Drug and Protein Interaction Network Construction for Drug Repurposing in Alzheimer’s Disease
Abstract
:1. Introduction
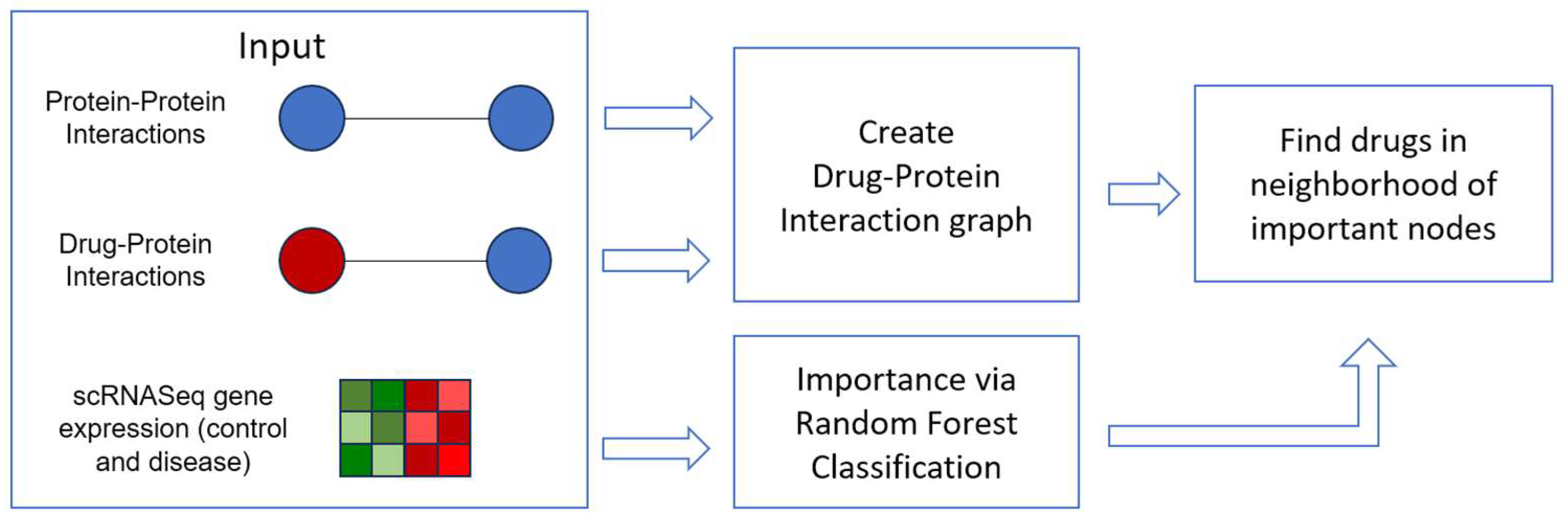
2. Materials and Methods
2.1. Graph Data
2.2. Gene Expression Data
2.3. Drug Repurposing via the Drug-Protein Network
3. Results
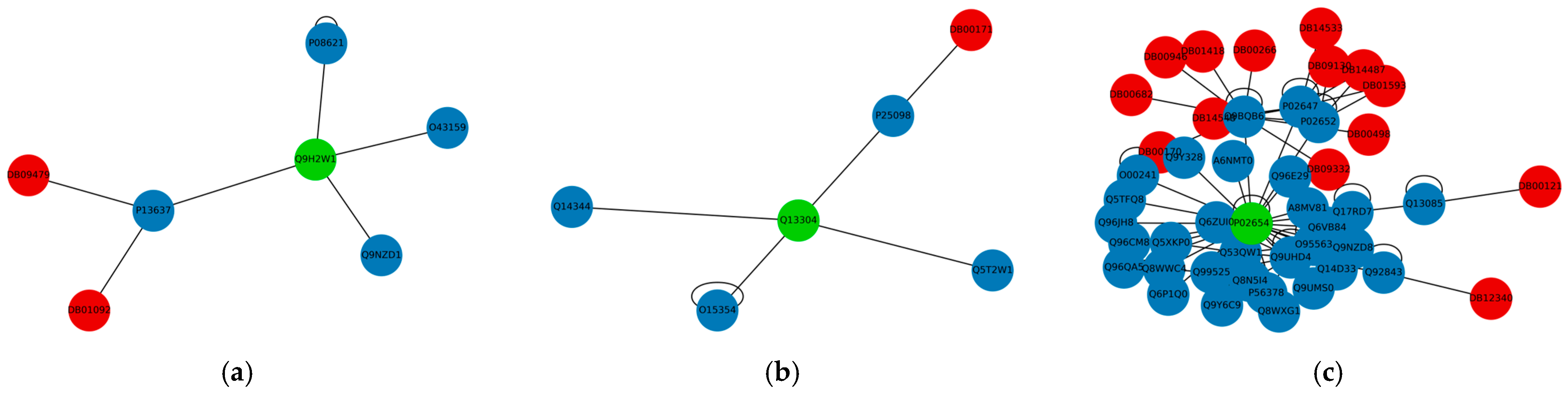
3.1. Important Genes
3.2. Drug Repurposing for AD
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Deng, Y.; Wang, H.; Gu, K.; Song, P. Alzheimer’s disease with frailty: Prevalence, screening, assessment, intervention strategies and challenges. BioSci. Trends 2023, 17, 283–292. [Google Scholar] [CrossRef] [PubMed]
- Tahami Monfared, A.A.; Byrnes, M.J.; White, L.A.; Zhang, Q. The humanistic and economic burden of Alzheimer’s disease. Neurol. Ther. 2022, 11, 525–551. [Google Scholar] [CrossRef] [PubMed]
- Vrahatis, A.G.; Skolariki, K.; Krokidis, M.G.; Lazaros, K.; Exarchos, T.P.; Vlamos, P. Revolutionizing the Early Detection of Alzheimer’s Disease through Non-Invasive Biomarkers: The Role of Artificial Intelligence and Deep Learning. Sensors 2023, 23, 4184. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, S.; Hug, C.; Todorov, P.; Moret, N.; Boswell, S.A.; Evans, K.; Sokolov, A. Machine learning identifies candidates for drug repurposing in Alzheimer’s disease. Nat. Commun. 2021, 12, 1033. [Google Scholar] [CrossRef] [PubMed]
- Somolinos, F.J.; León, C.; Guerrero-Aspizua, S. Drug repurposing using biological networks. Processes 2021, 9, 1057. [Google Scholar] [CrossRef]
- Polamreddy, P.; Gattu, N. The drug repurposing landscape from 2012 to 2017: Evolution, challenges, and possible solutions. Drug Discov. Today 2019, 24, 789–795. [Google Scholar] [CrossRef] [PubMed]
- Gligorijević, V.; Malod-Dognin, N.; Pržulj, N. Integrative methods for analyzing big data in precision medicine. Proteomics 2016, 16, 741–758. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Li, Z.; Chen, G.; Yin, Y.; Chen, C.Y.C. Hybrid neural network approaches to predict drug–target binding affinity for drug repurposing: Screening for potential leads for Alzheimer’s disease. Front. Mol. Biosci. 2023, 10, 1227371. [Google Scholar] [CrossRef]
- Aslanis, I.; Krokidis, M.G.; Dimitrakopoulos, G.N.; Vrahatis, A.G. Identifying Network Biomarkers for Alzheimer’s Disease Using Single-Cell RNA Sequencing Data. In Proceedings of the Worldwide Congress on “Genetics, Geriatrics and Neurodegenerative Diseases Research”, Zakinthos, Greece, 20–22 October 2022; Springer International Publishing: New York, NY, USA, 2022; pp. 207–214. [Google Scholar]
