Abstract
The availability of systematic drug response registries for hundreds cell lines, coupled with the comprehensive profiling of their genomes/transcriptomes enabled the development of computational methods that investigate the molecular basis of drug responsiveness. Herein, we propose an automated, multi-omics systems pharmacology method that identifies genomic markers of anti-cancer drug response. Given a cancer type and a therapeutic compound, the method builds two cell line groups on the antipodes of the drug response spectrum, based on the outer quartiles of the maximum micromolar screening concentration. The method intersects cell lines that share common features in their mutation status, gene expression levels or copy number variants, and a pool of drug response biomarkers (core genes) is built, using genes with mutually exclusive alterations in the two cell line groups. The relevance with the drug target pathways is then quantified, using the combined interaction score of the core genes and an accessory protein network having strong, physical/functional interactions. We demonstrate the applicability and effectiveness of our methodology in three use cases that end up in known drug-gene interactions. The method steps into explainable bioinformatics approaches for novel anticancer drug-gene interactions, offering high accuracy and increased interpretability of the analysis results. Availability: https://github.com/PGxAUTH/PGxGDSC.
1. Introduction
The advances in -omics technologies have led to the creation of high-throughput and high-content resources, as essential tools upon experimentation in cancer research, implementing systems pharmacology and molecular medicine approaches. The molecular profiling of cells incorporates data at various levels of biological knowledge. The proper integration of these structured data to uncover semantic similarities, complex relationships, and patterns of pharmacological, pharmacogenomic and therapeutic relevance is stressfully needed to pave precision medicine therapeutic decisions in cancer therapy and innovative drug development [1,2].
In parallel with the advances been witnessed by the research community in genomic technologies, during the last decades the research in anticancer drug discovery has led to the approval of more than 100 agents by the US Food and Drug Administration (FDA) [3]. These agents have improved the overall patient survival, yet cancer remains the leading cause of deaths, and the burden of cancer incidences and mortality is rapidly increasing worldwide [4]. One of the reasons is that the clinical benefit of anti-cancer agents is limited by the presence of genomic alterations at functionally key loci that block the cascading molecular events triggered by the administered drug and cause adverse drug reactions or aberrant responses. Next-generation sequencing enabled the development of predictive models that incorporate genomic and gene expression data in order to assess the therapeutic potential of anti-cancer drugs [5]. These pharmacogenomic models have proven highly accurate in several cases, implying the prevalence of strong interactions between genomic profiles and drug sensitivity in the pre-clinical setting. However, to exploit this evidence in a clinical environment, we first need to estimate the clinical risk-benefit in standardized pharmacogenomically informed clinical procedures [6]. Additional obstacles, such as the lack of extensive experimental validation and biological reasoning are also limiting factors towards precision medicine. In the absence of wide-ranging in vivo studies, researchers focus on the analysis of large-scale data that provide a public baseline of utmost importance for multi-omics portraits of cancer cell lines in relation with their response to multiple anticancer drugs. The Genomics of Drug Sensitivity in Cancer (GDSC) [7], the Cancer Cell Line Encyclopedia (CCLE) [8] and the Cancer Therapeutics Response Portal (CTRP) [9] are among the most popular resources for drug–cell line interactions. The Connectivity Map (CMap), as part of the LINCS (Library of Integrated Network-based Cellular Signatures) consortium [10], provides comprehensive catalogs of >1.3 million cellular signatures representing systematic perturbation with genetic and pharmacologic traits, and the Cancer Dependency Map is a large-scale project integrating various types of information, such as gene expression, DNA methylation, loss-of-function screens, and drug sensitivity [11].
Combined these public resources enabled the development of analytical methods and predictive models for drug responses that most frequently involve specific drug-gene combinations [12,13,14], while fewer present generalizable computational methodologies for deciphering driver genes in pan-cancer drug resistance at large scale. Recently, Li et al. performed predictions of the IC50 values using cell line transcriptomes concluding in about half of the 473 GDSC drugs whose competence can be reasonably predicted using genetic algorithms [15]. In other pan-cancer studies, machine learning methods predicted drug sensitivity levels using gene expression data [16] and multi-omics data [17]. Tognetti et al. developed single-cell-based models for breast cancer cell lines that accurately predicted drug sensitivity and identified genomic features contributing to such behavior [18]. Kim el al. found kinase mutations in protein substructures that induce different drug responses, using GDSC and COSMIC (The Catalogue Of Somatic Mutations In Cancer) data [19]. A common obstacle in developing end-to-end solutions in pharmacogenomics is that computational methods often overlook the interpretability of the models’ outcome for the sake of improved prediction performance, while methodologies applied on specific compounds, or cancer types, usually fail to generalize, and are difficult to reproduce in similar research hypotheses.
