Harmful Cyanobacterial Blooms: Going beyond the “Green” to Monitor and Predict HCBs
Abstract
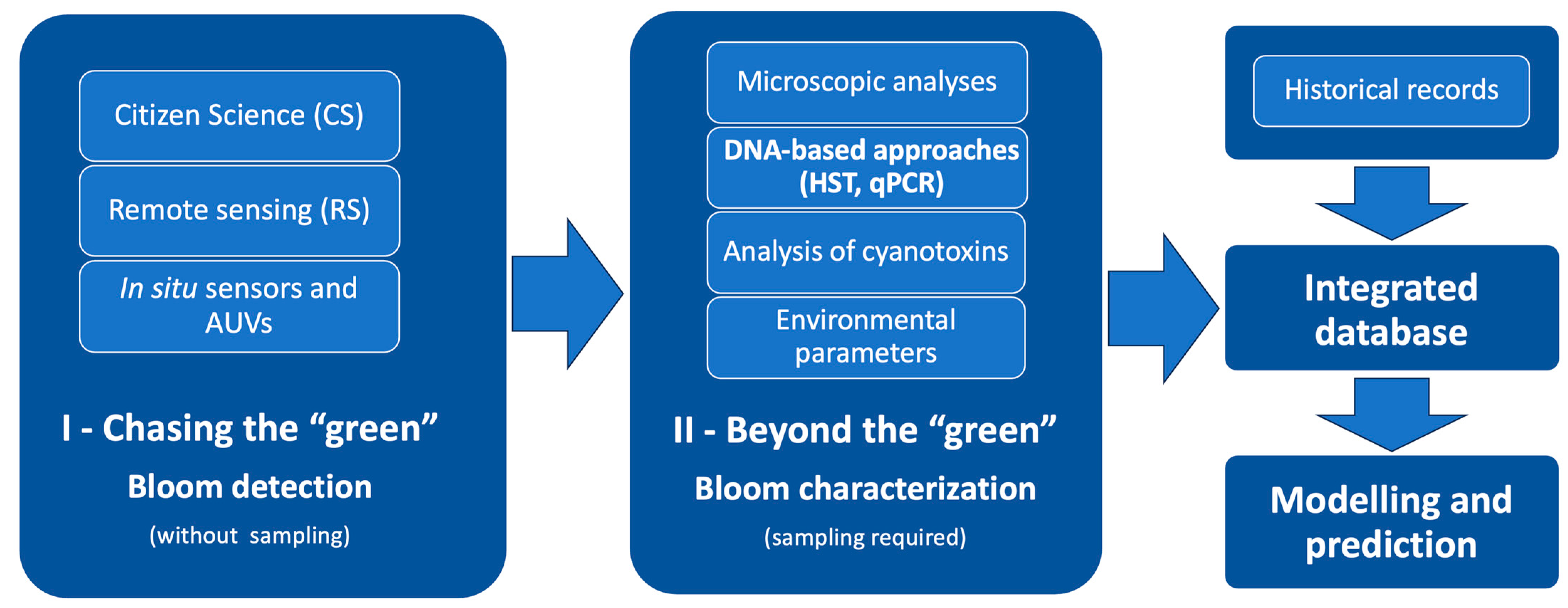
1. Introduction
2. Impact of Intraspecific Variability on Blooms’ Ecology and Toxicity
2.1. Classical Taxonomy vs. Phylogenetic Approaches
2.2. Co-Dominance and Dynamics of Cyanobacterial Blooms
2.3. Intraspecific Cyanotoxin Production Potential
3. Current Monitoring and Assessment Tools for Cyanobacterial Blooms
3.1. Methodologies Not Considering the Diversity of Cyanobacteria Nor Cyanotoxicity
3.1.1. Remote Sensing
3.1.2. In Situ Sensors and Automated Unmanned Vehicles (AUVs)
3.1.3. Turbidity and Visual Inspection—The Contribution from Citizen Science
3.2. Methodologies Considering the Diversity of Cyanobacteria or Cyanotoxins
3.2.1. Microscopic Identification and Cell Counting
3.2.2. DNA-Based Molecular Approaches
3.2.3. Detection and Quantification of Cyanotoxins
4. Integrative Perspective for an Effective Monitoring and Modelling of HCBs
Funding
Data Availability Statement
Conflicts of Interest
References
- Xu, H.-F.; Raanan, H.; Dai, G.-Z.; Oren, N.; Berkowicz, S.; Murik, O.; Kaplan, A.; Qiu, B.-S. Reading and Surviving the Harsh Conditions in Desert Biological Soil Crust: The Cyanobacterial Viewpoint. FEMS Microbiol. Rev. 2021, 45, fuab036. [Google Scholar] [CrossRef]
- Jasser, I.; Panou, M.; Khomutovska, N.; Sandzewicz, M.; Panteris, E.; Niyatbekov, T.; Łach, Ł.; Kwiatowski, J.; Kokociński, M.; Gkelis, S. Cyanobacteria in Hot Pursuit: Characterization of Cyanobacteria Strains, Including Novel Taxa, Isolated from Geothermal Habitats from Different Ecoregions of the World. Mol. Phylogenet. Evol. 2022, 170, 107454. [Google Scholar] [CrossRef] [PubMed]
- Perera, I.; Subashchandrabose, S.R.; Venkateswarlu, K.; Naidu, R.; Megharaj, M. Consortia of Cyanobacteria/Microalgae and Bacteria in Desert Soils: An Underexplored Microbiota. Appl. Microbiol. Biotechnol. 2018, 102, 7351–7363. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Pichel, F. Cyanobacteria. In Encyclopedia of Microbiology; Schaechter, M., Ed.; Academic Press: Cambridge, MA, USA, 2009; pp. 107–124. [Google Scholar]
- de Figueiredo, D.R.; Azeiteiro, U.M.; Esteves, S.M.; Gonçalves, F.J.M.; Pereira, M.J. Microcystin-Producing Blooms—A Serious Global Public Health Issue. Ecotoxicol. Environ. Saf. 2004, 59, 151–163. [Google Scholar] [CrossRef] [PubMed]
- Wood, R. Acute Animal and Human Poisonings from Cyanotoxin Exposure—A Review of the Literature. Environ. Int. 2016, 91, 276–282. [Google Scholar] [CrossRef] [PubMed]
- Cotterill, V.; Hamilton, D.P.; Puddick, J.; Suren, A.; Wood, S.A. Phycocyanin Sensors as an Early Warning System for Cyanobacteria Blooms Concentrations: A Case Study in the Rotorua Lakes. N. Z. J. Mar. Freshw. Res. 2019, 53, 555–570. [Google Scholar] [CrossRef]
- Hilborn, E.D.; Beasley, V.R. One Health and Cyanobacteria in Freshwater Systems: Animal Illnesses and Deaths Are Sentinel Events for Human Health Risks. Toxins 2015, 7, 1374–1395. [Google Scholar] [CrossRef]
- Carmichael, W.W.; Boyer, G.L. Health Impacts from Cyanobacteria Harmful Algae Blooms: Implications for the North American Great Lakes. Harmful Algae 2016, 54, 194–212. [Google Scholar] [CrossRef]
- Śliwińska-Wilczewska, S.; Maculewicz, J.; Felpeto, A.B.; Latała, A. Allelopathic and Bloom-Forming Picocyanobacteria in a Changing World. Toxins 2018, 10, 48. [Google Scholar] [CrossRef]
- Janssen, E.M.L. Cyanobacterial Peptides beyond Microcystins—A Review on Co-Occurrence, Toxicity, and Challenges for Risk Assessment. Water Res. 2019, 151, 488–499. [Google Scholar] [CrossRef]
- Redouane, E.M.; El Amrani Zerrifi, S.; El Khalloufi, F.; Oufdou, K.; Oudra, B.; Lahrouni, M.; Campos, A.; Vasconcelos, V. Mode of Action and Fate of Microcystins in the Complex Soil-Plant Ecosystems. Chemosphere 2019, 225, 270–281. [Google Scholar] [CrossRef] [PubMed]
- Vasconcelos, V. Cyanobacteria Toxins: Diversity and Ecological Effects. Limnetica 2001, 20, 45–58. [Google Scholar] [CrossRef]
- Chorus, I.; Welker, M. Toxic Cyanobacteria in Water: A Guide to Their Public Health Consequences, Monitoring and Management, 2nd ed.; Chorus, I., Welker, M., Eds.; CRC Press: Boca Raton, FL, USA, 2021; ISBN 9788490225370. [Google Scholar]
- Cheung, M.Y.; Liang, S.; Lee, J. Toxin-Producing Cyanobacteria in Freshwater: A Review of the Problems, Impact on Drinking Water Safety, and Efforts for Protecting Public Health. J. Microbiol. 2013, 51, 1–10. [Google Scholar] [CrossRef]
- Paerl, H.W.; Fulton, R.S.; Moisander, P.H.; Dyble, J. Harmful Freshwater Algal Blooms, with an Emphasis on Cyanobacteria. Sci. World J. 2001, 1, 76–113. [Google Scholar] [CrossRef] [PubMed]
- Paerl, H.W.; Barnard, M.A. Mitigating the Global Expansion of Harmful Cyanobacterial Blooms: Moving Targets in a Human- and Climatically-Altered World. Harmful Algae 2020, 96, 101845. [Google Scholar] [CrossRef] [PubMed]
- Huisman, J.; Codd, G.A.; Paerl, H.W.; Ibelings, B.W.; Verspagen, J.M.H.; Visser, P.M. Cyanobacterial Blooms. Nat. Rev. Microbiol. 2018, 16, 471–483. [Google Scholar] [CrossRef] [PubMed]
- Paerl, H.W.; Otten, T.G. Duelling “CyanoHABs”: Unravelling the Environmental Drivers Controlling Dominance and Succession among Diazotrophic and Non-N2-Fixing Harmful Cyanobacteria. Environ. Microbiol. 2016, 18, 316–324. [Google Scholar] [CrossRef] [PubMed]
- Havens, K.E.; Paerl, H.W. Climate Change at a Crossroad for Control of Harmful Algal Blooms. Environ. Sci. Technol. 2015, 49, 12605–12606. [Google Scholar] [CrossRef]
- Scholz, S.N.; Esterhuizen-Londt, M.; Pflugmacher, S. Rise of Toxic Cyanobacterial Blooms in Temperate Freshwater Lakes: Causes, Correlations and Possible Countermeasures. Toxicol. Environ. Chem. 2017, 99, 543–577. [Google Scholar] [CrossRef]
- O’Neil, J.M.; Davis, T.W.; Burford, M.A.; Gobler, C.J. The Rise of Harmful Cyanobacteria Blooms: The Potential Roles of Eutrophication and Climate Change. Harmful Algae 2012, 14, 313–334. [Google Scholar] [CrossRef]
- Mehinto, A.C.; Smith, J.; Wenger, E.; Stanton, B.; Linville, R.; Brooks, B.W.; Sutula, M.A.; Howard, M.D.A. Synthesis of Ecotoxicological Studies on Cyanotoxins in Freshwater Habitats—Evaluating the Basis for Developing Thresholds Protective of Aquatic Life in the United States. Sci. Total Environ. 2021, 795, 148864. [Google Scholar] [CrossRef] [PubMed]
- Weralupitiya, C.; Wanigatunge, R.P.; Gunawardana, D.; Vithanage, M.; Magana-Arachchi, D. Cyanotoxins Uptake and Accumulation in Crops: Phytotoxicity and Implications on Human Health. Toxicon 2022, 211, 21–35. [Google Scholar] [CrossRef]
- Paerl, H.W.; Otten, T.G. Harmful Cyanobacterial Blooms: Causes, Consequences, and Controls. Microb. Ecol. 2013, 65, 995–1010. [Google Scholar] [CrossRef] [PubMed]
- Corbel, S.; Mougin, C.; Bouaïcha, N. Cyanobacterial Toxins: Modes of Actions, Fate in Aquatic and Soil Ecosystems, Phytotoxicity and Bioaccumulation in Agricultural Crops. Chemosphere 2014, 96, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Ger, K.A.; Urrutia-Cordero, P.; Frost, P.C.; Hansson, L.A.; Sarnelle, O.; Wilson, A.E.; Lürling, M. The Interaction between Cyanobacteria and Zooplankton in a More Eutrophic World. Harmful Algae 2016, 54, 128–144. [Google Scholar] [CrossRef]
