Modeling and Visualizing the Dynamic Spread of Epidemic Diseases—The COVID-19 Case
Abstract
:1. Introduction
2. Materials and Methods
2.1. Context
- It considers the initial distribution of the infections, through the lattice population.
- We can visualize this spread through time and space, making the whole concept easier to understand.
- In most of the cases, accurate initial distribution of the infected individuals in the lattice cannot be observed through the existing data, so any considerations are based on random distribution (uniform). To make clear the importance of this property, we return to the simple 3 × 3 square lattice, dividing population into infected (I-nodes) and susceptible (S-nodes) as presented in Figure 2. In the following example, both lattices (Figure 2a,b) represent a population of nine nodes–individuals. Two of them are infected (I0 = 2) and seven are susceptible (S0 = 7). We note that their distribution is not the same. Both diagrams could appear equally probable but may lead to different paths of the virus spread, because of their initial distribution. SIR models cannot conceive such information. It is obvious that the initial distribution of infected individuals through the lattice determines the number of possible contagions. This way, it affects the reproduction number of the virus. For example, we can verify that the central node of Figure 2a can transmit the virus to up to four neighboring nodes, while the bottom right node of Figure 2b can only affect a maximum of one node. Additionally, if contagions represented by arrows happen with a probability less than 1, then susceptible individuals have different probabilities of becoming infected. For example, the susceptible nodes of the second column of Figure 2a would have different probabilities of becoming infected in such a case. The first-row node is exposed to two separate contagious individuals, while the third-row node is exposed to one contagious individual.
- A second realistic stochasticity is related to susceptible individuals who can contact infected ones after every sweep (period). Some contacts may meet while others will not (considering restrictions on mobility, self-protection measures, isolation etc.). We only allow for a percentage of those possible contacts to transform susceptible individuals to exposed, while the rest emerge with no contagion. We use a random process, so that contact is enabled only for two neighboring nodes of each infectious node. In the example presented in Figure 3, only up and right neighboring nodes can become infected. In the first case (Figure 3a) both interactions lead to infections, while in the second case (Figure 3b), one of them is already infected so only the other interaction leads to a new infection.
- The third realistic stochasticity presented in our model allows some exposed individuals to evolve to be asymptomatic, while the rest evolve to be infected (symptomatic). Both situations lead to immunity after some days (sweeps).

2.2. Algorithm and Experimentation
- become able to estimate the number of infected and susceptible individuals and represent those states clearly, by replacing charge with flag values. We used 1 for infected and 0 for susceptible. Diffusion of charge is replaced by contagion of virus to neighboring nodes/individuals,
- apply to an extended SIR model (we experimented using SEAIR). In addition to values 1 and 0, for infected (I) and susceptible (S), respectively, we used 0.5 for exposed (E), 0.75 for asymptomatic (A) and −0.25 and −0.5 for asymptomatic who recovered (AR) and infected who recovered (R), respectively,
- visualize results. Our model’s results can be depicted in short videos, presenting the lattice from day one (initial condition) to the final day. An observer can identify the exact coordinates of each infection and recovery and understand patterns of contagion and immunity building in the population.
- All nodes/individuals are considered as susceptible. Then, according to the initial conditions, an amount of I0, E0, and A0 nodes turn into infected, exposed and asymptomatic. Their positions are chosen through a random process (thus any repetition of the experiment is different).
- During the first step (period one):
- ○
- The algorithm recognizes all infected nodes/individuals and chooses randomly n out of 4 neighboring nodes as possible infectious connections. If any of the nodes chosen are susceptible, they may transform to exposed with a probability p that allows us to reproduce n × p infectious contacts per individual per period. If the nodes chosen are not susceptible, nothing changes.
- ○
- The same process with different parameters holds for asymptomatic nodes/individuals.
- ○
- The algorithm recognizes exposed nodes and may transform them into asymptomatic or infected. The exact period and the type of transformation are defined by a stochastic process, according to the parameters.
- ○
- The algorithm recognizes asymptomatic nodes and may transform them into asymptomatic recovered if the period of asymptomatic infection is completed.
- ○
- The algorithm recognizes infected nodes and may transform them into recovered if the period of symptomatic infection is completed.