To facilitate the extraction of molecular information associated with drug efficacy in new drug development research efforts and consequently in the clinical setting, an emphasis has been given to develop and integrate bioinformatics approaches, applicable to systems pharmacology and precision medicine. In this work, we propose an automated methodology that relies on public resources to explain the causal relationships between genomic variants, copy number variations and gene expression profiles with drug efficacy over various cancer cell lines and tumor types. The proposed design and implementation rely on the differential analysis of multi-omics data from cell lines characterized by extreme responses to an anticancer therapeutic compound resulting in mutually exclusive genes that are prioritized based on the interaction level with the drug targets. The relevance of each gene candidate with the drug compound is assessed using the core genes and their accessory protein network, i.e., the protein products on a stringent interacting network of the core gene set. Contrary to the drug sensitivity prediction methods, this work aims at deciphering explainable and biologically meaningful associations of the molecular features that drive improved drug responses. Using three use cases, we demonstrate that our method is able to prioritize key markers of known significance in various pairs of therapeutic compounds and cancer types. The method is generalizable to a wide range of cancer types and chemical compounds and offers a straightforward roadmap to detect biomarkers of anticancer drug responsiveness.
2. Materials and Methods
The building blocks and connections of the proposed methodology are shown in Figure 1.
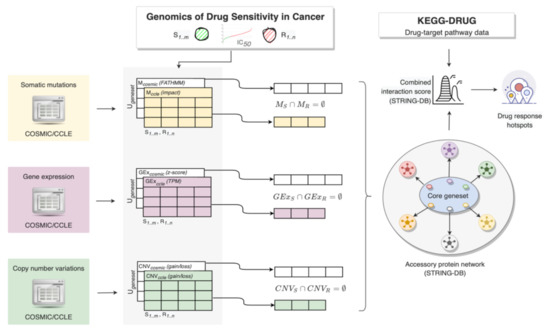
Figure 1.
Schematic representation of the methodology. Given a pair of tumor type and chemical compound, the cancer cell lines with extreme responses are first selected from GDSC1/GDSC2 and the mutation (M), gene expression (GEx) and copy number (CNV) profiles from COSMIC and CCLE are built into gene-level matrices for the sensitive (S) and resistant (R) cell line models. A vector of genes with mutually exclusive alterations per data module and data source (M, GEx and CNV) is built, and then merged into the core gene set containing rigorously defined dysregulated genes that are shared among all S cell lines and are absent in the R cell models, and vice versa. On the periphery on the core gene set, a broader spectrum of protein products (accessory protein network) is built to enclose connected molecules that are known to interact with at least one protein of the core set. The STRING interactions of the core and accessory gene products are assessed against the genes of the drug target pathways (KEGG-DRUG).
2.1. Cancer Cell Line Selection
Cancer cell line and drug screening data were downloaded from GDSC, using phase 1 (GDSC1) and phase 2 (GDSC2) assays, keeping the data from the updated experimental setup (GDSC2), when a drug is screened in both versions (Figure 1) [7]. Given the sensitivity measures expressed in IC50 (half-maximal inhibitory concentration of cell growth) and a research hypothesis involving an anticancer drug compound and a target tumor type, we first build two vectors of length m and n for the cancer-specific sensitive (S) and resistant (R) cell lines, respectively, based on the maximal micromolar concentration (maxCo). The R group is built on the cell line models that belong to the upper quartile of the IC50 > maxCo, while the S group includes the cell lines with IC50 in the lower quartile of the cancer cell lines with IC50 ≤ maxCo. At least three cell lines must fulfill the selection criteria in each group.
2.2. Dinstict Mutation, Gene Expression and Copy Number Profiles
Drug responses are then associated with the genomic and transcriptomic profiles of the S and R cell models, using whole exome screening data and array-based molecular characterization from the COSMIC cell lines project v94 [20] and mutation, gene expression and copy number variation (CNV) data from CCLE [8].
2.2.1. Mutation Profiles
The COSMIC single-point variants and short insertions/deletions at gene level that consistently identified in all m or in all n subgroups of MS or MR matrices, respectively, and those that are mutually exclusive between S and R models are considered of potential significance for drug response. An estimation of the damaging alleles burden is assigned to each mutation, as a measure of its functional impact, using the FATHMM-MKL algorithm [21]. Genes with mutually exclusive alterations in MS and MR with at least one polymorphic site of FATHMM-MKL score > 0.5 are considered pathogenic. For less stringent analyses, the FATHMM criterion may be ignored in the subsequent drug–gene associations. The impact of the CCLE mutations (DepMap, Public 21Q3) [22] is assessed based on the categorical values assigned to each polymorphic site, i.e., the damaging (nonsense, frameshift, splice site, de novo out-of-frame start and SNP/insertion/deletions at start codon), non-conserving (missense, in-frame deletions/insertions), and silent mutations. As for the COSMIC data, the genes with mutually exclusive alterations that have at least one damaging mutation in all S or R models are considered potential gene markers with functional relevance to drug response.