- Paerl, H.W.; Gardner, W.S.; Havens, K.E.; Joyner, A.R.; McCarthy, M.J.; Newell, S.E.; Qin, B.; Scott, J.T. Mitigating Cyanobacterial Harmful Algal Blooms in Aquatic Ecosystems Impacted by Climate Change and Anthropogenic Nutrients. Harmful Algae 2016, 54, 213–222. [Google Scholar] [CrossRef]
- Paerl, H.W. Mitigating Toxic Planktonic Cyanobacterial Blooms in Aquatic Ecosystems Facing Increasing Anthropogenic and Climatic Pressures. Toxins 2018, 10, 76. [Google Scholar] [CrossRef]
- Visser, P.M.; Verspagen, J.M.H.; Sandrini, G.; Stal, L.J.; Matthijs, H.C.P.; Davis, T.W.; Paerl, H.W.; Huisman, J. How Rising CO2 and Global Warming May Stimulate Harmful Cyanobacterial Blooms. Harmful Algae 2016, 54, 145–159. [Google Scholar] [CrossRef]
- Watson, S.B.; Miller, C.; Arhonditsis, G.; Boyer, G.L.; Carmichael, W.; Charlton, M.N.; Confesor, R.; Depew, D.C.; Höök, T.O.; Ludsin, S.A.; et al. The Re-Eutrophication of Lake Erie: Harmful Algal Blooms and Hypoxia. Harmful Algae 2016, 56, 44–66. [Google Scholar] [CrossRef]
- Bullerjahn, G.S.; McKay, R.M.; Davis, T.W.; Baker, D.B.; Boyer, G.L.; D’Anglada, L.V.; Doucette, G.J.; Ho, J.C.; Irwin, E.G.; Kling, C.L.; et al. Global Solutions to Regional Problems: Collecting Global Expertise to Address the Problem of Harmful Cyanobacterial Blooms. A Lake Erie Case Study. Harmful Algae 2016, 54, 223–238. [Google Scholar] [CrossRef]
- Stewart, I.; Webb, P.M.; Schluter, P.J.; Shaw, G.R. Recreational and Occupational Field Exposure to Freshwater Cyanobacteria—A Review of Anecdotal and Case Reports, Epidemiological Studies and the Challenges for Epidemiologic Assessment. Environ. Health 2006, 5, 6. [Google Scholar] [CrossRef] [PubMed]
- Carmichael, W.W. Cyanobacteria Secondary Metabolites—The Cyanotoxins. J. Appl. Bacteriol. 1992, 72, 445–459. [Google Scholar] [CrossRef] [PubMed]
- Manning, S.R.; Nobles, D.R. Impact of Global Warming on Water Toxicity: Cyanotoxins. Curr. Opin. Food Sci. 2017, 18, 14–20. [Google Scholar] [CrossRef]
- Huang, I.S.; Zimba, P.V. Cyanobacterial Bioactive Metabolites—A Review of Their Chemistry and Biology. Harmful Algae 2019, 83, 42–94. [Google Scholar] [CrossRef]
- Wiegand, C.; Pflugmacher, S. Ecotoxicological Effects of Selected Cyanobacterial Secondary Metabolites a Short Review. Toxicol. Appl. Pharmacol. 2005, 203, 201–218. [Google Scholar] [CrossRef]
- Smith, J.L.; Boyer, G.L.; Zimba, P. V A Review of Cyanobacterial Odorous and Bioactive Metabolites: Impacts and Management Alternatives in Aquaculture. Aquaculture 2008, 208, 5–20. [Google Scholar] [CrossRef]
- Svirčev, Z.; Lalić, D.; Bojadžija Savić, G.; Tokodi, N.; Drobac Backović, D.; Chen, L.; Meriluoto, J.; Codd, G.A. Global Geographical and Historical Overview of Cyanotoxin Distribution and Cyanobacterial Poisonings; Springer: Berlin/Heidelberg, Germany, 2019; Volume 93, ISBN 0123456789. [Google Scholar]
- He, X.; Liu, Y.L.; Conklin, A.; Westrick, J.; Weavers, L.K.; Dionysiou, D.D.; Lenhart, J.J.; Mouser, P.J.; Szlag, D.; Walker, H.W. Toxic Cyanobacteria and Drinking Water: Impacts, Detection, and Treatment. Harmful Algae 2016, 54, 174–193. [Google Scholar] [CrossRef]
- Cao, L.; Massey, I.Y.; Feng, H.; Yang, F. A Review of Cardiovascular Toxicity of Microcystins. Toxins 2019, 11, 507. [Google Scholar] [CrossRef]
- Svirčev, Z.; Drobac, D.; Tokodi, N.; Mijović, B.; Codd, G.A.; Meriluoto, J. Toxicology of Microcystins with Reference to Cases of Human Intoxications and Epidemiological Investigations of Exposures to Cyanobacteria and Cyanotoxins. Arch. Toxicol. 2017, 91, 621–650. [Google Scholar] [CrossRef]
- Schreidah, C.M.; Ratnayake, K.; Senarath, K.; Karunarathne, A. Microcystins: Biogenesis, Toxicity, Analysis, and Control. Chem. Res. Toxicol. 2020, 33, 2225–2246. [Google Scholar] [CrossRef]
- Du, X.; Liu, H.; Yuan, L.; Wang, Y.; Ma, Y.; Wang, R.; Chen, X.; Losiewicz, M.D.; Guo, H.; Zhang, H. The Diversity of Cyanobacterial Toxins on Structural Characterization, Distribution and Identification: A Systematic Review. Toxins 2019, 11, 530. [Google Scholar] [CrossRef] [PubMed]
- Koreiviene, J.; Anne, O.; Kasperovičiene, J.; Burškyte, V. Cyanotoxin Management and Human Health Risk Mitigation in Recreational Waters. Environ. Monit. Assess. 2014, 186, 4443–4459. [Google Scholar] [CrossRef] [PubMed]
- Testai, E.; Scardala, S.; Vichi, S.; Buratti, F.M.; Funari, E. Risk to Human Health Associated with the Environmental Occurrence of Cyanobacterial Neurotoxic Alkaloids Anatoxins and Saxitoxins. Crit. Rev. Toxicol. 2016, 46, 385–419. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, K.; Musgrave, I.F.; Humpage, A. Low Dose Extended Exposure to Saxitoxin and Its Potential Neurodevelopmental Effects: A Review. Environ. Toxicol. Pharmacol. 2016, 48, 7–16. [Google Scholar] [CrossRef]
- Moreira, C.; Azevedo, J.; Antunes, A.; Vasconcelos, V. Cylindrospermopsin: Occurrence, Methods of Detection and Toxicology. J. Appl. Microbiol. 2013, 114, 605–620. [Google Scholar] [CrossRef] [PubMed]
- Scarlett, K.R.; Kim, S.; Lovin, L.M.; Chatterjee, S.; Scott, J.T.; Brooks, B.W. Global Scanning of Cylindrospermopsin: Critical Review and Analysis of Aquatic Occurrence, Bioaccumulation, Toxicity and Health Hazards. Sci. Total Environ. 2020, 738, 139807. [Google Scholar] [CrossRef] [PubMed]
- Hinojosa, M.G.; Prieto, A.I.; Gutiérrez-Praena, D.; Moreno, F.J.; Cameán, A.M.; Jos, A. Neurotoxic Assessment of Microcystin-LR, Cylindrospermopsin and Their Combination on the Human Neuroblastoma SH-SY5Y Cell Line. Chemosphere 2019, 224, 751–764. [Google Scholar] [CrossRef] [PubMed]
- Hinojosa, M.G.; Gutiérrez-Praena, D.; Prieto, A.I.; Guzmán-Guillén, R.; Jos, A.; Cameán, A.M. Neurotoxicity Induced by Microcystins and Cylindrospermopsin: A Review. Sci. Total Environ. 2019, 668, 547–565. [Google Scholar] [CrossRef]
- Chorus, I.; Fastner, J.; Welker, M. Cyanobacteria and Cyanotoxins in a Changing Environment: Concepts, Controversies, Challenges. Water 2021, 13, 2463. [Google Scholar] [CrossRef]
- Edwards, C.; Graham, D.; Fowler, N.; Lawton, L.A. Biodegradation of Microcystins and Nodularin in Freshwaters. Chemosphere 2008, 73, 1315–1321. [Google Scholar] [CrossRef]
- Bi, X.; Dai, W.; Wang, X.; Dong, S.; Zhang, S.; Zhang, D.; Wu, M. Microcystins Distribution, Bioaccumulation, and Microcystis Genotype Succession in a Fish Culture Pond. Sci. Total Environ. 2019, 688, 380–388. [Google Scholar] [CrossRef]
- Onyango, D.M.; Orina, P.S.; Ramkat, R.C.; Kowenje, C.; Githukia, C.M.; Lusweti, D.; Lung’ayia, H.B.O. Review of Current State of Knowledge of Microcystin and Its Impacts on Fish in Lake Victoria. Lakes Reserv. 2020, 25, 350–361. [Google Scholar] [CrossRef]
- Vareli, K.; Zarali, E.; Zacharioudakis, G.S.A.; Vagenas, G.; Varelis, V.; Pilidis, G.; Briasoulis, E.; Sainis, I. Microcystin Producing Cyanobacterial Communities in Amvrakikos Gulf (Mediterranean Sea, NW Greece) and Toxin Accumulation in Mussels (Mytilus galloprovincialis). Harmful Algae 2012, 15, 109–118. [Google Scholar] [CrossRef]
