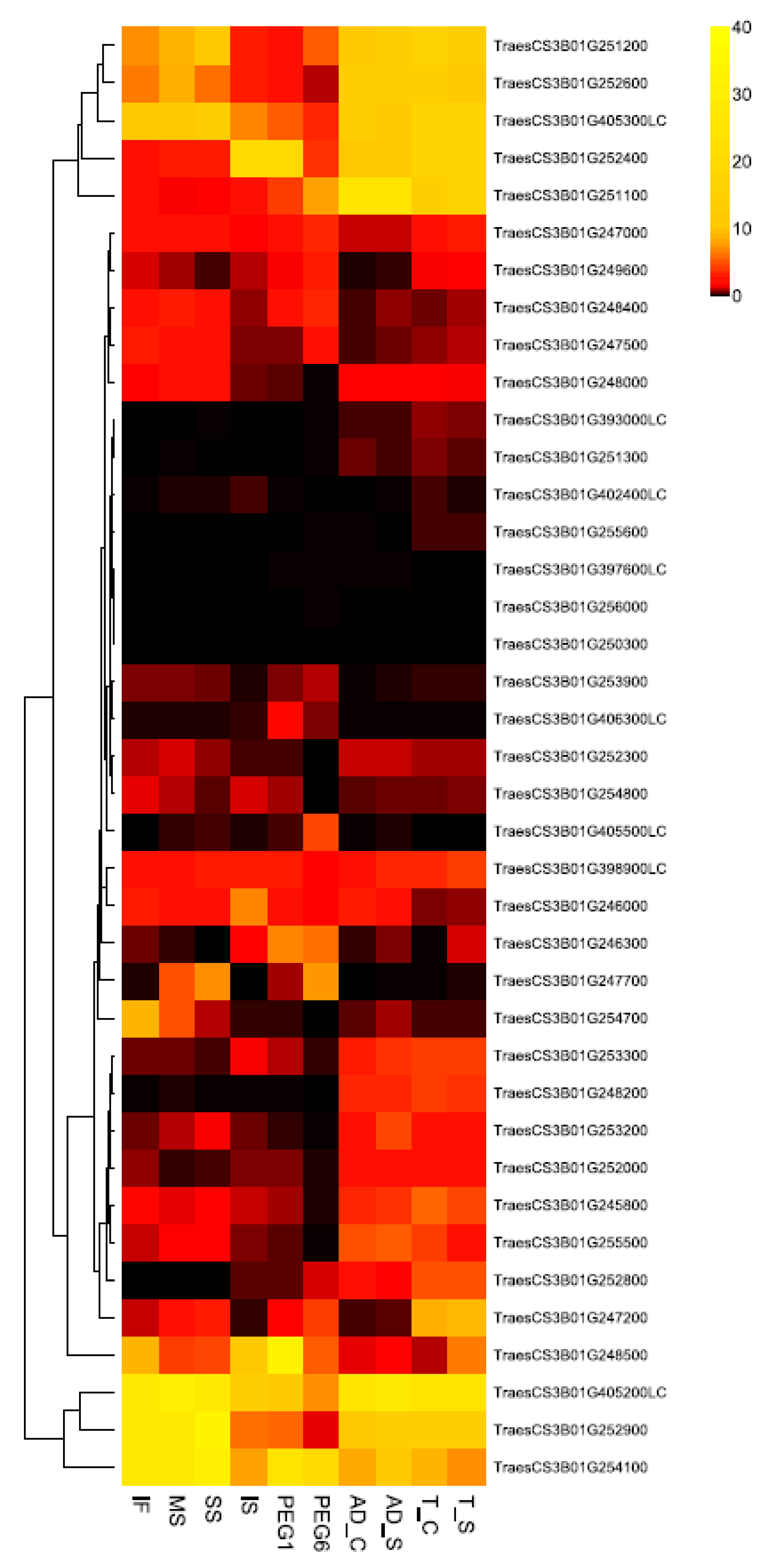
2.3. Gene Expression Analysis
3. Results and Discussion
Author Contributions
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| HRM | High Resolution Melting |
| MQTL | Meta-Quantitative Trait Loci |
| TE | Transposable Element |
| ISBP | Insertion Site-Based Polymorphism |
| bp | base pairs |
| Mb | megabase |
References
- Briggle, L.W.; Curtis, B.C. Wheat Worldwide. In Wheat and Wheat Improvement, 2nd ed.; Heyne, E.G., Ed.; American Socierty of Agronomy: Madison, WI, USA, 1988; Volume 146, pp. 292–293. [Google Scholar] [CrossRef]
- Rampino, P.; Pataleo, S.; Gerardi, C.; Mita, G.; Perrotta, C. Drought stress response in wheat: Physiological and molecular analysis of resistant and sensitive genotypes. Plant Cell Environ. 2006, 29, 2143–2152. [Google Scholar] [CrossRef]
- Sallam, A.; Alqudah, A.M.; Dawood, M.F.A.; Baenziger, P.S.; Börner, A. Drought stress tolerance in wheat and barley: Advances in physiology, breeding and genetics research. Int. J. Mol. Sci. 2019, 20, 3137. [Google Scholar] [CrossRef]
- Wang, W.; Vinocur, B.; Altman, A. Plant responses to drought, salinity and extreme temperatures: Towards genetic engineering for stress tolerance. Planta 2003, 218, 1–14. [Google Scholar] [CrossRef]
- Wang, W.; Vinocur, B.; Shoseyov, O.; Altman, A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004, 9, 244–252. [Google Scholar] [CrossRef] [PubMed]
- IPCC Assessment Report. 2020. Available online: https//www.ipccch/srccl/ (accessed on 31 March 2020).