2.2.2. Gene Expression Profiles
Gene expression analysis follows a similar two-step procedure. The normalized gene expression levels from the COSMIC cell line repository are measured by Z-scores. Genes with Z-scores greater than 2 and less than −2 are considered over- and under-expressed, respectively. Over- and under-expressed genes that characterize exclusively all S or all R models meet the selection criteria. The mRNA expression profiles from CCLE are quantified as log2(TPM) (Transcripts per Million) values from RNA-seq data. The expression cutoffs are set to 10 and 0.02 for the over- and under-expressed genes, respectively. Of these genes, only those with average value across group’s cell lines above the 0.95 and below the 0.5 quantile for the over-expressed and under-expressed groups, respectively, are finally selected as candidate expression-based gene markers of drug response given that these are shared among all R or S cancer cell lines. Then, the gene expression matrices of the sensitive GExS and resistant cell lines GExR are built, and those fulfilling the criterion GExS ∩ GExR = Ø for each data source are considered high-confidence factors for drug response.
2.2.3. Copy Number Profiles
Gene level copy number data of about 1000 cancer cell lines were downloaded from COSMIC and CCLE. To define gene amplifications and losses, we followed the definition of the COSMIC annotation. For CCLE, copy number data were log2 transformed with a pseudo count of 1. We used 0.5 as a higher cutoff for copy number loss, and 5 as a lower cutoff for copy number gain. The selection of the gene-level copy number alterations that characterize the sensitive and resistant cell models follow the same steps, as for the mutation and gene expression profiles.
2.3. Interaction Network with Anticancer Drug Targets
The outcome of the three data modules is a set of genes with distinctive mutation, expression or copy number profiles between R and S models. These genes constitute the core set of clinically pertinent markers that are further evaluated for their relevance with the target pathways of the drug compound (KEGG-DRUG), using the STRING combined interaction score [23]. The core genes are accompanied by a high confidence set of accessory proteins that are known to be in strong interaction with the protein products of the core genes (interaction score > 0.9). Three levels of interactions are then defined in decreasing strength: (a) a core gene is part of at least one target pathway of the drug, (b) a protein coded by a core gene has high confidence interactions (interaction score > 0.9) with at least one protein coded by a gene of the drug target pathways, and (c) an accessory protein has high confidence interactions (interaction score > 0.95) with at least one protein coded by a gene of the drug target pathways.
3. Results
We provide use cases for three pairs of drug and cancer types. Afatinib’s responsiveness is investigated against breast cancer cell lines, glioblastoma multiforme cell lines are used to identify trametinib response markers, and dabrafenib is assessed against skin cutaneous melanoma cell lines.
3.1. Breast Cancer and Afatinib
Afatinib (KEGG:D09724) is a second-generation, irreversible tyrosine kinase inhibitor that downregulates the ERBB signaling pathway by covalently bound to epidermal growth factor receptor (EGFR), human EGFRs, HER2, and HER4. It has been initially approved in 2013 as first-line treatment of patients with metastatic non-small cell lung cancer (NSCLC) harboring known driving mutations of EGFR [24,25,26]. Afatinib is further investigated for the treatment of multiple malignancies, e.g., head–neck squamous cell carcinoma (HNSCC) and breast cancer. According to the KEGG-DRUG database, afatinib acts over the MAPK (hsa04010) and ERBB (hsa04012) signaling pathways, and pathways in cancer (hsa05200) [27].
3.1.1. Breast Cancer Cell Lines with Extreme Responses
The list of sensitive and resistant cell lines was selected, using the GDSC2 version of the drug sensitivity data. Four resistant cell lines met the criteria, namely CAL-120, MDA-MB-157, MDA-MB-436 and BT-483 and eight sensitive cell lines (OCUB-M, ZR-75-30, AU565, BT-474, MDA-MB-330, HCC2218, MDA-MB-175-VII, UACC-893). Among the sensitive cell lines, MDA-MB-175-VII was excluded from the COSMIC gene expression analysis and OCUB-M and MDA-MB-330 from the CCLE gene expression analysis, due to the lack of relevant data. CAL-120 mutation profiles were also not available in COSMIC.
3.1.2. High-Confidence Drug-Response Gene Markers
The analysis of each data module resulted in a set of candidate biomarkers that were further evaluated for their relevance with the afatinib molecular targets (Supplementary Table S1). The interaction network analysis filtered out irrelevant or loosely connected proteins and exported two high confidence genes that most likely control drug sensitivity, ERBB2 (HER2) and RAC3 (Figure 2). ERBB2 is a significant gene that is part of the afatinib target pathways and, following the stringent cutoffs of the methodology, is over-expressed in all sensitive cell lines, contrary to the resistant cell lines. Importantly, RAC3 mutations are identified in all resistant cell lines in COSMIC, while all sensitive cell lines lack any type of mutation in this gene. RAC3 is part of the MAPK signaling pathway and other pathways contributing to cancer progression. RAC3 activity is also in strong interaction with the ERBB signaling pathway through four gene members, PAK1, PAK2, PAK4 and SRC (Figure 2). Moreover, three other genes, PRLR, GRB7 and RGL2, are also mutually exclusively over-expressed in all sensitive cell lines; these genes also strongly interact with genes of the ERBB pathway.