- ○
- The algorithm recognizes immunized (asymptomatic recovered and recovered) nodes/individuals and may transform them into susceptible if the immunity period is completed.
- The process is repeated for all following periods, keeping track of the data.
3. Results
3.1. 6 × 6 Lattice
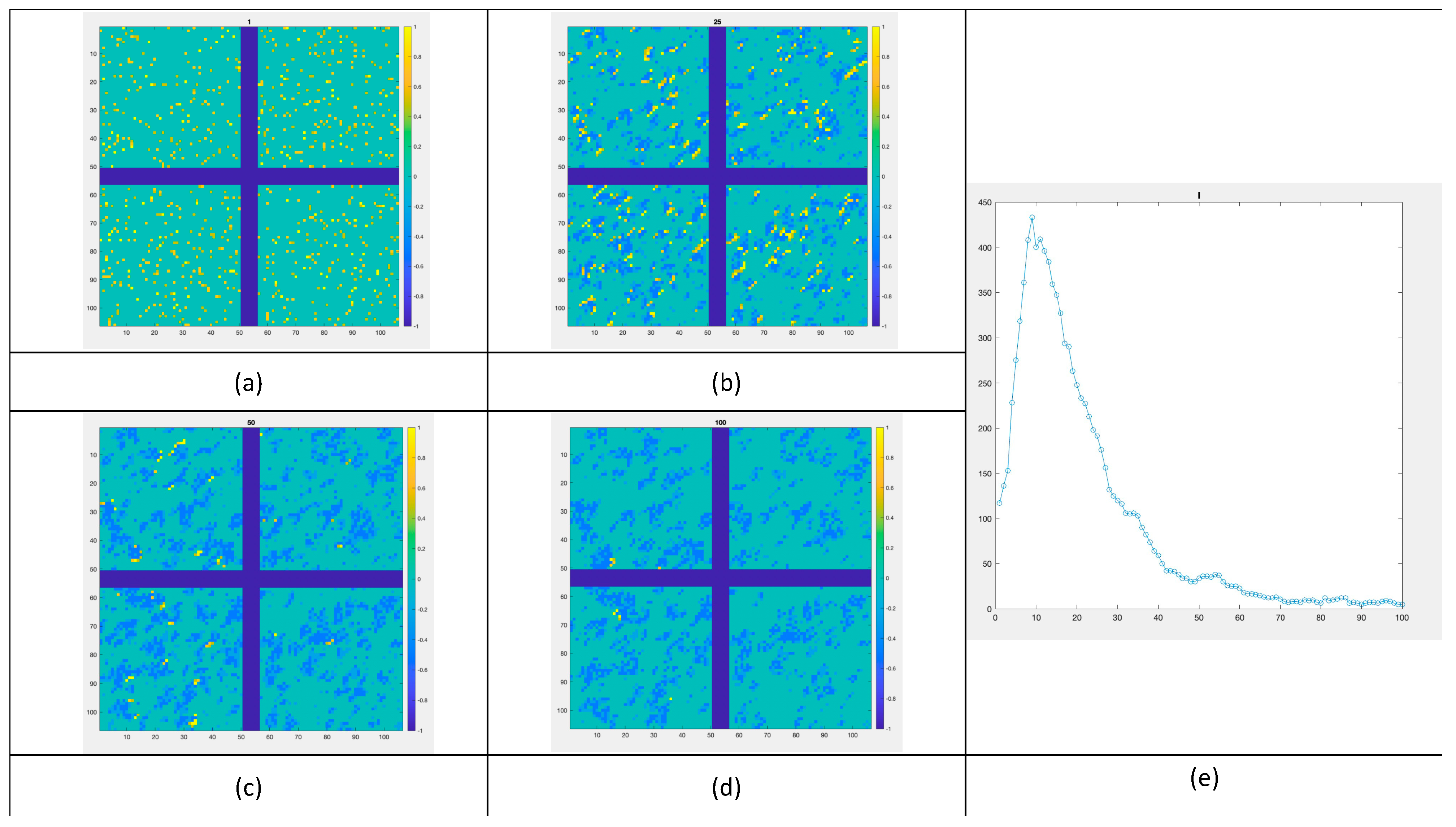
3.2. 100 × 100 Lattice
3.3. 3282 × 3282 Lattice
3.4. Applying Borders
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. MATLAB Code Used for Simulations
| %Initial Conditions L=3282; n=input("sweeps"); infected=117; cnact=1; rt=6; imm=90; bis=input(“bis”); bas=input(“bas”); daysas=5; daysis=3; EtoI=2.65; AtoI=2.84; filename = ‘SEAIR3282.xlsx’; sim=input("simulations"); for SIMS=1:sim LB=L sitold=zeros(LB,LB); sitnew=zeros(LB,LB); timer=zeros(LB,LB); exposed=infected*EtoI; asymptomatic=infected*AtoI; sumI=0; sumE=0; sumA=0; sumS=0; sumAR=0; sumR=0; %infected initial for i=1:infected test=0; while test<1 rndx1(i)=rand; ix(i)=fix(1+rndx1(i)*LB); rndy1(i)=rand; iy(i)=fix(1+rndy1(i)*LB); if timer(ix(i),iy(i))==0 sitold(ix(i),iy(i))=sitold(ix(i),iy(i))+1; sitnew(ix(i),iy(i))=sitnew(ix(i),iy(i))+1; timer(ix(i),iy(i))=daysis+randi([1 rt]); test=1; sumI=sumI+1; end end end %asymptomatic initial for i=1:asymptomatic test=0; while test<1 rndx1(i)=rand; ix(i)=fix(1+rndx1(i)*LB); rndy1(i)=rand; iy(i)=fix(1+rndy1(i)*LB); if timer(ix(i),iy(i))==0 sitold(ix(i),iy(i))=sitold(ix(i),iy(i))+0.75; sitnew(ix(i),iy(i))=sitnew(ix(i),iy(i))+0.75; timer(ix(i),iy(i))=daysas+randi([1 rt]); test=1; sumA=sumA+1; end end end for i=1:exposed test=0; while test<1 rndx1(i)=rand; ix(i)=fix(1+rndx1(i)*LB); rndy1(i)=rand; iy(i)=fix(1+rndy1(i)*LB); if timer(ix(i),iy(i))==0 