- Lesk, C.; Rowhani, P.; Ramankutty, N. Influence of extreme weather disasters on global crop production. Nature 2016, 529, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Cattivelli, L.; Rizza, F.; Badeck, F.W.; Mazzucotelli, E.; Mastrangelo, A.M.; Francia, E.; Marè, C.; Tondelli, A.; Stanca, A.M. Drought tolerance improvement in crop plants: An integrated view from breeding to genomics. Field Crops Res. 2008, 105, 1–14. [Google Scholar] [CrossRef]
- Mérida-García, R.; Gálvez, S.; Paux, E.; Dorado, G.; Pascual, L.; Giraldo, P.; Hernandez, P. High resolution melting and insertion site-based polymorphism markers for wheat variability analysis and candidate genes selection at drought and heat MQTL loci. Agronomy 2020, 10, 1294. [Google Scholar] [CrossRef]
- Paux, E.; Faure, S.; Choulet, F.; Roger, D.; Gauthier, V.; Martinant, J.P.; Sourdille, P.; Balfourier, F.; Le Paslier, M.C.; Chauveau, A.; et al. Insertion site-based polymorphism markers open new perspectives for genome saturation and marker-assisted selection in wheat. Plant Biotechnol. J. 2010, 8, 196–210. [Google Scholar] [CrossRef]
- Paux, E.; Gao, L.; Faure, S.; Choulet, F.; Roger, D.; Chevalier, K.; Saintenac, C.; Balfourier, F.; Paux, K.; Cakir, M.; et al. Insertion Site-Based Polymorphism: A Swiss Army Knife for Wheat Genomics; Sydney University Press: Sydney, Australia, 2008; pp. 4–6. [Google Scholar]
- Paux, E.; Roger, D.; Badaeva, E.; Gay, G.; Bernard, M.; Sourdille, P.; Feuillet, C. Characterizing the composition and evolution of homoeologous genomes in hexaploid wheat through BAC-end sequencing on chromosome 3B. Plant J. 2006, 48, 463–474. [Google Scholar] [CrossRef] [PubMed]
- Lucas, S.J.; Šimková, H.; Šafář, J.; Jurman, I.; Cattonaro, F.; Vautrin, S.; Bellec, A.; Berges, H.; Doležel, J.; Budak, H. Functional features of a single chromosome arm in wheat (1AL) determined from its structure. Funct. Integr. Genom. 2012, 12, 173–182. [Google Scholar] [CrossRef]
- Sehgal, S.K.; Li, W.; Rabinowicz, P.D.; Chan, A.; Šimková, H.; Doležel, J.; Gill, B.S. Chromosome arm-specific BAC end sequences permit comparative analysis of homoeologous chromosomes and genomes of polyploid wheat. BMC Plant Biol. 2012, 12, 64. [Google Scholar] [CrossRef]
- IWGSC; Appels, R.; Eversole, K.; Stein, N.; Feuillet, C.; Keller, B.; Rogers, J.; Pozniak, C.J.; Choulet, F.; Distelfeld, A.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361. [Google Scholar] [CrossRef]
- Salina, E.A.; Nesterov, M.A.; Frenkel, Z.; Kiseleva, A.A.; Timonova, E.M.; Magni, F.; Vrána, J.; Šafár, J.; Šimková, H.; Doležel, J.; et al. Features of the organization of bread wheat chromosome 5BS based on physical mapping. BMC Genom. 2018, 19. [Google Scholar] [CrossRef]
- Dong, C.; Vincent, K.; Sharp, P. Simultaneous mutation detection of three homoeologous genes in wheat by high resolution melting analysis and mutation Surveyor®. BMC Plant Biol. 2009, 9, 143. [Google Scholar] [CrossRef]
- Mondini, L.; Nachit, M.M.; Porceddu, E.; Pagnotta, M.A. HRM technology for the identification and characterization of INDEL and SNPs mutations in genes involved in drought and salt tolerance of durum wheat. Plant Genet. Resour. Characterisation Util. 2011, 9, 166–169. [Google Scholar] [CrossRef]