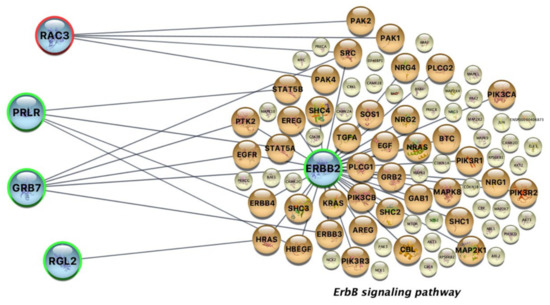
Figure 2.
High confidence pharmacogenes (blue nodes) and their interacting proteins of the afatinib’s target ERBB signaling pathway. Orange nodes are actively involved in the interaction network, and yellow nodes are other target pathway proteins not directly interacting with the high confidence gene products exported by our analysis. ERBB2 is a mutual exclusive gene that is over-expressed in all sensitive cell lines (green contour) and is also a member of the afatinib’s target pathway. PRLR, GRB7 and RGL2 are mutually exclusive genes that are over-expressed in all sensitive cell lines and strongly interact with genes of the ERBB pathway. RAC3 is mutated in all resistant cell lines (red contour) in a mutually exclusive manner and is a strong interactor with proteins of the same pathway.
3.1.3. Interpretability of the Afatinib-Gene Interactions
By assessing preclinical data, the clinical significance of our method in unraveling drug–gene pharmacogenomic correlations in the ERBB signaling pathway was verified in the case of afatinib in breast cancer cell lines. The pharmacogenes and their directly interacting genes of the afatinib’s target ERBB signaling pathway identified in this analysis (Figure 2) correlate well with existing data, showing that overexpression of ERBB2 in cancerous cell lines is positively correlated with increased sensitivity to afatinib treatment [24,25,26]. Similar data for HNSCC, has shown that afatinib’s increased efficacy was correlated with EGFR amplification [28]. Furthermore, the importance of the heparin-binding EGF-like growth factor (HBEGF) that acts through binding to EGFR, ERBB2 and ERBB4 has been revealed in eliciting afatinib’s antimetastatic effects in gastric cell lines [29]. It is also important that the acquired HER2T798I-mediated neratinib resistance upon the tumor progression in a patient harboring an initial activating HER2L869R mutation could be avoided by afatinib [30]. Moreover, Tamura et al. proposed that the in vitro responses to afatinib and dacomitinib in bladder cancer can be pre-clinically evaluated by assessing the sensitivity drug profiles in relevant cell lines [31].
Another crucial issue is that the small GTPase RAC3 gene, found to be mutated in all resistant to afatinib breast cancer cell lines, has been previously shown to control invasion and metastasis in breast cancer, by controlling adhesion and matrix degradation [32]. Moreover, the observation that in all sensitive to afatinib breast cancer cell lines, the prolactin receptor (PRLR) gene has been found to be overexpressed, further validates the obtained findings, since recent evidence supports that the PRLR and EGFR/HER2 signaling crosstalk play a crucial role in the development of mammary glands and breast carcinogenesis, whereas their combined actions may be responsible for endocrine resistance in breast cancer patients resulting in tumor relapse [33]. Similarly, the analysis of TCGA molecular data revealed that the EGFR family MAP kinase adaptor GRB7 gene is co-amplified with ERBB2/HER2 in five different tumor types e.g., GRB7 was amplified in 10 and 20 copies in lung adenocarcinoma and breast cancer, respectively [34]. Furthermore, regarding the third gene Ral Guanine Nucleotide Dissociation Stimulator Like 2 (RGL2) gene that is found to be overexpressed in all the sensitive to afatinib breast cancer cell lines, it has been recently shown that RGL2 modulates β-catenin and KRAS, by promoting colorectal cancer cells metastasis [35]. Among all pharmacogenes targets, SRC is the only gene in the ERBB signaling pathway whose protein product strongly interacts with proteins coded by both resistant and sensitive gene markers (Figure 2). Specifically, the SRC protein is in strong interaction with the protein coded by RAC3, which was found mutated in all resistant cell lines in a mutually exclusive manner, and the proteins coded by ERBB2 and GRB7, which are mutually exclusively over-expressed in all sensitive cell lines. SRC could therefore to play a mediating role in afatinib’s response through a different course of action. Up to now, there has been significant evidence suggesting that SRC is a mediator of resistance to afatinib and other HER-family member inhibitors [36,37,38,39].
3.2. Glioblastoma Multiforme and Trametinib
3.2.1. Glioblastoma Resistant and Sensitive Cancer Cell Lines
Sensitive and resistant cancer cell lines with extreme responses to trametinib (KEGG:D10175) were selected based on the GDSC1 version of the drug sensitivity data. Three resistant cell lines met the criteria (CCF-STTG1, LN-405, AM-38) and six sensitive cell lines (Becker, DK-MG, KS-1, D-263MG, D-566MG and LN-229). Becker, DK-MG were excluded from the COSMIC mutation analysis, due to the lack of relevant data.