- Ballard, C.; Aarsland, D.; Cummings, J.; O’Brien, J.; Mills, R.; Molinuevo, J.L.; Fladby, T.; Williams, G.; Doherty, P.; Corbett, A.; et al. Drug Repositioning and Repurposing for Alzheimer. Nat. Rev. Neurol. 2020, 16, 661. [Google Scholar] [CrossRef]
- La Manna, S.; Leone, M.; Iacobucci, I.; Annuziata, A.; Di Natale, C.; Lagreca, E.; Malfitano, A.M.; Ruffo, F.; Merlino, A.; Monti, M.; et al. Glucosyl Platinum(II) Complexes Inhibit Aggregation of the C-Terminal Region of the Aβ Peptide. Inorg. Chem. 2022, 61, 3540–3552. [Google Scholar] [CrossRef]
- Florio, D.; Malfitano, A.M.; Di Somma, S.; Mügge, C.; Weigand, W.; Ferraro, G.; Iacobucci, I.; Monti, M.; Morelli, G.; Merlino, A.; et al. Platinum(II) O,S Complexes Inhibit the Aggregation of Amyloid Model Systems. Int. J. Mol. Sci. 2019, 20, 829. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Desai, R.J.; Handy, D.E.; Wang, R.; Schneeweiss, S.; Barabási, A.L.; Loscalzo, J. Network-based approach to prediction and population-based validation of in silico drug repurposing. Nat. Commun. 2018, 9, 2691. [Google Scholar] [CrossRef] [PubMed]
- Dimitrakopoulos, G.N.; Klapa, M.I.; Moschonas, N.K. How far are we from the completion of the human protein interactome reconstruction? Biomolecules 2022, 12, 140. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Wang, C.; Qi, R.; Fu, H.; Ma, Q. scREAD: A single-cell RNA-Seq database for Alzheimer’s disease. Iscience 2020, 23, 11. [Google Scholar] [CrossRef] [PubMed]
- Leng, K.; Li, E.; Eser, R.; Piergies, A.; Sit, R.; Tan, M.; Kampmann, M. Molecular characterization of selectively vulnerable neurons in Alzheimer’s disease. Nat. Neurosci. 2021, 24, 276–287. [Google Scholar] [CrossRef] [PubMed]
- Hie, B.; Bryson, B.; Berger, B. Efficient integration of heterogeneous single-cell transcriptomes using Scanorama. Nat. Biotechnol. 2019, 37, 685–691. [Google Scholar] [CrossRef] [PubMed]
- Wolf, F.A.; Angerer, P.; Theis, F.J. SCANPY: Large-scale single-cell gene expression data analysis. Genome Biol. 2018, 19, 1–5. [Google Scholar] [CrossRef]
- Qi, Y. Random forest for bioinformatics. In Ensemble Machine Learning: Methods and Applications; Springer: Berlin/Heidelberg, Germany, 2012; pp. 307–323. [Google Scholar]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Verkhratsky, A.; Olabarria, M.; Noristani, H.N.; Yeh, C.Y.; Rodriguez, J.J. Astrocytes in Alzheimer’s disease. Neurotherapeutics 2010, 7, 399–412. [Google Scholar] [CrossRef]
- Preman, P.; Alfonso-Triguero, M.; Alberdi, E.; Verkhratsky, A.; Arranz, A.M. Astrocytes in Alzheimer’s disease: Pathological significance and molecular pathways. Cells 2021, 10, 540. [Google Scholar] [CrossRef]
- Jope, R.S.; Mines, M.A.; Beurel, E. Regulation of Cell Survival Mechanisms in Alzheimer’s Disease by Glycogen Synthase Kinase-3. Int. J. Alzheimer’s Dis. 2011, 2011, 861072. [Google Scholar] [CrossRef]