- Preece, E.P.; Hardy, F.J.; Moore, B.C.; Bryan, M. A Review of Microcystin Detections in Estuarine and Marine Waters: Environmental Implications and Human Health Risk. Harmful Algae 2017, 61, 31–45. [Google Scholar] [CrossRef]
- Zhu, J.; Ren, X.; Liu, H.; Liang, C. Effect of Irrigation with Microcystins-Contaminated Water on Growth and Fruit Quality of Cucumis sativus L. and the Health Risk. Agric. Water Manag. 2018, 204, 91–99. [Google Scholar] [CrossRef]
- Romero-Oliva, C.S.; Contardo-Jara, V.; Block, T.; Pflugmacher, S. Accumulation of Microcystin Congeners in Different Aquatic Plants and Crops—A Case Study from Lake Amatitlán, Guatemala. Ecotoxicol. Environ. Saf. 2014, 102, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.L.; Utsumi, M. An Overview of the Accumulation of Microcystins in Aquatic Ecosystems. J. Environ. Manag. 2018, 213, 520–529. [Google Scholar] [CrossRef]
- Weirich, C.A.; Miller, T.R. Freshwater Harmful Algal Blooms: Toxins and Children’s Health. Curr. Probl. Pediatr. Adolesc. Health Care 2014, 44, 2–24. [Google Scholar] [CrossRef]
- Otten, T.G.; Paerl, H.W. Health Effects of Toxic Cyanobacteria in U.S. Drinking and Recreational Waters: Our Current Understanding and Proposed Direction. Curr. Environ. Health Rep. 2015, 2, 75–84. [Google Scholar] [CrossRef]
- Österholm, J.; Popin, R.V.; Fewer, D.P.; Sivonen, K. Phylogenomic Analysis of Secondary Metabolism in the Toxic Cyanobacterial Genera Anabaena, Dolichospermum and Aphanizomenon. Toxins 2020, 12, 248. [Google Scholar] [CrossRef]
- Jiang, Y.; Song, G.; Pan, Q.; Yang, Y.; Li, R. Identification of Genes for Anatoxin-a Biosynthesis in Cuspidothrix issatschenkoi. Harmful Algae 2015, 46, 43–48. [Google Scholar] [CrossRef]
- Cirés, S.; Ballot, A. A Review of the Phylogeny, Ecology and Toxin Production of Bloom-Forming Aphanizomenon spp. and Related Species within the Nostocales (Cyanobacteria). Harmful Algae 2016, 54, 21–43. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Dreher, T.W.; Li, R. An Overview of Diversity, Occurrence, Genetics and Toxin Production of Bloom-Forming Dolichospermum (Anabaena) Species. Harmful Algae 2016, 54, 54–68. [Google Scholar] [CrossRef] [PubMed]
- Wood, S.A.; Puddick, J.; Fleming, R.; Heussner, A.H. Detection of Anatoxin-Producing Phormidium in a New Zealand Farm Pond and an Associated Dog Death. N. Z. J. Bot. 2017, 55, 36–46. [Google Scholar] [CrossRef]
- Chernova, E.; Sidelev, S.; Russkikh, I.; Voyakina, E.; Babanazarova, O.; Romanov, R.; Kotovshchikov, A.; Mazur-Marzec, H. Dolichospermum and Aphanizomenon as Neurotoxins Producers in Some Russian Freshwaters. Toxicon 2017, 130, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Capelli, C.; Ballot, A.; Cerasino, L.; Papini, A.; Salmaso, N. Biogeography of Bloom-Forming Microcystin Producing and Non-Toxigenic Populations of Dolichospermum lemmermannii (Cyanobacteria). Harmful Algae 2017, 67, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Antunes, J.T.; Leão, P.N.; Vasconcelos, V.M. Cylindrospermopsis raciborskii: Review of the Distribution, Phylogeography, and Ecophysiology of a Global Invasive Species. Front. Microbiol. 2015, 6, 473. [Google Scholar] [CrossRef]
- Ballot, A.; Scherer, P.I.; Wood, S.A. Variability in the Anatoxin Gene Clusters of Cuspidothrix issatschenkoi from Germany, New Zealand, China and Japan. PLoS ONE 2018, 13, e0200774. [Google Scholar] [CrossRef]
- Wilhelm, S.W.; Bullerjahn, G.S.; McKay, R.M.L. The Complicated and Confusing Ecology of Microcystis Blooms. mBio 2020, 11, 1–5. [Google Scholar] [CrossRef]
- Buratti, F.M.; Manganelli, M.; Vichi, S.; Stefanelli, M.; Scardala, S.; Testai, E.; Funari, E. Cyanotoxins: Producing Organisms, Occurrence, Toxicity, Mechanism of Action and Human Health Toxicological Risk Evaluation. Arch. Toxicol. 2017, 91, 1049–1130. [Google Scholar] [CrossRef]
- Deng, J.; Qin, B.; Paerl, H.W.; Zhang, Y.; Ma, J.; Chen, Y. Earlier and Warmer Springs Increase Cyanobacterial (Microcystis spp.) Blooms in Subtropical Lake Taihu, China. Freshw. Biol. 2014, 59, 1076–1085. [Google Scholar] [CrossRef]
- de Figueiredo, D.R.; Reboleira, A.S.S.P.; Antunes, S.C.; Abrantes, N.; Azeiteiro, U.; Gonçalves, F.; Pereira, M.J.; Gonçalves, F.; Figueiredo, D.R. The Effect of Environmental Parameters and Cyanobacterial Blooms on Phytoplankton Dynamics of a Portuguese Temperate Lake. Hydrobiologia 2006, 568, 145–157. [Google Scholar] [CrossRef]
- Wang, P.; Ma, J.; Wang, X.; Tan, Q. Rising Atmospheric CO2 Levels Result in an Earlier Cyanobacterial Bloom-Maintenance Phase with Higher Algal Biomass. Water Res. 2020, 185, 116267. [Google Scholar] [CrossRef] [PubMed]
- Vanderley, R.F.; Ger, K.A.; Becker, V.; Bezerra, M.G.T.A.; Panosso, R. Abiotic Factors Driving Cyanobacterial Biomass and Composition under Perennial Bloom Conditions in Tropical Latitudes. Hydrobiologia 2021, 848, 943–960. [Google Scholar] [CrossRef]
- Qin, B.; Deng, J.; Shi, K.; Wang, J.; Brookes, J.; Zhou, J.; Zhang, Y.; Zhu, G.; Paerl, H.W.; Wu, L. Extreme Climate Anomalies Enhancing Cyanobacterial Blooms in Eutrophic Lake Taihu, China. Water Resour. Res. 2021, 57, e2020WR029371. [Google Scholar] [CrossRef]
- Hayes, N.M.; Haig, H.A.; Simpson, G.L.; Leavitt, P.R. Effects of Lake Warming on the Seasonal Risk of Toxic Cyanobacteria Exposure. Limnol. Oceanogr. Lett. 2020, 5, 393–402. [Google Scholar] [CrossRef]
- Liu, X.; Lu, X.; Chen, Y. The Effects of Temperature and Nutrient Ratios on Microcystis Blooms in Lake Taihu, China: An 11-Year Investigation. Harmful Algae 2011, 10, 337–343. [Google Scholar] [CrossRef]
- Rzymski, P.; Brygider, A.; Kokociński, M. On the Occurrence and Toxicity of Cylindrospermopsis raciborskii in Poland. Limnol. Rev. 2017, 17, 23–29. [Google Scholar] [CrossRef]
- Moreira, C.; Fathalli, A.; Vasconcelos, V.; Antunes, A. Genetic Diversity and Structure of the Invasive Toxic Cyanobacterium Cylindrospermopsis raciborskii. Curr. Microbiol. 2011, 62, 1590–1595. [Google Scholar] [CrossRef]
- Kokociński, M.; Gagala, I.; Jasser, I.; Karosiene, J.; Kasperovičiene, J.; Kobos, J.; Koreiviene, J.; Soininen, J.; Szczurowska, A.; Woszczyk, M.; et al. Distribution of Invasive Cylindrospermopsis raciborskii in the East-Central Europe Is Driven by Climatic and Local Environmental Variables. FEMS Microbiol. Ecol. 2017, 93, fix035. [Google Scholar] [CrossRef]
- Kokocinski, M.; Dziga, D.; Spoof, L.; Stefaniak, K.; Jurczak, T.; Mankiewicz-Boczek, J.; Meriluoto, J. First Report of the Cyanobacterial Toxin Cylindrospermopsin in the Shallow, Eutrophic Lakes of Western Poland. Chemosphere 2009, 74, 669–675. [Google Scholar] [CrossRef] [PubMed]
- Fastner, J.; Rücker, J.; Stüken, A.; Preußel, K.; Nixdorf, B.; Chorus, I.; Köhler, A.; Wiedner, C. Occurrence of the Cyanobacterial Toxin Cylindrospermopsin in Northeast Germany. Environ. Toxicol. 2007, 22, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Stüken, A.; Rücker, J.; Endrulat, T.; Preussel, K.; Hemm, M.; Nixdorf, B.; Karsten, U.; Wiedner, C. Distribution of Three Alien Cyanobacterial Species (Nostocales) in Northeast Germany: Cylindrospermopsis raciborskii, Anabaena bergii and Aphanizomenon aphanizomenoides. Phycologia 2006, 45, 696–703. [Google Scholar] [CrossRef]