3.2.2. High-Confidence Trametinib-Response Gene Markers
For trametinib, there were three mutually exclusive genes (Supplementary Table S2). The copy number loss of CDKN2A was a common alteration in sensitive cell lines, while this alteration does not exist in any resistant cell line. CDKN2A is part of the melanoma pathway (hsa05218) and the pathways in cancer (hsa05200) which, according to the KEGG-DRUG database, are target pathways of trametinib and thus could act as regulators of drug response. In a case study for recurrent/progressive pediatric low-grade glioma, a patient with loss of a copy of CDKN2A gene had responded to trametinib treatment, with 32.4% tumor reduction, being the best response, and his treatment continued for almost 30 months. These results were better than the other patients’ results [40]. Another study used a luciferase-modified cell line from a CDKN2A deficient murine high-grade glioma where trametinib was delivered orally daily. MAPK pathway was suppressed effectively and resulted in a durable anti-growth tumor cell effect, as well as a significant survival benefit of animals [41].
In the expression analysis, overexpression of the FLOT1 gene was found to be a common alteration in the resistant to trametinib cancer cell lines, whereas there was no such alteration in any of the sensitive ones (Figure 3). FLOT1 is not involved in the drug target pathways, but the gene’s protein is in direct interaction with the CDH1 gene (E-cadherin protein), which belongs to the target pathways of trametinib. Pudewell et al. showed that a large number of proteins, called “accessory proteins”, take part in liquid–liquid phase separation [42]. Accessory proteins connect with at least two components to regulate their spatiotemporal localization and enhance their assembly by reducing the dimensionality of interactions and/or increasing local concentrations of interacting proteins. One subset of these are the anchoring proteins, such as the FLOT1 protein. In this case, the PHB domain of FLOT1 protein is found to be membrane-associated, as it is post-translationally modified by palmitoylation and binds lipid rafts of phagosomes and the plasma membrane. Thus, the components of individual signal transduction pathways are linked. Finally, in the mutation analysis, plectin (PLEC) gene was found mutated in all the resistant cell lines, whereas there was no alteration in any of the sensitive ones (Figure 3). Even though the mutation effect is considered neutral, it could be an important biomarker, implying drug resistance. PLEC did not participate in the pathways of cancer, yet plectin is in direct interaction with caspase 8; this finding is in line with previous studies showing that the induction of apoptosis by chemical factors leads to active caspase 8 being translocated to plectin [43].
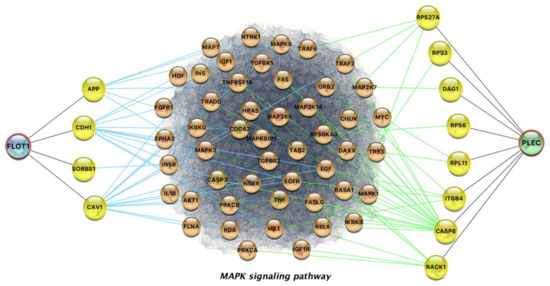
Figure 3.
Network representation of the indirect interactions of FLOT1 and PLEC with proteins involved in the MAPK signaling pathway. Overexpressed FLOT1 and mutant PLEC are found in all resistant glioblastoma multiforme cell lines, contrary to sensitive cell lines.
3.3. Skin Cutaneous Melanoma and Dabrafenib
3.3.1. Dabrafenib’s Sensitive and Resistant Skin Cutaneous Melanoma Cell Lines
Dabrafenib is a BRAF kinase inhibitor that targets pathways in cancer and the MAPK and ERBB signaling pathways. Skin cutaneous melanoma cell lines were selected from the pool of GDSC2 cancer cell lines, following the selection criteria for IC50 extreme drug responses against dabrafenib (KEGG:D10064). Three skin cutaneous melanoma cell lines were found to be resistant to dabrafenib (SK-MEL-2, COLO-792, Mewo) and five sensitive (M14, A375, C32, SH-4, IGR-37). M14 was excluded from the CCLE gene expression analysis due to the lack of relevant data.
3.3.2. High-Confidence Dabrafenib-Response Gene Markers
The analysis resulted in a set of core and accessory genes that may act as potential regulators of dabrafenib response (Table 1, Supplementary Table S3). Fibroblast Growth Factor Receptor 1 (FGFR1) was found mutated exclusively in all resistant cell lines in both CCLE and COSMIC. FGFR1 mutations are considered pathogenic in COSMIC, while CCLE includes missense and silent mutations of FGFR1 in the three resistant cell lines, classified as non-conserving/silent. FGFR and especially FGFR1 is a transmembrane receptor connected with one FGF receptor, whereas FGF/FGFR signaling contributes to intratumoral angiogenesis, melanoma cell survival, and development of resistance to therapeutics. Therefore, inhibitors of aberrant FGF/FGFR signaling are considered as potential anticancer drugs in combination treatment schemes. As a result, it seems that the FGFR1 mutations revealed by our method, that induce the resistance to dabrafenib, are mutations that enhance the FGF/FGFR signaling pathway. The reduced drug sensitivity of melanomas has been associated with increased expression or activities of multi-RTKs, including FGFR1 [44,45]. FGFR1 can limit the antiproliferative effects of vemurafenib, which is accompanied by the reduced ability of vemurafenib to inhibit pMEK. This effect is reversed by the FGFR inhibitor PD173074 [46].