sitold(ix(i),iy(i))=sitold(ix(i),iy(i))+0.5; sitnew(ix(i),iy(i))=sitnew(ix(i),iy(i))+0.5; timer(ix(i),iy(i))=randi([1 daysas]); test=1; sumE=sumE+1; end end end for i=1:LB for j=1:LB if sitold(i,j) == 0 sumS=sumS+1; elseif sitold(i,j) == −0.25 sumAR=sumAR+1; elseif sitold(i,j) == −0.5 sumR=sumR+1; end end end I(1)=sumI; A(1)=sumA; E(1)=sumE; S(1)=sumS; AR(1)=sumAR; R(1)=sumR; tiledlayout(1,2) nexttile plot(I,‘-o’); axis square title("I") nexttile clims=[−1 1]; imagesc(sitold,clims); axis square; colorbar; title(1); set(gcf, ‘Position’, [50, 50, 1300, 600]); drawnow F(1) = getframe(gcf); %infections for k=2:n for i=1:LB for j=1:LB if timer(i,j) > 0 && sitold(i,j) == 0.75 rnd(i,j)=rand; if rnd(i,j) >= (1− (bas/4)) if i > 1 && sitold(i−1,j) == 0 sitnew(i−1,j)=0.5; timer(i−1,j)=1; end if j > 1 && sitold(i,j−1) == 0 sitnew(i,j−1)=0.5; timer(i,j−1)=1; end end if rnd(i,j) < (bas/4) if i < LB && sitold(i+1,j) == 0 sitnew(i+1,j)=0.5; timer(i+1,j)=1; end if j < LB && sitold(i,j+1) == 0 sitnew(i,j+1)=0.5; timer(i,j+1)=1; end end end if timer(i,j) > 0 && sitold(i,j) == 1 rnd(i,j)=rand; if rnd(i,j) >= (1− (bis/4)) if i > 1 && sitold(i−1,j) == 0 sitnew(i−1,j)=0.5; timer(i−1,j)=1; end if j > 1 && sitold(i,j−1) == 0 sitnew(i,j−1)=0.5; timer(i,j−1)=1; end end if rnd(i,j) < (bis/4) if i < LB && sitold(i+1,j) == 0 sitnew(i+1,j)=0.5; timer(i+1,j)=1; end if j < LB && sitold(i,j+1) == 0 sitnew(i,j+1)=0.5; timer(i,j+1)=1; end end end end end %count infected and timer/transition sumI=0; sumA=0; sumE=0; sumS=0; sumAR=0; sumR=0; for i=1:LB for j=1:LB if sitnew(i,j) >= cnact if timer(i,j) < daysis + rt timer(i,j)=timer(i,j)+1; else sitnew(i,j)= −0.5; timer(i,j)= −imm; end end if sitnew(i,j) == 0.75 if timer(i,j) < daysas + rt