- Kaštovský, J.; Hauer, T.; Mareš, J.; Krautová, M.; Bešta, T.; Komárek, J.; Desortová, B.; Heteša, J.; Hindáková, A.; Houk, V.; et al. A Review of the Alien and Expansive Species of Freshwater Cyanobacteria and Algae in the Czech Republic. Biol. Invasions 2010, 12, 3599–3625. [Google Scholar] [CrossRef]
- de Figueiredo, D.R.; Lopes, A.R.; Pereira, M.J.; Polónia, A.R.; Castro, B.B.; Gonçalves, F.; Gomes, N.C.M.; Cleary, D.F.R. Bacterioplankton Community Shifts during a Spring Bloom of Aphanizomenon gracile and Sphaerospermopsis aphanizomenoides at a Temperate Shallow Lake. Hydrobiology 2022, 1, 499–517. [Google Scholar] [CrossRef]
- Kim, Y.-J.; Park, H.-K.; Kim, I.-S. Invasion and Toxin Production by Exotic Nostocalean cyanobacteria (Cuspidothrix, Cylindrospermopsis, and Sphaerospermopsis) in the Nakdong River, Korea. Harmful Algae 2020, 100, 101954. [Google Scholar] [CrossRef] [PubMed]
- Ramos, T.K.; Costa, L.D.F.; Yunes, J.S.; Resgalla, C.; Barufi, J.B.; de Oliveira Bastos, E.; Horta, P.A.; Rörig, L.R. Saxitoxins from the Freshwater Cyanobacterium Raphidiopsis raciborskii Can Contaminate Marine Mussels. Harmful Algae 2021, 103, 102004. [Google Scholar] [CrossRef]
- Tawong, W.; Pongcharoen, P.; Nishimura, T.; Adachi, M. Molecular Characterizations of Thai Raphidiopsis raciborskii (Nostocales, Cyanobacteria) Based on 16s rDNA, RbcLX, and Cylindrospermopsin Synthetase Genes. Plankton Benthos Res. 2019, 14, 211–223. [Google Scholar] [CrossRef]
- Cirés, S.; Wörmer, L.; Ballot, A.; Agha, R.; Wiedner, C.; Velázquez, D.; Casero, M.C.; Quesada, A. Phylogeography of Cylindrospermopsin and Paralytic Shellfish Toxin-Producing Nostocales Cyanobacteria from Mediterranean Europe (Spain). Appl. Environ. Microbiol. 2014, 80, 1359–1370. [Google Scholar] [CrossRef]
- Wood, S.A.; Rasmussen, J.P.; Holland, P.T.; Campbell, R.; Crowe, A.L.M. First Report of the Cyanotoxin Anatoxin-a from Aphanizomenon issatschenkoi (Cyanobacteria). J. Phycol. 2007, 43, 356–365. [Google Scholar] [CrossRef]
- Palinska, K.A.; Surosz, W. Taxonomy of Cyanobacteria: A Contribution to Consensus Approach. Hydrobiologia 2014, 740, 1–11. [Google Scholar] [CrossRef]
- Hindák, F. Morphological Variation of Four Planktic Nostocalean Cyanophytes—Members of the Genus Aphanizomenon or Anabaena? Hydrobiologia 2000, 438, 107–116. [Google Scholar] [CrossRef]
- Li, R.; Carmichael, W.W.; Liu, Y.; Watanabe, M.M. Taxonomic Re-Evaluation of Aphanizomenon flos-aquae NH-5 Based on Morphology and 16S rRNA Gene Sequences. Hydrobiologia 2000, 438, 99–105. [Google Scholar] [CrossRef]
- Komárek, J.; Zapomelová, E. Planktic Morphospecies of the Cyanobacterial Genus Anabaena = Subg. Dolichospermum—2. Part: Straight Types. Fottea 2008, 8, 1–14. [Google Scholar]
- Zapomělová, E.; Skácelová, O.; Pumann, P.; Kopp, R.; Janeček, E. Polyphasic Characterization of Three Strains of Anabaena reniformis and Aphanizomenon aphanizomenoides (Cyanobacteria) and Their Reclassification To Sphaerospermum gen. nov. (Incl. Anabaena kisseleviana) (45:1363–73). J. Phycol. 2010, 46, 415. [Google Scholar] [CrossRef] [PubMed]
- Zapomělová, E.; Hrouzek, P.; Řezanka, T.; Jezberová, J.; Řeháková, K.; Hisem, D.; Komárková, J. Polyphasic Characterization of Dolichospermum spp. and Sphaerospermopsis spp. (Nostocales, Cyanobacteria): Morphology, 16S rRNA Gene Sequences and Fatty Acid and Secondary Metabolite Profiles. J. Phycol. 2011, 47, 1152–1163. [Google Scholar] [CrossRef] [PubMed]
- Gama, W.A.; Rigonato, J.; Fiore, M.F.; Sant’Anna, C.L. New Insights into Chroococcus (Cyanobacteria) and Two Related Genera: Cryptococcum gen. nov. and Inacoccus gen. nov. Eur. J. Phycol. 2019, 54, 315–325. [Google Scholar] [CrossRef]
- Komárek, J. A Polyphasic Approach for the Taxonomy of Cyanobacteria: Principles and Applications. Eur. J. Phycol. 2016, 51, 346–353. [Google Scholar] [CrossRef]
- Komárková, J.; Jezberová, J.; Komárek, O.; Zapomělová, E. Variability of Chroococcus (Cyanobacteria) Morphospecies with Regard to Phylogenetic Relationships. Hydrobiologia 2010, 639, 69–83. [Google Scholar] [CrossRef]
- Zapomělová, E.; Jezberová, J.; Hrouzek, P.; Hisem, D.; Řeháková, K.; Komárková, J. Polyphasic Characterization of Three Strains of Anabaena reniformis and Aphanizomenon aphanizomenoides (Cyanobacteria) and Their Reclassification to Sphaerospermum gen. nov. (Incl. Anabaena kisseleviana). J. Phycol. 2009, 45, 1363–1373. [Google Scholar] [CrossRef]
- Komárek, J. The Cyanobacterial Genus Macrospermum. Fottea 2008, 8, 79–86. [Google Scholar] [CrossRef]
- Zapomělová, E.; Skácelová, O.; Pumann, P.; Kopp, R.; Janeček, E. Biogeographically Interesting Planktonic Nostocales (Cyanobacteria) in the Czech Republic and Their Polyphasic Evaluation Resulting in Taxonomic Revisions of Anabaena bergii Ostenfeld 1908 (Chrysosporum gen. nov.) and A. tenericaulis Nygaard 1949 (Dolichospermum tenericaule comb. nova). Hydrobiologia 2012, 698, 353–365. [Google Scholar] [CrossRef]
- Zapomělová, E.; Řeháková, K.; Jezberová, J.; Komárková, J. Polyphasic Characterization of Eight Planktonic Anabaena Strains (Cyanobacteria) with Reference to the Variability of 61 Anabaena Populations Observed in the Field. Hydrobiologia 2010, 639, 99–113. [Google Scholar] [CrossRef]
- Aguilera, A.; Gómez, E.B.; Kaštovský, J.; Echenique, R.O.; Salerno, G.L. The Polyphasic Analysis of Two Native Raphidiopsis Isolates Supports the Unification of the Genera Raphidiopsis and Cylindrospermopsis (Nostocales, Cyanobacteria). Phycologia 2018, 57, 130–146. [Google Scholar] [CrossRef]
- Rajaniemi, P.; Komárek, J.; Willame, R.; Hrouzek, P.; Kaštovská, K.; Hoffmann, L.; Sivonen, K. Taxonomic Consequences from the Combined Molecular and Phenotype Evaluation of Selected Anabaena and Aphanizomenon Strains. Algol. Stud. Arch. Für Hydrobiol. 2005, 117, 371–391. [Google Scholar] [CrossRef]
- Choi, H.J.; Joo, J.H.; Kim, J.H.; Wang, P.; Ki, J.S.; Han, M.S. Morphological Characterization and Molecular Phylogenetic Analysis of Dolichospermum hangangense (Nostocales, Cyanobacteria) sp. nov. from Han River, Korea. Algae 2018, 33, 143–156. [Google Scholar] [CrossRef]
- Johansen, J.R.; Kovacik, L.; Casamatta, D.A.; Fučiková, K.; Kaštovský, J. Utility of 16S-23S ITS Sequence and Secondary Structure for Recognition of Intrageneric and Intergeneric Limits within Cyanobacterial Taxa: Leptolyngbya corticola sp. nov. (Pseudanabaenaceae, Cyanobacteria). Nova Hedwig. 2011, 92, 283–302. [Google Scholar] [CrossRef]
- Moore, D.; McGregor, G.B.; Shaw, G. Morphological Changes during Akinete Germination in Cylindrospermopsis raciborskii (Nostocales, Cyanobacteria). J. Phycol. 2004, 40, 1098–1105. [Google Scholar] [CrossRef]
- Ryu, H.S.; Shin, R.Y.; Lee, J.H. Morphology and Taxonomy of the Aphanizomenon spp. (Cyanophyceae) and Related Species in the Nakdong River, South Korea. J. Ecol. Environ. 2016, 41, 6. [Google Scholar] [CrossRef]
- de Figueiredo, D.R.; Gonçalves, A.M.M.; Castro, B.B.; Gonçalves, F.; Pereira, M.J.; Correia, A. Differential Inter-and Intra-Specific Responses of Aphanizomenon Strains to Nutrient Limitation and Algal Growth Inhibition. J. Plankton Res. 2011, 33, 1606–1616. [Google Scholar] [CrossRef]