Table 1.
High confidence pharmacogenes that are part of (a), or strongly interact (b) with, the dabrafenib’s target pathways.
In melanoma cells, the RTK expression was reduced due to the strong endogenous BRAF/NRAS mutation-driven activation of the MAPK-ERK signaling pathway, a fact that prevents cell senescence; the opposite happens in cells with activated PTK signaling. Additionally, FGFR1 has been listed among the resistance-associated genes that are temporarily expressed in melanoma cells. Moreover, the FGFR1 over-expression allows melanoma cells to react immediately on ligands that are induced by anticancer drugs, resulting in resistance to the chemotherapeutic treatment [47].
4. Discussion
The pharmacological effects are elicited in the body through the interaction of drug molecules with their targets at the molecular level through a cascade of signaling pathways relevant to the therapeutic outcome, as well as any accompanying adverse drug reactions [48]. Given the existing molecular and cellular metabolism heterogeneity in diseases pathophysiology, e.g., cancer, it is conceivable that we are capable to analyze and clinically translate the relevant molecular information in terms of drug efficacy and safety [49,50]. Especially in oncology, such efforts are urgently needed since molecular traits are driving forces of tumor cell development and progression [51]. The massive load of molecular oncology data being available for analysis, as well as the interdisciplinary nature of the efforts needed to be undertaken, stress out the importance for the development of suitable bioinformatic methods towards pharmacogenomics and precision cancer medicine. To this end, the research approaches aim to provide practicability of the translated molecular knowledge, both at the new drug development era and the clinical setting [1,52,53]. Our work focuses on unraveling drug–gene interactions from pharmacogenomic data to advance systems pharmacology decisions in tumor therapeutics. The implementation of our workflow also provides an efficient manner to extract pharmacologically and clinically relevant knowledge from the generated -omics massive data deposited in biorepositories. Multiscale systems–pharmacology models designed to predict efficacy of novel anti-cancer drugs could be heavily influenced by approaches of this type [54,55]. Gene regulators of anti-cancer drug response identified through our methodology can be used as predictive biomarkers by such models in simulations exploring strategies for increasing the activity of drugs to guide future clinical studies and drive targeted clinical decision making. Consequently, the advancements provided by such systems pharmacology methodologies can be used to improve drug development productivity, as well as drug delivery profiles [1,15,56,57,58,59].
The established cancer cell lines represent a cornerstone in oncology since their application as suitable models mimicking the tumor development and progression mainly support the preclinical new anticancer drug environment. Nowadays, the high-throughput methodologies applied in the molecular analysis further permit the pharmacological exploitation of multi-omics data generated from the large number of cell-based anticancer therapy research and clinical efforts. To this end, hypothesis-driven bioinformatics analysis, capable to implement the methodology in the cell-based pharmacological experimentation, largely reinforce the pre-clinical data productivity, the better clinical translation of the generated data, as well as a more efficient clinical validation for molecular knowledge to enter the clinical setting. The need, however, for better understanding of how accurately the cell models represent the molecular features of patient tumors remains an unsolved task in oncology. It is expected, however, that the application of high-throughput computational approaches and appropriate bioinformatics pipelines will enable the systematic comparisons of the molecular features needed to better understand cell models behavior versus patient tumor types, as well as to identify the druggable potential traits from the current cancer preclinical models [1,2,54,55,60,61]
The pharmacogenomics-guided therapeutic decisions are the goal of precision medicine in oncology, as a means to tailor therapeutics delivery for each patient individually using genomic profiling data. Towards this direction, the pharmacogenomics datasets from cancer cell lines are considered valuable resources for the prediction of drug sensitivity capacity or complementary of drug resistance behavior, the latter representing a crucial therapy task of precision oncology practice [51,57]. Moreover, the massive genomic and transcriptomic screening of cancer cell lines provides a unique opportunity for targeted therapeutic decisions, but also for deciphering the complete picture of a drug’s molecular interactions. In this study, we provide novel insights into drug-gene interactions that exploits our current knowledge of the underlying target pathways and protein–protein interactions, in order to provide the basis for implementing novel experimental research hypotheses. The methodology was tested for specific pairs of cancer types and drugs. Yet, the interaction between cancer drugs and cell line profiles could be studied out of the tissue of origin, enabling deeper investigation of the drug induced molecular reactions. The latter is very important, since the so-called tumor-agnostic therapeutics have already entered the market in oncology therapy, by targeting genetic features regardless of the tissue involved in the tumor development and progression [62,63].
Still, the wide adoption of such methodologies is prone to the non-uniform genomic and transcriptomic profiling of cancer cell lines across different pharmacogenomics platforms and to the poorly described data for other molecular regulators of drug response. Recent advances on variant calling in difficult-to-map regions, such as regions with segmental duplications and in HLA types of the major histocompatibility complex, are promising towards in-depth and explainable pharmacogenomic studies [64]. To this direction, epigenetic modifications are also important regulators of gene expression and drug response. Although the involvement of DNA methylation to drug pharmacological effects remains unclear, there is strong evidence associating cancer treatment resistance in altered DNA methylation profiles [65,66]. Systematic analysis of DNA methylation profiles across cancer cell lines will provide a valuable resource for comprehensive insights of the gene-drug interactions. As a matter of fact, by suitably exploiting the -omics cancer cell line datasets, the landscape of precision cancer medicine is enriched and progressed, implementing the clinical setting to optimize treatments, and thus ensuring efficiency and safety outcomes in tumor patients.