timer(i,j)=timer(i,j)+1; else sitnew(i,j)= −0.25; timer(i,j)= −imm; end end if sitnew(i,j) == 0.5 if timer(i,j) >= daysas sitnew(i,j)=0.75; timer(i,j)=timer(i,j)+1; elseif timer(i,j) == daysis test=rand; if (bis/(bis+bas)) >= test sitnew(i,j)=1; end timer(i,j)=timer(i,j)+1; else timer(i,j)=timer(i,j)+1; end end if timer(i,j) < −1 timer(i,j)=timer(i,j)+1; elseif timer(i,j) == −1 timer(i,j)=timer(i,j)+1; sitnew(i,j)=0; end end end for i=1:LB for j=1:LB if sitnew(i,j) == 0 sumS=sumS+1; elseif sitnew(i,j) == −0.25 sumAR=sumAR+1; elseif sitnew(i,j) == −0.5 sumR=sumR+1; elseif sitnew(i,j)==0.5 sumE=sumE+1; elseif sitnew(i,j)==0.75 sumA=sumA+1; elseif sitnew(i,j)==1 sumI=sumI+1; end end end %density and renew Q(k)=sumI/(L^2); I(k)=sumI; A(k)=sumA; E(k)=sumE; S(k)=sumS; AR(k)=sumAR; R(k)=sumR; for i=1:LB for j=1:LB sitold(i,j)=sitnew(i,j); end end %Graphs tiledlayout(1,2) nexttile plot(I,‘-o’); axis square title("I") nexttile clims=[−1 1]; imagesc(sitold,clims); axis square; colorbar; title(k); set(gcf, ‘Position’, [50, 50, 1300, 600]); drawnow F(k) = getframe(gcf); end FN=strcat(‘covidas3282_’,num2str(SIMS),‘.avi’) video = VideoWriter(FN, ‘Uncompressed AVI’); open(video) writeVideo(video,F) close(video) cell=strcat(‘A’,num2str(SIMS)) writematrix(I,filename,‘Sheet’,"I",‘Range’,cell) writematrix(A,filename,‘Sheet’,"A",‘Range’,cell) writematrix(E,filename,‘Sheet’,"E",‘Range’,cell) writematrix(S,filename,‘Sheet’,"S",‘Range’,cell) writematrix(AR,filename,‘Sheet’,"AR",‘Range’,cell) writematrix(R,filename,‘Sheet’,"R",‘Range’,cell) clear I clear A clear E end |
References
- World Health Organization. Coronavirus Disease (COVID-19). Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (accessed on 30 June 2023).