- Komárek, J. Modern Taxonomic Revision of Planktic Nostocacean Cyanobacteria: A Short Review of Genera. Hydrobiologia 2010, 639, 231–243. [Google Scholar] [CrossRef]
- Komárek, J. Cyanobacterial Taxonomy: Current Problems and Prospects for the Integration of Traditional and Molecular Approaches. Algae 2006, 21, 349–375. [Google Scholar] [CrossRef]
- Komárek, J. Quo Vadis, Taxonomy of Cyanobacteria (2019). Fottea 2020, 20, 104–110. [Google Scholar] [CrossRef]
- Sanchis, D.; Carrasco, D.; Quesada, A. The Genus Microcystis (Microcystaceae/Cyanobacteria) from a Spanish Reservoir: A Contribution to the Definition of Morphological Variations. Nova Hedwig. 2004, 79, 479–495. [Google Scholar] [CrossRef]
- de Figueiredo, D.R.; Alves, A.; Pereira, M.J.; Correia, A. Molecular Characterization of Bloom-Forming Aphanizomenon Strains Isolated from Vela Lake (Western Central Portugal). J. Plankton Res. 2010, 32, 239–252. [Google Scholar] [CrossRef]
- Werner, V.R.; Laughinghouse, H.D., IV; Fiore, M.F.; Sant’anna, C.L.; Hoff, C.; de Souza Santos, K.R.; Neuhaus, E.B.; Molica, R.J.R.; Honda, R.Y.; Echenique, R.O.; et al. Morphological and Molecular Studies of Sphaerospermopsis torques-reginae (Cyanobacteria, Nostocales) from South American Water Blooms. Phycologia 2012, 51, 228–238. [Google Scholar] [CrossRef]
- Lv, X.; Cheng, Y.; Zhang, S.; Hu, Z.; Xiao, P.; Zhang, H.; Geng, R.; Li, R. Polyphasic Characterization and Taxonomic Evaluation of a Bloom-Forming Strain Morphologically Resembling Radiocystis fernandoi (Chroococcales, Cyanobacteria) from Lake Erhai, China. Diversity 2022, 14, 816. [Google Scholar] [CrossRef]
- Tanvir, R.U.; Hu, Z.; Zhang, Y.; Lu, J. Cyanobacterial Community Succession and Associated Cyanotoxin Production in Hypereutrophic and Eutrophic Freshwaters. Environ. Pollut. 2021, 290, 118056. [Google Scholar] [CrossRef]
- Gobler, C.J.; Burkholder, J.A.M.; Davis, T.W.; Harke, M.J.; Johengen, T.; Stow, C.A.; Van de Waal, D.B. The Dual Role of Nitrogen Supply in Controlling the Growth and Toxicity of Cyanobacterial Blooms. Harmful Algae 2016, 54, 87–97. [Google Scholar] [CrossRef]
- Ma, J.; Wang, P. Effects of Rising Atmospheric CO2 Levels on Physiological Response of Cyanobacteria and Cyanobacterial Bloom Development: A Review. Sci. Total Environ. 2021, 754, 141889. [Google Scholar] [CrossRef]
- Symes, E.; van Ogtrop, F.F. The Effect of Pre-Industrial and Predicted Atmospheric CO2 Concentrations on the Development of Diazotrophic and Non-Diazotrophic Cyanobacterium: Dolichospermum circinale and Microcystis aeruginosa. Harmful Algae 2019, 88, 101536. [Google Scholar] [CrossRef] [PubMed]
- Budzyńska, A.; Gołdyn, R. Domination of Invasive Nostocales (Cyanoprokaryota) at 52°N Latitude. Phycol. Res. 2017, 65, 322–332. [Google Scholar] [CrossRef]
- Budzyńska, A.; Rosińska, J.; Pełechata, A.; Toporowska, M.; Napiórkowska-Krzebietke, A.; Kozak, A.; Messyasz, B.; Pęczuła, W.; Kokociński, M.; Szeląg-Wasielewska, E.; et al. Environmental Factors Driving the Occurrence of the Invasive Cyanobacterium Sphaerospermopsis aphanizomenoides (Nostocales) in Temperate Lakes. Sci. Total Environ. 2019, 650, 1338–1347. [Google Scholar] [CrossRef] [PubMed]
- Kokociński, M.; Soininen, J. New Insights into the Distribution of Alien Cyanobacterium Chrysosporum bergii (Nostocales, Cyanobacteria). Phycol. Res. 2019, 67, 208–214. [Google Scholar] [CrossRef]
- Zheng, L.; Liu, Y.; Li, R.; Yang, Y.; Jiang, Y. Recent Advances in the Ecology of Bloom-Forming Raphidiopsis (Cylindrospermopsis) raciborskii: Expansion in China, Intraspecific Heterogeneity and Critical Factors for Invasion. Int. J. Environ. Res. Public Health 2023, 20, 1984. [Google Scholar] [CrossRef] [PubMed]
- Galvanese, E.F.; Padial, A.A.; Aubriot, L. Acclimation at High Temperatures Increases the Ability of Raphidiopsis raciborskii (Cyanobacteria) to Withstand Phosphate Deficiency and Reveals Distinct Strain Responses. Eur. J. Phycol. 2019, 54, 359–368. [Google Scholar] [CrossRef]
- Joung, S.-H.; Oh, H.-M.; Ko, S.-R.; Ahn, C.-Y. Correlations between Environmental Factors and Toxic and Non-Toxic Microcystis Dynamics during Bloom in Daechung Reservoir, Korea. Harmful Algae 2011, 10, 188–193. [Google Scholar] [CrossRef]
- Yoshida, M.; Yoshida, T.; Satomi, M.; Takashima, Y.; Hosoda, N.; Hiroishi, S. Intra-Specific Phenotypic and Genotypic Variation in Toxic Cyanobacterial Microcystis Strains. J. Appl. Microbiol. 2008, 105, 407–415. [Google Scholar] [CrossRef]
- Haande, S.; Rohrlack, T.; Ballot, A.; Røberg, K.; Skulberg, R.; Beck, M.; Wiedner, C. Genetic Characterisation of Cylindrospermopsis raciborskii (Nostocales, Cyanobacteria) Isolates from Africa and Europe. Harmful Algae 2008, 7, 692–701. [Google Scholar] [CrossRef]
- Gugger, M.; Molica, R.; Le Berre, B.; Dufour, P.; Bernard, C.; Humbert, J.-F. Genetic Diversity of Cylindrospermopsis Strains (Cyanobacteria) Isolated from Four Continents. Appl. Environ. Microbiol. 2005, 71, 1097–1100. [Google Scholar] [CrossRef]
- Hur, M.; Lee, I.; Tak, B.-M.; Lee, H.J.; Yu, J.J.; Cheon, S.U.; Kim, B.-S. Temporal Shifts in Cyanobacterial Communities at Different Sites on the Nakdong River in Korea. Water Res. 2013, 47, 6973–6982. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, S.W.; Farnsley, S.E.; LeCleir, G.R.; Layton, A.C.; Satchwell, M.F.; DeBruyn, J.M.; Boyer, G.L.; Zhu, G.; Paerl, H.W. The Relationships between Nutrients, Cyanobacterial Toxins and the Microbial Community in Taihu (Lake Tai), China. Harmful Algae 2011, 10, 207–215. [Google Scholar] [CrossRef]
- Davis, T.W.; Berry, D.L.; Boyer, G.L.; Gobler, C.J. The Effects of Temperature and Nutrients on the Growth and Dynamics of Toxic and Non-Toxic Strains of Microcystis during Cyanobacteria Blooms. Harmful Algae 2009, 8, 715–725. [Google Scholar] [CrossRef]
- Kurmayer, R.; Christiansen, G.; Fastner, J.; Börner, T. Abundance of Active and Inactive Microcystin Genotypes in Populations of the Toxic Cyanobacterium Planktothrix spp. Environ. Microbiol. 2004, 6, 831–841. [Google Scholar] [CrossRef]
- Johansson, E.; Legrand, C.; Björnerås, C.; Godhe, A.; Mazur-Marzec, H.; Säll, T.; Rengefors, K. High Diversity of Microcystin Chemotypes within a Summer Bloom of the Cyanobacterium Microcystis botrys. Toxins 2019, 11, 698. [Google Scholar] [CrossRef] [PubMed]
- Pearson, L.A.; Dittmann, E.; Mazmouz, R.; Ongley, S.E.; D’Agostino, P.M.; Neilan, B.A. The Genetics, Biosynthesis and Regulation of Toxic Specialized Metabolites of Cyanobacteria. Harmful Algae 2016, 54, 98–111. [Google Scholar] [CrossRef] [PubMed]
- Rantala-Ylinen, A.; Känä, S.; Wang, H.; Rouhiainen, L.; Wahlsten, M.; Rizzi, E.; Berg, K.; Gugger, M.; Sivonen, K. Anatoxin-a Synthetase Gene Cluster of the Cyanobacterium Anabaena sp. strain 37 and Molecular Methods to Detect Potential Producers. Appl. Environ. Microbiol. 2011, 77, 7271–7278. [Google Scholar] [CrossRef] [PubMed]
- Méjean, A.; Paci, G.; Gautier, V.; Ploux, O. Biosynthesis of Anatoxin-a and Analogues (Anatoxins) in Cyanobacteria. Toxicon 2014, 91, 15–22. [Google Scholar] [CrossRef]