- Duan, K.; Ma, Y.; Tan, J.; Miao, Y.; Zhang, Q. Identification of genetic molecular markers and immune infiltration characteristics of Alzheimer’s disease through weighted gene co-expression network analysis. Front. Neurol. 2022, 13, 947781. [Google Scholar] [CrossRef] [PubMed]
- Bousleiman, J.; Pinsky, A.; Ki, S.; Su, A.; Morozova, I.; Kalachikov, S.; Austin, R.N. Function of metallothionein-3 in neuronal cells: Do metal ions alter expression levels of MT3? Int. J. Mol. Sci. 2017, 18, 1133. [Google Scholar] [CrossRef] [PubMed]
- Nathan, C. The Moving Frontier in Nitric Oxide–Dependent Signaling. Sci. STKE 2004, 2004, pe52. [Google Scholar] [CrossRef] [PubMed]
- Chatziantoniou, A.; Zaravinos, A. Signatures of Co-Deregulated Genes and Their Transcriptional Regulators in Lung Cancer. Int. J. Mol. Sci. 2022, 23, 10933. [Google Scholar] [CrossRef] [PubMed]
- Foster, M.W.; Hess, D.T.; Stamler, J.S. Protein S-Nitrosylation in Health and Disease: A Current Perspective. Trends Mol. Med. 2009, 15, 391. [Google Scholar] [CrossRef] [PubMed]
- Karch, C.M.; Jeng, A.T.; Nowotny, P.; Cady, J.; Cruchaga, C.; Goate, A.M. Expression of novel Alzheimer’s disease risk genes in control and Alzheimer’s disease brains. PLoS ONE 2012, 7, e50976. [Google Scholar] [CrossRef] [PubMed]
- Ciana, P.; Fumagalli, M.; Trincavelli, M.L.; Verderio, C.; Rosa, P.; Lecca, D.; Ferrario, S.; Parravicini, C.; Capra, V.; Gelosa, P.; et al. The Orphan Receptor GPR17 Identified as a New Dual Uracil Nucleotides/Cysteinyl-Leukotrienes Receptor. EMBO J. 2006, 25, 4615. [Google Scholar] [CrossRef]
- Lecca, D.; Trincavelli, M.L.; Gelosa, P.; Sironi, L.; Ciana, P.; Fumagalli, M.; Villa, G.; Verderio, C.; Grumelli, C.; Guerrini, U.; et al. The Recently Identified P2Y-Like Receptor GPR17 Is a Sensor of Brain Damage and a New Target for Brain Repair. PLoS ONE 2008, 3, e3579. [Google Scholar] [CrossRef]
- Jin, S.Y.; Wang, X.; Xiang, X.T.; Wu, Y.M.; Hu, J.; Li, Y.Y.; Lin Dong, Y.; Tan, Y.Q.; Wu, X. Inhibition of GPR17 with Cangrelor Improves Cognitive Impairment and Synaptic Deficits Induced by Aβ1–42 through Nrf2/HO-1 and NF-ΚB Signaling Pathway in Mice. Int. Immunopharmacol. 2021, 101, 108335. [Google Scholar] [CrossRef]
- Zhou, Q.; Zhao, F.; Lv, Z.P.; Zheng, C.G.; Zheng, W.D.; Sun, L.; Yang, Z. Association between APOC1 polymorphism and Alzheimer’s disease: A case-control study and meta-analysis. PLoS ONE 2014, 9, e87017. [Google Scholar] [CrossRef] [PubMed]