5. Conclusions
The present systems pharmacology methodology is particularly suitable when increased interpretability of the molecular evidence is needed to decipher the underlying paths of drug responsiveness. This is offered by incorporating the information of the molecular pathways of both the chemical compound and the candidate gene markers and by filtering out low impact and poorly discriminated genomic features of cancer cell lines exhibiting extreme responses. The methodology can be used to validate known pharmacogenomic interactions, but also to build novel research hypotheses on currently unknown pharmacogenomic gene-drug associations.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/futurepharmacol2010003/s1, Table S1: Complete list of high-confidence genes with direct and indirect interactions with the afatinib’s target pathways. Table S2: Complete list of high-confidence genes with direct and indirect interactions with the trametinib’s target pathways. Table S3: Complete list of high-confidence genes with direct and indirect interactions with the dabrafenib’s target pathways. Suppl_File_S4: Instructions on how to run the source code available at GitHub.
Author Contributions
Conceptualization, A.M. and I.S.V.; methodology, A.M.; software, K.K.; validation, I.A., E.K. and E.S.; writing—original draft preparation, A.M., I.S.V., K.K., I.A., E.K. and E.S.; supervision, A.M. All authors contributed to preparing the original draft, writing and review of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The methodology is implemented in R and the source code is available at GitHub (https://github.com/PGxAUTH/PGxGDSC).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Vizirianakis, I.S.; Miliotou, A.N.; Mystridis, G.A.; Andriotis, E.G.; Andreadis, I.I.; Papadopoulou, L.C.; Fatouros, D.G. Tackling pharmacological response heterogeneity by PBPK modeling to advance precision medicine productivity of nanotechnology and genomics therapeutics. Expert Rev. Precis. Med. Drug Dev. 2019, 4, 139–151. [Google Scholar] [CrossRef]
- Vizirianakis, I.; Fatouros, D.G. Personalized nanomedicine: Paving the way to the practical clinical utility of genomics and nanotechnology advancements. Adv. Drug Deliv. Rev. 2012, 64, 1359–1362. [Google Scholar] [CrossRef] [PubMed]
- Kinch, M.S. An analysis of FDA-approved drugs for oncology. Drug Discov. Today 2014, 19, 1831–1835. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Russell, L.E.; Zhou, Y.; Almousa, A.A.; Sodhi, J.K.; Nwabufo, C.K.; Lauschke, V.M. Pharmacogenomics in the Era of next Generation Sequencing–from Byte to Bedside. Drug Metab. Rev. 2021, 53, 253–278. [Google Scholar] [CrossRef]
- Pritchard, D.E.; Moeckel, F.; Villa, M.S.; Housman, L.T.; McCarty, C.A.; McLeod, H.L. Strategies for integrating personalized medicine into healthcare practice. Pers. Med. 2017, 14, 141–152. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.; Soares, J.; Greninger, P.; Edelman, E.J.; Lightfoot, H.; Forbes, S.; Bindal, N.; Beare, D.; Smith, J.A.; Thompson, I.R.; et al. Genomics of Drug Sensitivity in Cancer (GDSC): A resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2012, 41, D955–D961. [Google Scholar] [CrossRef] [Green Version]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehár, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef]
- Rees, M.G.; Seashore-Ludlow, B.; Cheah, J.H.; Adams, D.J.; Price, E.V.; Gill, S.; Javaid, S.; Coletti, M.E.; Jones, V.L.; Bodycombe, N.E.; et al. Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat. Chem. Biol. 2016, 12, 109–116. [Google Scholar] [CrossRef]
- Subramanian, A.; Narayan, R.; Corsello, S.M.; Peck, D.D.; Natoli, T.E.; Lu, X.; Gould, J.; Davis, J.F.; Tubelli, A.A.; Asiedu, J.K.; et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 2017, 171, 1437–1452. [Google Scholar] [CrossRef]
- Wang, W.; Malyutina, A.; Pessia, A.; Saarela, J.; Heckman, C.A.; Tang, J. Combined gene essentiality scoring improves the prediction of cancer dependency maps. EBioMedicine 2019, 50, 67–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, P.; Li, H.; Li, S.; Leung, K.-S. Improving prediction of phenotypic drug response on cancer cell lines using deep convolutional network. BMC Bioinform. 2019, 20, 408–414. [Google Scholar] [CrossRef] [PubMed]