- Burford, M.A.; Davis, T.W.; Orr, P.T.; Sinha, R.; Willis, A.; Neilan, B. Nutrient-Related Changes in the Toxicity of Field Blooms of the Cyanobacterium, Cylindrospermopsis raciborskii. FEMS Microbiol. Ecol. 2014, 89, 135–148. [Google Scholar] [CrossRef]
- Lei, L.; Lei, M.; Cheng, N.; Chen, Z.; Xiao, L.; Han, B.P.; Lin, Q. Nutrient Regulation of Relative Dominance of Cylindrospermopsin-Producing and Non-Cylindrospermopsin-Producing Raphidiopsis raciborskii. Front. Microbiol. 2021, 12, 793544. [Google Scholar] [CrossRef]
- Shimizu, K.; Maseda, H.; Okano, K.; Itayama, T.; Kawauchi, Y.; Chen, R.; Utsumi, M.; Zhang, Z.; Sugiura, N. How Microcystin-Degrading Bacteria Express Microcystin Degradation Activity. Lakes Reserv. 2011, 16, 169–178. [Google Scholar] [CrossRef]
- Kormas, K.A.; Lymperopoulou, D.S. Cyanobacterial Toxin Degrading Bacteria: Who Are They? Biomed. Res. Int. 2013, 2013, 1463894. [Google Scholar] [CrossRef] [PubMed]
- Dziallas, C.; Grossart, H.P. Increasing Oxygen Radicals and Water Temperature Select for Toxic Microcystis sp. PLoS ONE 2011, 6, e25569. [Google Scholar] [CrossRef] [PubMed]
- Omidi, A.; Esterhuizen-Londt, M.; Pflugmacher, S. Still Challenging: The Ecological Function of the Cyanobacterial Toxin Microcystin–What We Know so Far. Toxin Rev. 2018, 37, 87–105. [Google Scholar] [CrossRef]
- Ogashawara, I.; Mishra, D.; Mishra, S.; Curtarelli, M.; Stech, J. A Performance Review of Reflectance Based Algorithms for Predicting Phycocyanin Concentrations in Inland Waters. Remote Sens. 2013, 5, 4774–4798. [Google Scholar] [CrossRef]
- Borges, H.D.; Cicerelli, R.E.; De Almeida, T.; Roig, H.L.; Olivetti, D. Monitoring Cyanobacteria Occurrence in Freshwater Reservoirs Using Semi-Analytical Algorithms and Orbital Remote Sensing. Mar. Freshw. Res. 2020, 71, 569–578. [Google Scholar] [CrossRef]
- Greenwold, M.J.; Cunningham, B.R.; Lachenmyer, E.M.; Pullman, J.M.; Richardson, T.L.; Dudycha, J.L. Diversification of Light Capture Ability Was Accompanied by the Evolution of Phycobiliproteins in Cryptophyte Algae. Proc. R. Soc. B Biol. Sci. 2019, 286, 20190655. [Google Scholar] [CrossRef]
- Ogashawara, I. Determination of Phycocyanin from Space—A Bibliometric Analysis. Remote Sens. 2020, 12, 567. [Google Scholar] [CrossRef]
- Mishra, S.; Mishra, D.R.; Lee, Z.; Tucker, C.S. Quantifying Cyanobacterial Phycocyanin Concentration in Turbid Productive Waters: A Quasi-Analytical Approach. Remote Sens. Environ. 2013, 133, 141–151. [Google Scholar] [CrossRef]
- Li, L.; Li, L.; Song, K. Remote Sensing of Freshwater Cyanobacteria: An Extended IOP Inversion Model of Inland Waters (IIMIW) for Partitioning Absorption Coefficient and Estimating Phycocyanin. Remote Sens. Environ. 2015, 157, 9–23. [Google Scholar] [CrossRef]
- Yan, Y.; Bao, Z.; Shao, J. Phycocyanin Concentration Retrieval in Inland Waters: A Comparative Review of the Remote Sensing Techniques and Algorithms. J. Great Lakes Res. 2018, 44, 748–755. [Google Scholar] [CrossRef]
- Witter, D.L.; Ortiz, J.D.; Palm, S.; Heath, R.T.; Budd, J.W. Assessing the Application of SeaWiFS Ocean Color Algorithms to Lake Erie. J. Great Lakes Res. 2009, 35, 361–370. [Google Scholar] [CrossRef]
- Pirasteh, S.; Mollaee, S.; Fatholahi, S.N.; Li, J. Estimation of Phytoplankton Chlorophyll-a Concentrations in the Western Basin of Lake Erie Using Sentinel-2 and Sentinel-3 Data|Estimation Des Concentrations de Chlorophylle-a Du Phytoplancton Dans Le Bassin Ouest Du Lac Érié à l’aide Des Données Sentin. Can. J. Remote Sens. 2020, 46, 585–602. [Google Scholar] [CrossRef]
- Binding, C.E.; Greenberg, T.A.; Bukata, R.P. The MERIS Maximum Chlorophyll Index; Its Merits and Limitations for Inland Water Algal Bloom Monitoring. J. Great Lakes Res. 2013, 39, 100–107. [Google Scholar] [CrossRef]
- Wynne, T.T.; Stumpf, R.P.; Tomlinson, M.C.; Fahnenstiel, G.L.; Dyble, J.; Schwab, D.J.; Joshi, S.J. Evolution of a Cyanobacterial Bloom Forecast System in Western Lake Erie: Development and Initial Evaluation. J. Great Lakes Res. 2013, 39, 90–99. [Google Scholar] [CrossRef]
- Vincent, R.K.; Qin, X.; McKay, R.M.L.; Miner, J.; Czajkowski, K.; Savino, J.; Bridgeman, T. Phycocyanin Detection from LANDSAT TM Data for Mapping Cyanobacterial Blooms in Lake Erie. Remote Sens. Environ. 2004, 89, 381–392. [Google Scholar] [CrossRef]
- Yin, Z.; Li, J.; Liu, Y.; Xie, Y.; Zhang, F.; Wang, S.; Sun, X.; Zhang, B. Water Clarity Changes in Lake Taihu over 36 Years Based on Landsat TM and OLI Observations. Int. J. Appl. Earth Obs. Geoinf. 2021, 102, 102457. [Google Scholar] [CrossRef]
- Isenstein, E.M.; Kim, D.; Park, M.H. Modeling for Multi-Temporal Cyanobacterial Bloom Dominance and Distributions Using Landsat Imagery. Ecol. Inform. 2020, 59, 101119. [Google Scholar] [CrossRef]
- Shi, K.; Zhang, Y.; Qin, B.; Zhou, B. Remote Sensing of Cyanobacterial Blooms in Inland Waters: Present Knowledge and Future Challenges. Sci. Bull. 2019, 64, 1540–1556. [Google Scholar] [CrossRef]
- Simis, S.G.H.; Peters, S.W.M.; Gons, H.J. Remote Sensing of the Cyanobacterial Pigment Phycocyanin in Turbid Inland Water. Limnol. Oceanogr. 2005, 50, 237–245. [Google Scholar] [CrossRef]
- Ruiz-Verdú, A.; Simis, S.G.H.; de Hoyos, C.; Gons, H.J.; Peña-Martínez, R. An Evaluation of Algorithms for the Remote Sensing of Cyanobacterial Biomass. Remote Sens. Environ. 2008, 112, 3996–4008. [Google Scholar] [CrossRef]
- Liu, M.; Ling, H.; Wu, D.; Su, X.; Cao, Z. Sentinel-2 and Landsat-8 Observations for Harmful Algae Blooms in a Small Eutrophic Lake. Remote Sens. 2021, 13, 4479. [Google Scholar] [CrossRef]
- Veerman, J.; Kumar, A.; Mishra, D.R. Exceptional Landscape-Wide Cyanobacteria Bloom in Okavango Delta, Botswana in 2020 Coincided with a Mass Elephant Die-off Event. Harmful Algae 2022, 111, 102145. [Google Scholar] [CrossRef] [PubMed]
- Alcantara, E.; Coimbra, K.; Ogashawara, I.; Rodrigues, T.; Mantovani, J.; Rotta, L.H.; Park, E.; Fernandes Cunha, D.G. A Satellite-Based Investigation into the Algae Bloom Variability in Large Water Supply Urban Reservoirs during COVID-19 Lockdown. Remote Sens. Appl. 2021, 23, 100555. [Google Scholar] [CrossRef]
- Sòria-Perpinyà, X.; Vicente, E.; Urrego, P.; Pereira-Sandoval, M.; Ruíz-Verdú, A.; Delegido, J.; Soria, J.M.; Moreno, J. Remote Sensing of Cyanobacterial Blooms in a Hypertrophic Lagoon (Albufera of València, Eastern Iberian Peninsula) Using Multitemporal Sentinel-2 Images. Sci. Total Environ. 2020, 698, 134305. [Google Scholar] [CrossRef] [PubMed]
- Niroumand-Jadidi, M.; Bovolo, F. Deep Learning-Based Retrieval of an Orange Band Sensitive to Cyanobacteria for Landsat-8/9 and Sentinel-2. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2023, 16, 3929–3937. [Google Scholar] [CrossRef]
- Pérez-González, R.; Sòria-Perpinyà, X.; Soria, J.M.; Delegido, J.; Urrego, P.; Sendra, M.D.; Ruíz-Verdú, A.; Vicente, E.; Moreno, J. Phycocyanin Monitoring in Some Spanish Water Bodies with Sentinel-2 Imagery. Water 2021, 13, 2866. [Google Scholar] [CrossRef]
- Zamyadi, A.; Choo, F.; Newcombe, G.; Stuetz, R.; Henderson, R.K. A Review of Monitoring Technologies for Real-Time Management of Cyanobacteria: Recent Advances and Future Direction. TrAC-Trends Anal. Chem. 2016, 85, 83–96. [Google Scholar] [CrossRef]