- Kulminski, A.M.; Philipp, I.; Shu, L.; Culminskaya, I. Definitive roles of TOMM40-APOE-APOC1 variants in the Alzheimer’s risk. Neurobiol. Aging 2022, 110, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Carecchio, M.; Zorzi, G.; Ragona, F.; Zibordi, F.; Nardocci, N. ATP1A3-Related Disorders: An Update. Eur. J. Paediatr. Neurol. 2018, 22, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Obrenovich, M.E.; Smith, M.A.; Siedlak, S.L.; Chen, S.G.; Torre, J.C.; Perry, G.; Aliev, G. Overexpression of GRK2 in Alzheimer Disease and in a Chronic Hypoperfusion Rat Model Is an Early Marker of Brain Mitochondrial Lesions. Neurotox. Res. 2006, 10, 43–56. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Li, J.; Bao, Z.; Ruan, Q.; Yu, Z. Serum Levels of ApoA1 and ApoA2 Are Associated with Cognitive Status in Older Men. BioMed Res. Int. 2015, 2015, 481621. [Google Scholar] [CrossRef]
- Mur, J.; McCartney, D.L.; Chasman, D.I.; Visscher, P.M.; Muniz-Terrera, G.; Cox, S.R.; Russ, T.C.; Marioni, R.E. Variation in VKORC1 Is Associated with Vascular Dementia. J. Alzheimer’s Dis. 2021, 80, 1329. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Wang, Y.; Ogawa, O.; Lee, H.G.; Raina, A.K.; Siedlak, S.L.; Harris, P.L.R.; Fujioka, H.; Shimohama, S.; Tabaton, M.; et al. Neuroprotective Properties of Bcl-w in Alzheimer Disease. J. Neurochem. 2004, 89, 1233–1240. [Google Scholar] [CrossRef] [PubMed]
- Currais, A.; Huang, L.; Goldberg, J.; Petrascheck, M.; Ates, G.; Pinto-Duarte, A.; Shokhirev, M.N.; Schubert, D.; Maher, P. Elevating Acetyl-CoA Levels Reduces Aspects of Brain Aging. eLife 2019, 8, e47866. [Google Scholar] [CrossRef]
- Li, W.X.; Li, G.H.; Tong, X.; Yang, P.P.; Huang, J.F.; Xu, L.; Dai, S.X. Systematic metabolic analysis of potential target, therapeutic drug, diagnostic method and animal model applicability in three neurodegenerative diseases. Aging 2020, 12, 9882. [Google Scholar] [CrossRef]
- Bunch, T.J.; May, H.; Cutler, M.; Woller, S.C.; Jacobs, V.; Stevens, S.M.; Anderson, J.L. Impact of anticoagulation therapy on the cognitive decline and dementia in patients with non-valvular atrial fibrillation (cognitive decline and dementia in patients with non-valvular atrial fibrillation [CAF] trial). J. Arrhythmia 2022, 38, 997–1008. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, Y.; Lian, X.; Li, F.; Wang, C.; Zhu, F.; Chen, Y. Therapeutic target database update 2022: Facilitating drug discovery with enriched comparative data of targeted agents. Nucleic Acids Res. 2022, 50, D1398–D1407. [Google Scholar] [CrossRef]

| Code | Term | p-Value |
|---|---|---|
| GO:0014002 | astrocyte development | 1.23 × 10−4 |
| GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway | 5.30 × 10−4 |
| GO:0070488 | neutrophil aggregation | 0.0019 |
| GO:0032119 | sequestering of zinc ion | 0.0037 |