- Matsuda, R.; Iehisa, J.C.M.; Takumi, S. Application of real-time PCR-based SNP detection for mapping of Net2, a causal D-genome gene for hybrid necrosis in interspecific crosses between tetraploidwheat and Aegilops tauschii. Genes Genet. Syst. 2012, 87, 137–143. [Google Scholar] [CrossRef]
- Lehmensiek, A.; Sutherland, M.W.; McNamara, R.B. The use of high resolution melting (HRM) to map single nucleotide polymorphism markers linked to a covered smut resistance gene in barley. Theor. Appl. Genet. 2008, 117, 721–728. [Google Scholar] [CrossRef] [PubMed]
- Shatalina, M.; Messmer, M.; Feuillet, C.; Mascher, F.; Paux, E.; Choulet, F.; Wicker, T.; Keller, B. High-resolution analysis of a QTL for resistance to Stagonospora nodorum glume blotch in wheat reveals presence of two distinct resistance loci in the target interval. Theor. Appl. Genet. 2014, 127, 573–586. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Acuña-Galindo, M.A.; Mason, R.E.; Subramanian, N.K.; Hays, D.B. Meta-analysis of wheat QTL regions associated with adaptation to drought and heat stress. Crop Sci. 2015, 55, 477–492. [Google Scholar] [CrossRef]
- Goffinet, B.; Gerber, S. Quantitative trait loci: A meta-analysis. Genetics 2000, 155, 463–473. [Google Scholar] [CrossRef] [PubMed]
- Soriano, J.M.; Alvaro, F. Discovering consensus genomic regions in wheat for root-related traits by QTL meta-analysis. Sci. Rep. 2019, 9, 10537. [Google Scholar] [CrossRef]
- Avni, R.; Oren, L.; Shabtay, G.; Assili, S.; Pozniak, C.; Hale, I.; Ben-David, R.; Peleg, Z.; Distelfeld, A. Genome based meta-QTL analysis of grain weight in tetraploid wheat identifies rare alleles of GRF4 associated with larger grains. Genes 2018, 9, 636. [Google Scholar] [CrossRef]
- Swamy, B.M.; Vikram, P.; Dixit, S.; Ahmed, H.U.; Kumar, A. Meta-analysis of grain yield QTL identified during agricultural drought in grasses showed consensus. BMC Genom. 2011, 12. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Saripalli, G.; Jan, I.; Kumar, K.; Sharma, P.K.; Balyan, H.S.; Gupta, P.K. Meta-QTL analysis and identification of candidate genes for drought tolerance in bread wheat (Triticum aestivum L.). Physiol. Mol. Biol. Plants 2020, 26, 1713–1725. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, P.; Martis, M.; Dorado, G.; Pfeifer, M.; Gálvez, S.; Schaaf, S.; Jouve, N.; Šimková, H.; Valárik, M.; Doležel, J.; et al. Next-generation sequencing and syntenic integration of flow-sorted arms of wheat chromosome 4A exposes the chromosome structure and gene content. Plant J. 2012, 69, 377–386. [Google Scholar] [CrossRef]
- Liu, Z.; Xin, M.; Qin, J.; Peng, H.; Ni, Z.; Yao, Y.; Sun, Q. Temporal transcriptome profiling reveals expression partitioning of homeologous genes contributing to heat and drought acclimation in wheat (Triticum aestivum L.). BMC Plant Biol. 2015, 15, 152. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Li, R.; Wang, H.; Li, D.; Wang, X.; Zhang, Y.; Zhen, W.; Duan, H.; Yan, G.; Li, Y. Transcriptomics analyses reveal wheat responses to drought stress during reproductive stages under field conditions. Front. Plant Sci. 2017, 8, 1–13. [Google Scholar] [CrossRef]
- Gálvez, S.; Mérida-García, R.; Camino, C.; Borrill, P.; Abrouk, M.; Ramírez-González, R.H.; Biyiklioglu, S.; Amil-Ruiz, F.; Dorado, G.; Budak, H.; et al. Hotspots in the genomic architecture of field drought responses in wheat as breeding targets. Funct. Integr. Genom. 2019, 19, 295–309. [Google Scholar] [CrossRef]