- Ma, L.; Moradinejad, S.; Guerra Maldonado, J.F.; Zamyadi, A.; Dorner, S.; Prévost, M. Factors Affecting the Interpretation of Online Phycocyanin Fluorescence to Manage Cyanobacteria in Drinking Water Sources. Water 2022, 14, 3749. [Google Scholar] [CrossRef]
- Macário, I.P.; Castro, B.B.; Nunes, I.M.S.; Antunes, S.C.S.C.; Pizarro, C.; Coelho, C.; Gonçalves, F.; de Figueiredo, D.R. New Insights towards the Establishment of Phycocyanin Concentration Thresholds Considering Species-Specific Variability of Bloom-Forming Cyanobacteria. Hydrobiologia 2015, 757, 155–165. [Google Scholar] [CrossRef]
- Bunyon, C.L.; Fraser, B.T.; McQuaid, A.; Congalton, R.G. Using Imagery Collected by an Unmanned Aerial System to Monitor Cyanobacteria in New Hampshire, USA, Lakes. Remote Sens. 2023, 15, 2839. [Google Scholar] [CrossRef]
- Pershin, S.M.; Katsnelson, B.G.; Grishin, M.Y.; Lednev, V.N.; Zavozin, V.A.; Ostrovsky, I. Laser Remote Sensing of Lake Kinneret by Compact Fluorescence LiDAR. Sensors 2022, 22, 7307. [Google Scholar] [CrossRef]
- Wu, D.; Li, R.; Zhang, F.; Liu, J. A Review on Drone-Based Harmful Algae Blooms Monitoring. Environ. Monit. Assess. 2019, 191, 211. [Google Scholar] [CrossRef]
- Becker, R.H.; Sayers, M.; Dehm, D.; Shuchman, R.; Quintero, K.; Bosse, K.; Sawtell, R. Unmanned Aerial System Based Spectroradiometer for Monitoring Harmful Algal Blooms: A New Paradigm in Water Quality Monitoring. J. Great Lakes Res. 2019, 45, 444–453. [Google Scholar] [CrossRef]
- Johansen, R.A.; Beck, R.; Stumpf, R.; Lekki, J.; Tokars, R.; Tolbert, C.; McGhan, C.; Black, T.; Ma, O.; Xu, M.; et al. HABSat-1: Assessing the Feasibility of Using CubeSats for the Detection of Cyanobacterial Harmful Algal Blooms in Inland Lakes and Reservoirs. Lake Reserv. Manag. 2019, 35, 193–207. [Google Scholar] [CrossRef]
- Azevedo, R.; Rodriguez, E.; Figueiredo, D.; Peixoto, F.; Santos, C. Methodologies for the Study of Filamentous Cyanobacteria by Flow Cytometry. Fresenius Environ. Bull. 2012, 21, 679–684. [Google Scholar]
- Dashkova, V.; Malashenkov, D.; Poulton, N.; Vorobjev, I.; Barteneva, N.S. Imaging Flow Cytometry for Phytoplankton Analysis. Methods 2017, 112, 188–200. [Google Scholar] [CrossRef]
- Mirasbekov, Y.; Zhumakhanova, A.; Zhantuyakova, A.; Sarkytbayev, K.; Malashenkov, D.V.; Baishulakova, A.; Dashkova, V.; Davidson, T.A.; Vorobjev, I.A.; Jeppesen, E.; et al. Semi-Automated Classification of Colonial Microcystis by FlowCAM Imaging Flow Cytometry in Mesocosm Experiment Reveals High Heterogeneity during Seasonal Bloom. Sci. Rep. 2021, 11, 9377. [Google Scholar] [CrossRef]
- Kraft, K.; Seppälä, J.; Hällfors, H.; Suikkanen, S.; Ylöstalo, P.; Anglès, S.; Kielosto, S.; Kuosa, H.; Laakso, L.; Honkanen, M.; et al. First Application of IFCB High-Frequency Imaging-in-Flow Cytometry to Investigate Bloom-Forming Filamentous Cyanobacteria in the Baltic Sea. Front. Mar. Sci. 2021, 8, 594144. [Google Scholar] [CrossRef]
- George, G.; Menon, N.N.; Abdulaziz, A.; Brewin, R.J.W.; Pranav, P.; Gopalakrishnan, A.; Mini, K.G.; Kuriakose, S.; Sathyendranath, S.; Platt, T. Citizen Scientists Contribute to Real-Time Monitoring of Lake Water Quality Using 3D Printed Mini Secchi Disks. Front. Water 2021, 3, 662142. [Google Scholar] [CrossRef]
- Ho, S.Y.F.; Xu, S.J.; Lee, F.W.F. Citizen Science: An Alternative Way for Water Monitoring in Hong Kong. PLoS ONE 2020, 15, e0238349. [Google Scholar] [CrossRef]
- Siano, R.; Chapelle, A.; Antoine, V.; Michel-Guillou, E.; Rigaut-Jalabert, F.; Guillou, L.; Hégaret, H.; Leynaert, A.; Curd, A. Citizen Participation in Monitoring Phytoplankton Seawater Discolorations. Mar. Policy 2020, 117, 103039. [Google Scholar] [CrossRef]
- Cunha, D.G.F.; Casali, S.P.; de Falco, P.B.; Thornhill, I.; Loiselle, S.A. The Contribution of Volunteer-Based Monitoring Data to the Assessment of Harmful Phytoplankton Blooms in Brazilian Urban Streams. Sci. Total Environ. 2017, 584–585, 586–594. [Google Scholar] [CrossRef]
- Sanseverino, I.; Conduto António, D.; Loos, R.; Lettieri, T. Cyanotoxins: Methods and Approaches for Their Analysis and Detection; Joint Research Centre: Luxembourg, 2017. [Google Scholar]
- Komárek, J.; Mareš, J. An Update to Modern Taxonomy (2011) of Freshwater Planktic Heterocytous Cyanobacteria. Hydrobiologia 2012, 698, 327–351. [Google Scholar] [CrossRef]
- Thiel, S.U.; Wiltshire, R.J.; Davies, L.J. Automated Object Recognition of Blue-Green Algae for Measuring Water Quality—A Preliminary Study; Elsevier: Amsterdam, The Netherlands, 1995; Volume 29. [Google Scholar]
- Almesjö, L.; Rolff, C. Automated Measurements of Filamentous Cyanobacteria by Digital Image Analysis. Limnol. Oceanogr. Methods 2007, 5, 217–224. [Google Scholar] [CrossRef]
- Geronimo, J.O.-N.; Arguelles, E. Automated Classification and Identification System for Freshwater Algae Using Convolutional Neural Networks. Philipine J. Sci. 2023, 152, 325–335. [Google Scholar] [CrossRef]
- Chong, J.W.R.; Khoo, K.S.; Chew, K.W.; Vo, D.V.N.; Balakrishnan, D.; Banat, F.; Munawaroh, H.S.H.; Iwamoto, K.; Show, P.L. Microalgae Identification: Future of Image Processing and Digital Algorithm. Bioresour. Technol. 2023, 369, 128418. [Google Scholar] [CrossRef]
- Gaur, A.; Pant, G.; Jalal, A.S. Computer-Aided Cyanobacterial Harmful Algae Blooms (CyanoHABs) Studies Based on Fused Artificial Intelligence (AI) Models. Algal Res. 2022, 67, 102842. [Google Scholar] [CrossRef]
- Kraft, K.; Velhonoja, O.; Eerola, T.; Suikkanen, S.; Tamminen, T.; Haraguchi, L.; Ylöstalo, P.; Kielosto, S.; Johansson, M.; Lensu, L.; et al. Towards Operational Phytoplankton Recognition with Automated High-Throughput Imaging, near-Real-Time Data Processing, and Convolutional Neural Networks. Front. Mar. Sci. 2022, 9, 867695. [Google Scholar] [CrossRef]
- Nübel, U.; Garcia-Pichel, F.; Muyzer, G. PCR Primers to Amplify 16S rRNA Genes from Cyanobacteria. Appl. Environ. Microbiol. 1997, 63, 3327–3332. [Google Scholar] [CrossRef]
- Biegala, I.C.; Raimbault, P. High Abundance of Diazotrophic Picocyanobacteria (<3 μm) in a Southwest Pacific Coral Lagoon. Aquat. Microb. Ecol. 2008, 51, 45–53. [Google Scholar] [CrossRef]
- Wang, X.; Sun, M.; Wang, J.; Yang, L.; Luo, L.; Li, P.; Kong, F. Microcystis Genotype Succession and Related Environmental Factors in Lake Taihu during Cyanobacterial Blooms. Microb. Ecol. 2012, 64, 986–999. [Google Scholar] [CrossRef]
- de Figueiredo, D.R.; Pereira, M.J.; Castro, B.B.; Correia, A. Bacterioplankton Community Composition in Portuguese Water Bodies under a Severe Summer Drought. Community Ecol. 2012, 13, 185–193. [Google Scholar] [CrossRef]
- Yu, Z.; Zhou, J.; Yang, J.; Yu, X.; Liu, L. Vertical Distribution of Diazotrophic Bacterial Community Associated with Temperature and Oxygen Gradients in a Subtropical Reservoir. Hydrobiologia 2014, 741, 69–77. [Google Scholar] [CrossRef]
- Touzet, N.; McCarthy, D.; Gill, A.; Fleming, G.T.A. Comparative Summer Dynamics of Surface Cyanobacterial Communities in Two Connected Lakes from the West of Ireland. Sci. Total Environ. 2016, 553, 416–428. [Google Scholar] [CrossRef]