| GO:0018119 | peptidyl-cysteine S-nitrosylation | 0.0046 |
| GO:0035425 | autocrine signaling | 0.0065 |
| GO:0002544 | chronic inflammatory response | 0.0092 |
| GO:0002523 | leukocyte migration involved in inflammatory response | 0.0120 |
| GO:0045087 | innate immune response | 0.0191 |
| GO:0002526 | acute inflammatory response | 0.0193 |
| GO:0051493 | regulation of cytoskeleton organization | 0.0229 |
| GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway | 0.0293 |
| GO:0048144 | fibroblast proliferation | 0.0311 |
| GO:0035924 | cellular response to vascular endothelial growth factor stimulus | 0.0311 |
| GO:0050832 | defense response to fungus | 0.0320 |
| Protein | Gene | Importance | Drugs—Direct Interaction | Drugs—Immediate Interaction |
|---|---|---|---|---|
| P06702 | S100A9 | 0.030 | 5 | 103 |
| Q9UBX3 | SLC25A10 | 0.027 | 1 | 8 |
| P33552 | CKS2 | 0.026 | 0 | 202 |
| P02654 | APOC1 | 0.025 | 0 | 14 |
| Q9Y6R7 | FCGBP | 0.022 | 0 | 0 |
| Q9H2W1 | MS4A6A | 0.020 | 0 | 2 |
| O95183 | VAMP5 | 0.020 | 0 | 58 |
| P30408 | TM4SF1 | 0.020 | 0 | 18 |
| P28562 | DUSP1 | 0.020 | 0 | 119 |
| Q9NY25 | CLEC5A | 0.019 | 0 | 0 |
| P13640 | MT1G | 0.018 | 2 | 72 |
| P01920 | HLA-DQB1 | 0.017 | 1 | 10 |
| Q93091 | RNASE6 | 0.017 | 0 | 0 |
| P61952 | GNG11 | 0.017 | 0 | 2 |
| Q13304 | GPR17 | 0.016 | 0 | 1 |
| O60356 | NUPR1 | 0.016 | 0 | 18 |
| P05109 | S100A8 | 0.016 | 6 | 213 |
| P08670 | VIM | 0.014 | 2 | 388 |
| P04792 | HSPB1 | 0.014 | 3 | 538 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dimitrakopoulos, G.N.; Vrahatis, A.G.; Exarchos, T.P.; Krokidis, M.G.; Vlamos, P. Drug and Protein Interaction Network Construction for Drug Repurposing in Alzheimer’s Disease. Future Pharmacol. 2023, 3, 731-741. https://doi.org/10.3390/futurepharmacol3040045
Dimitrakopoulos GN, Vrahatis AG, Exarchos TP, Krokidis MG, Vlamos P. Drug and Protein Interaction Network Construction for Drug Repurposing in Alzheimer’s Disease. Future Pharmacology. 2023; 3(4):731-741. https://doi.org/10.3390/futurepharmacol3040045
Chicago/Turabian StyleDimitrakopoulos, Georgios N., Aristidis G. Vrahatis, Themis P. Exarchos, Marios G. Krokidis, and Panagiotis Vlamos. 2023. "Drug and Protein Interaction Network Construction for Drug Repurposing in Alzheimer’s Disease" Future Pharmacology 3, no. 4: 731-741. https://doi.org/10.3390/futurepharmacol3040045
APA StyleDimitrakopoulos, G. N., Vrahatis, A. G., Exarchos, T. P., Krokidis, M. G., & Vlamos, P. (2023). Drug and Protein Interaction Network Construction for Drug Repurposing in Alzheimer’s Disease. Future Pharmacology, 3(4), 731-741. https://doi.org/10.3390/futurepharmacol3040045