- Akhunov, E.D.; Goodyear, A.W.; Geng, S.; Qi, L.L.; Echalier, B.; Gill, B.S.; Miftahudin, A.; Gustafson, J.P.; Lazo, G.; Chao, S.; et al. The organization and rate of evolution of wheat genomes are correlated with recombination rates along chromosomes arms. Genome Res. 2003, 13, 753–763. [Google Scholar] [CrossRef]
- Munkvold, J.D.; Greene, R.A.; Bermudez-Kandianis, C.E.; La Rota, C.M.; Edwards, H.; Sorrells, S.F.; Dake, T.; Benscher, D.; Kantety, R.; Linkiewicz, A.M.; et al. Group 3 chromosome bin maps of wheat and their relationship to rice chromosome 1. Genetics 2004, 168, 639–650. [Google Scholar] [CrossRef]
- Vogt, T.; Jones, P. Glycosyltransferases in plant-natural product synthesis: Characterization of a supergene family. Trends Plant Sci. 2000, 5, 380–386. [Google Scholar] [CrossRef]
- Kang, J.; Park, J.; Choi, H.; Burla, B.; Kretzschmar, T.; Lee, Y.; Martinoia, E. Plant ABC Transporters. Am. Soc. Plant Biol. 2011, 9, e0153. [Google Scholar] [CrossRef] [PubMed]
- Baldoni, E.; Genga, A.; Cominelli, E. Plant MYB transcription factors: Their role in drought response mechanisms. Int. J. Mol. Sci. 2015, 16, 15811–15851. [Google Scholar] [CrossRef]
- Katiyar, A.; Smita, S.; Lenka, S.K.; Rajwanshi, R.; Chinnusamy, V.; Bansal, K.C. Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genom. 2012, 13. [Google Scholar] [CrossRef]
- Kato, Y.; Sakamoto, W. FtsH protease in the thylakoid membrane: Physiological functions and the regulation of protease activity. Front. Plant Sci. 2018, 9, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ashoub, A.; Baeumlisberger, M.; Neupaertl, M.; Karas, M.; Brüggemann, W. Characterization of common and distinctive adjustments of wild barley leaf proteome under drought acclimation, heat stress and their combination. Plant Mol. Biol. 2015, 87, 459–471. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Li, K.; Ding, H.; Liao, C.; Li, X.; Yuan, L.; Li, C. Root morphological and proteomic responses to growth restriction in maize plants supplied with sufficient N. J. Plant Physiol. 2011, 168, 1067–1075. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mérida-García, R.; Galvez, S.; Paux, E.; Dorado, G.; Pascual, L.; Giraldo, P.; Hernandez, P. Cost-Effective Markers and Candidate Genes Analysis at Wheat MQTL Loci. Biol. Life Sci. Forum 2021, 4, 44. https://doi.org/10.3390/IECPS2020-08571
Mérida-García R, Galvez S, Paux E, Dorado G, Pascual L, Giraldo P, Hernandez P. Cost-Effective Markers and Candidate Genes Analysis at Wheat MQTL Loci. Biology and Life Sciences Forum. 2021; 4(1):44. https://doi.org/10.3390/IECPS2020-08571
Chicago/Turabian StyleMérida-García, Rosa, Sergio Galvez, Etienne Paux, Gabriel Dorado, Laura Pascual, Patricia Giraldo, and Pilar Hernandez. 2021. "Cost-Effective Markers and Candidate Genes Analysis at Wheat MQTL Loci" Biology and Life Sciences Forum 4, no. 1: 44. https://doi.org/10.3390/IECPS2020-08571
APA StyleMérida-García, R., Galvez, S., Paux, E., Dorado, G., Pascual, L., Giraldo, P., & Hernandez, P. (2021). Cost-Effective Markers and Candidate Genes Analysis at Wheat MQTL Loci. Biology and Life Sciences Forum, 4(1), 44. https://doi.org/10.3390/IECPS2020-08571