- Feng, L.; Liu, S.; Wu, W.; Ma, J.; Li, P.; Xu, H.; Li, N.; Feng, Y. Dominant Genera of Cyanobacteria in Lake Taihu and Their Relationships with Environmental Factors. J. Microbiol. 2016, 54, 468–476. [Google Scholar] [CrossRef]
- Kolmonen, E.; Sivonen, K.; Rapala, J.; Haukka, K. Diversity of Cyanobacteria and Heterotrophic Bacteria in Cyanobacterial Blooms in Lake Joutikas, Finland. Aquat. Microb. Ecol. 2004, 36, 201–211. [Google Scholar] [CrossRef][Green Version]
- Eiler, A.; Drakare, S.; Bertilsson, S.; Pernthaler, J.; Peura, S.; Rofner, C.; Simek, K.; Yang, Y.; Znachor, P.; Lindström, E.S. Unveiling Distribution Patterns of Freshwater Phytoplankton by a next Generation Sequencing Based Approach. PLoS ONE 2013, 8, e53516. [Google Scholar] [CrossRef]
- Steffen, M.M.; Li, Z.; Effler, T.C.; Hauser, L.J.; Boyer, G.L.; Wilhelm, S.W. Comparative Metagenomics of Toxic Freshwater Cyanobacteria Bloom Communities on Two Continents. PLoS ONE 2012, 7, e44002. [Google Scholar] [CrossRef]
- Steven, B.; McCann, S.; Ward, N.L. Pyrosequencing of Plastid 23S RRNA Genes Reveals Diverse and Dynamic Cyanobacterial and Algal Populations in Two Eutrophic Lakes. FEMS Microbiol. Ecol. 2012, 82, 607–615. [Google Scholar] [CrossRef]
- Stüken, A.; Campbell, R.J.; Quesada, A.; Sukenik, A.; Dadheech, P.K.; Wiedner, C. Genetic and Morphologic Characterization of Four Putative Cylindrospermopsin Producing Species of the Cyanobacterial Genera Anabaena and Aphanizomenon. J. Plankton Res. 2009, 31, 465–480. [Google Scholar] [CrossRef]
- Lezcano, M.Á.; Velázquez, D.; Quesada, A.; El-Shehawy, R. Diversity and Temporal Shifts of the Bacterial Community Associated with a Toxic Cyanobacterial Bloom: An Interplay between Microcystin Producers and Degraders. Water Res. 2017, 125, 52–61. [Google Scholar] [CrossRef]
- Chen, C.; Zhang, Z.; Ding, A.; Wu, J.; Xiao, J.; Sun, Y.; Cheng, C.; Zaichao, Z.; Aizhong, D.; Jiayan, W.; et al. Bar-Coded Pyrosequencing Reveals the Bacterial Community during Microcystis Water Bloom in Guanting Reservoir, Beijing. Procedia Eng. 2011, 18, 341–346. [Google Scholar] [CrossRef][Green Version]
- Salomon, P.S.; Janson, S.; Grane?li, E. Molecular Identification of Bacteria Associated with Filaments of Nodularia spumigena and Their Effect on the Cyanobacterial Growth. Harmful Algae 2003, 2, 261–272. [Google Scholar] [CrossRef]
- Berg, K.A.; Lyra, C.; Sivonen, K.; Paulin, L.; Suomalainen, S.; Tuomi, P.; Rapala, J. High Diversity of Cultivable Heterotrophic Bacteria in Association with Cyanobacterial Water Blooms. ISME J. 2008, 3, 314–325. [Google Scholar] [CrossRef]
- Shunyu, S.; Yongding, L.; Yinwu, S.; Genbao, L.; Dunhai, L. Lysis of Aphanizomenon flos-aquae (Cyanobacterium) by a Bacterium Bacillus cereus. Biol. Control. 2006, 39, 345–351. [Google Scholar] [CrossRef]
- Hennon, G.M.M.; Dyhrman, S.T. Progress and Promise of Omics for Predicting the Impacts of Climate Change on Harmful Algal Blooms. Harmful Algae 2020, 91, 101587. [Google Scholar] [CrossRef]
- Dziallas, C.; Pinnow, S.; Grossart, H.P. Quantification of Toxic and Toxin-Producing Cyanobacterial Cells by RING-FISH in Combination with Flow Cytometry. Limnol. Oceanogr. Methods 2011, 9, 67–73. [Google Scholar] [CrossRef]
- Legrand, B.; Lesobre, J.; Colombet, J.; Latour, D.; Sabart, M. Molecular Tools to Detect Anatoxin-a Genes in Aquatic Ecosystems: Toward a New Nested PCR-Based Method. Harmful Algae 2016, 58, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Caracciolo, A.B.; Dejana, L.; Fajardo, C.; Grenni, P.; Martin, M.; Mengs, G.; Sánchez-Fortún, S.; Lettieri, T.; Saccà, M.L.; Medlin, L.K. A New Fluorescent Oligonucleotide Probe for In-Situ Identification of Microcystis aeruginosa in Freshwater. Microchem. J. 2019, 148, 503–513. [Google Scholar] [CrossRef]
- Metcalf, J.S.; Reilly, M.; Young, F.M.; Codd, G.A. Localization of Microcystin Synthetase Genes in Colonies of the Cyanobacterium Microcystis Using Fluorescence in Situ Hybridization. J. Phycol. 2009, 45, 1400–1404. [Google Scholar] [CrossRef]
- Zeller, P.; Méjean, A.; Biegala, I.; Contremoulins, V.; Ploux, O. Fluorescence in Situ Hybridization of Microcystis Strains Producing Microcystin Using Specific mRNA Probes. Lett. Appl. Microbiol. 2016, 63, 376–383. [Google Scholar] [CrossRef]
- Brient, L.; Ben Gamra, N.; Periot, M.; Roumagnac, M.; Zeller, P.; Bormans, M.; Méjean, A.; Ploux, O.; Biegala, I.C. Rapid Characterization of Microcystin-Producing Cyanobacteria in Freshwater Lakes by TSA-FISH (Tyramid Signal Amplification-Fluorescent in Situ Hybridization). Front. Environ. Sci. 2017, 5, 43. [Google Scholar] [CrossRef]
- Casero, M.C.; Velázquez, D.; Medina-Cobo, M.; Quesada, A.; Cirés, S. Unmasking the Identity of Toxigenic Cyanobacteria Driving a Multi-Toxin Bloom by High-Throughput Sequencing of Cyanotoxins Genes and 16S rRNA Metabarcoding. Sci. Total Environ. 2019, 665, 367–378. [Google Scholar] [CrossRef]
- Cunha, I.; Biltes, R.; Sales, M.G.F.; Vasconcelos, V. Aptamer-Based Biosensors to Detect Aquatic Phycotoxins and Cyanotoxins. Sensors 2018, 18, 2367. [Google Scholar] [CrossRef]
- Macário, I.P.E.; Castro, B.B.; Nunes, I.M.S.; Pizarro, C.; Coelho, C.; Gonçalves, F.; de Figueiredo, D.R. Stepwise Strategy for Monitoring Toxic Cyanobacterial Blooms in Lentic Water Bodies. Environ. Monit. Assess. 2017, 189, 620. [Google Scholar] [CrossRef]
- Ho, J.C.; Michalak, A.M. Challenges in Tracking Harmful Algal Blooms: A Synthesis of Evidence from Lake Erie. J. Great Lakes Res. 2015, 41, 317–325. [Google Scholar] [CrossRef]
- Srivastava, A.; Singh, S.; Ahn, C.Y.; Oh, H.M.; Asthana, R.K. Monitoring Approaches for a Toxic Cyanobacterial Bloom. Environ. Sci. Technol. 2013, 47, 8999–9013. [Google Scholar] [CrossRef] [PubMed]
- Douglas Greene, S.B.; LeFevre, G.H.; Markfort, C.D. Improving the Spatial and Temporal Monitoring of Cyanotoxins in Iowa Lakes Using a Multiscale and Multi-Modal Monitoring Approach. Sci. Total Environ. 2020, 760, 143327. [Google Scholar] [CrossRef] [PubMed]
- Steffen, M.M.; Belisle, B.S.; Watson, S.B.; Boyer, G.L.; Bourbonniere, R.A.; Wilhelm, S.W. Metatranscriptomic Evidence for Co-Occurring Top-down and Bottom-up Controls on Toxic Cyanobacterial Communities. Appl. Environ. Microbiol. 2015, 81, 3268–3276. [Google Scholar] [CrossRef] [PubMed]
- Thompson, P.A.; Carstensen, J. Global Observing for Phytoplankton? A Perspective. J. Plankton Res. 2023, 45, 221–234. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Figueiredo, D.R. Harmful Cyanobacterial Blooms: Going beyond the “Green” to Monitor and Predict HCBs. Hydrobiology 2024, 3, 11-30. https://doi.org/10.3390/hydrobiology3010002
de Figueiredo DR. Harmful Cyanobacterial Blooms: Going beyond the “Green” to Monitor and Predict HCBs. Hydrobiology. 2024; 3(1):11-30. https://doi.org/10.3390/hydrobiology3010002
Chicago/Turabian Stylede Figueiredo, Daniela R. 2024. "Harmful Cyanobacterial Blooms: Going beyond the “Green” to Monitor and Predict HCBs" Hydrobiology 3, no. 1: 11-30. https://doi.org/10.3390/hydrobiology3010002
APA Stylede Figueiredo, D. R. (2024). Harmful Cyanobacterial Blooms: Going beyond the “Green” to Monitor and Predict HCBs. Hydrobiology, 3(1), 11-30. https://doi.org/10.3390/hydrobiology3010002