- Kermack, W.O.; McKendrick, A.G. A contribution to the mathematical theory of epidemics. Proc. R. Soc. Lond. Ser. A 1927, 115, 700–721. [Google Scholar] [CrossRef]
- Lajmanovich, A.; Yorke, J.A. A deterministic model for gonorrhea in a nonhomogeneous population. Math. Biosci. 1976, 28, 221–236. [Google Scholar] [CrossRef]
- Nallaswamy, R.; Shukla, J.B. Effects of dispersal on the stability of a gonorrhea endemic model. Math. Biosci. 1982, 61, 63–72. [Google Scholar] [CrossRef]
- Allen, L.J.; Kirupaharan, N.; Wilson, S.M. SIS epidemic models with multiple pathogen strains. J. Differ. Equ. Appl. 2004, 10, 53–75. [Google Scholar] [CrossRef]
- Xia, Z.Q.; Zhang, J.; Xue, Y.K.; Sun, G.Q.; Jin, Z. Modeling the transmission of Middle East respirator syndrome corona virus in the Republic of Korea. PLoS ONE 2015, 10, e0144778. [Google Scholar] [CrossRef] [PubMed]
- Kevrekidis, P.G.; Cuevas-Maraver, J.; Drossinos, Y.; Rapti, Z.; Kevrekidis, G.A. Reaction-diffusion spatial modeling of COVID-19: Greece and Andalusia as case examples. Physical Rev. E 2021, 104, 024412. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Deolia, P. COVID-19 outbreak: A predictive mathematical study incorporating shedding effect. J. Appl. Math. Comput. 2022, 69, 1239–1268. [Google Scholar] [CrossRef] [PubMed]
- Contoyiannis, Y.; Stavrinides, S.G.P.; Hanias, M.; Kampitakis, M.; Papadopoulos, P.; Picos, R.; Potirakis, M.S. A Universal Physics-Based Model Describing COVID-19 Dynamics in Europe. Int. J. Environ. Res. Public Health 2020, 17, 6525. [Google Scholar] [CrossRef] [PubMed]
- Ramani, A.; Carstea, A.S.; Willox, R.; Grammaticos, B. Oscillating epidemics: A discrete-time model. Physica A 2004, 333, 278–292. [Google Scholar] [CrossRef]
- Kadelka, C. Projecting social contact matrices to populations stratified by binary attributes with known homophily. arXiv 2022, arXiv:2207.12328. [Google Scholar] [CrossRef] [PubMed]
- MATLAB, Version 9.9.0; The MathWorks Inc.: Natick, MA, USA, 2020.
- Greek Legislation Concerning “Emergency Measures to Protect Public Health from the Risk of Further Spread of the COVID-19 Coronavirus”, FEK 1099/11.03.2022, B. Available online: https://www.aade.gr/sites/default/files/2022-03/1099fek.pdf (accessed on 29 November 2023).
- Argyrakis, P.; Kopelman, R.; Lindenberg, K. Diffusion-limited binary reactions: The hierarchy of nonclassical regimes for random initial conditions. Chem. Phys. 1993, 177, 693–707. [Google Scholar] [CrossRef]
- Kroese, D.P.; Brereton, T.J.; Taimre, T.; Botev, Z.I. Why the Monte Carlo method is so important today. Wiley Interdiscip. Rev. Comput. Stat. 2014, 6, 386–392. [Google Scholar] [CrossRef]
- Cuevas, E. An agent-based model to evaluate the COVID-19 transmission risks in facilities. Comput. Biol. Med. 2020, 121, 103827. [Google Scholar] [CrossRef] [PubMed]
- Bonabeau, E. Agent-based modeling: Methods and techniques for simulating human systems. Proc. Natl. Acad. Sci. USA 2002, 99 (Suppl. S3), 7280–7287. [Google Scholar] [CrossRef] [PubMed]










| Parameter | Value Used |
|---|---|
| Recovery time (in days) | 6 |
| Immunity duration (in days) | 90 |
| Transmission rate (per infected) | 0.31 |
| Transmission rate (per asymptomatic) | 0.21 |
| Latent period (E → A) (in days) | 3 |
| Incubation period (E → I) (in days) | 5 |
| 2.65 | |
| 2.84 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zachilas, L.; Benos, C. Modeling and Visualizing the Dynamic Spread of Epidemic Diseases—The COVID-19 Case. AppliedMath 2024, 4, 1-19. https://doi.org/10.3390/appliedmath4010001
Zachilas L, Benos C. Modeling and Visualizing the Dynamic Spread of Epidemic Diseases—The COVID-19 Case. AppliedMath. 2024; 4(1):1-19. https://doi.org/10.3390/appliedmath4010001
Chicago/Turabian StyleZachilas, Loukas, and Christos Benos. 2024. "Modeling and Visualizing the Dynamic Spread of Epidemic Diseases—The COVID-19 Case" AppliedMath 4, no. 1: 1-19. https://doi.org/10.3390/appliedmath4010001
APA StyleZachilas, L., & Benos, C. (2024). Modeling and Visualizing the Dynamic Spread of Epidemic Diseases—The COVID-19 Case. AppliedMath, 4(1), 1-19. https://doi.org/10.3390/appliedmath4010001