- Dorman, S.N.; Baranova, K.; Knoll, J.H.; Urquhart, B.L.; Mariani, G.; Carcangiu, M.L.; Rogan, P.K. Genomic signatures for paclitaxel and gemcitabine resistance in breast cancer derived by machine learning. Mol. Oncol. 2015, 10, 85–100. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Tang, T.; Zhang, B.; Li, G. A Prognostic Signature Based on Immunogenomic Profiling Offers Guidance for Esophageal Squamous Cell Cancer Treatment. Front. Oncol. 2021, 11, 603634. [Google Scholar] [CrossRef]
- Li, Y.; Umbach, D.M.; Krahn, J.M.; Shats, I.; Li, X.; Li, L. Predicting tumor response to drugs based on gene-expression biomarkers of sensitivity learned from cancer cell lines. BMC Genom. 2021, 22, 1–18. [Google Scholar] [CrossRef]
- Sotudian, S.; Paschalidis, I.C. Machine Learning for Pharmacogenomics and Personalized Medicine: A Ranking Model for Drug Sensitivity Prediction. IEEE/ACM Trans. Comput. Biol. Bioinform. 2021, 1. [Google Scholar] [CrossRef]
- Kusch, N.; Schuppert, A. Two-step multi-omics modelling of drug sensitivity in cancer cell lines to identify driving mechanisms. PLoS ONE 2020, 15, e0238961. [Google Scholar] [CrossRef]
- Tognetti, M.; Gabor, A.; Yang, M.; Cappelletti, V.; Windhager, J.; Rueda, O.M.; Charmpi, K.; Esmaeilishirazifard, E.; Bruna, A.; de Souza, N.; et al. Deciphering the signaling network of breast cancer improves drug sensitivity prediction. Cell Syst. 2021, 12, 401–418. [Google Scholar] [CrossRef]
- Kim, P.; Li, H.; Wang, J.; Zhao, Z. Landscape of drug-resistance mutations in kinase regulatory hotspots. Briefings Bioinform. 2021, 22, bbaa108. [Google Scholar] [CrossRef]
- Forbes, S.A.; Beare, D.; Gunasekaran, P.; Leung, K.; Bindal, N.; Boutselakis, H.; Ding, M.; Bamford, S.; Cole, C.; Ward, S.; et al. {COSMIC}: Exploring the World’s Knowledge of Somatic Mutations in Human Cancer. Nucleic Acids Res. 2015, 43, D805–D811. [Google Scholar] [CrossRef]
- A Shihab, H.; Gough, J.; Mort, M.; Cooper, D.N.; Day, I.N.; Gaunt, T.R. Ranking non-synonymous single nucleotide polymorphisms based on disease concepts. Hum. Genom. 2014, 8, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghandi, M.; Huang, F.W.; Jané-Valbuena, J.; Kryukov, G.V.; Lo, C.C.; McDonald, E.R., III; Barretina, J.; Gelfand, E.T.; Bielski, C.M.; Li, H.; et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 2019, 569, 503–508. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dungo, R.T.; Keating, G.M. Afatinib: First Global Approval. Drugs 2013, 73, 1503–1515. [Google Scholar] [CrossRef] [PubMed]
- Wind, S.; Schnell, D.; Ebner, T.; Freiwald, M.; Stopfer, P. Clinical Pharmacokinetics and Pharmacodynamics of Afatinib. Clin. Pharmacokinet. 2017, 56, 235–250. [Google Scholar] [CrossRef] [Green Version]
- Wecker, H.; Waller, C.F. Afatinib. Recent Results in Cancer Research; Martens, U., Ed.; Springer: Cham, Switzerland, 2018; Volume 211, pp. 199–215. [Google Scholar]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [Green Version]
- Young, N.R.; Soneru, C.; Liu, J.; Grushko, T.A.; Hardeman, A.; Olopade, O.I.; Baum, A.; Solca, F.; Cohen, E. Afatinib efficacy against squamous cell carcinoma of the head and neck cell lines in vitro and in vivo. Target. Oncol. 2015, 10, 501–508. [Google Scholar] [CrossRef] [Green Version]
- Ebert, K.; Zwingenberger, G.; Barbaria, E.; Keller, S.; Heck, C.; Arnold, R.; Hollerieth, V.; Mattes, J.; Geffers, R.; Raimúndez, E.; et al. Determining the effects of trastuzumab, cetuximab and afatinib by phosphoprotein, gene expression and phenotypic analysis in gastric cancer cell lines. BMC Cancer 2020, 20, 1–19. [Google Scholar] [CrossRef]
- Hanker, A.; Brewer, M.R.; Sheehan, J.H.; Koch, J.P.; Sliwoski, G.R.; Nagy, R.; Lanman, R.; Berger, M.F.; Hyman, D.M.; Solit, D.B.; et al. An Acquired HER2T798I Gatekeeper Mutation Induces Resistance to Neratinib in a Patient with HER2 Mutant–Driven Breast Cancer. Cancer Discov. 2017, 7, 575–585. [Google Scholar] [CrossRef] [Green Version]
- Tamura, S.; Wang, Y.; Veeneman, B.; Hovelson, D.; Bankhead, A.; Broses, L.J.; Hiles, G.L.; Liebert, M.; Rubin, J.R.; Day, K.C.; et al. Molecular Correlates of In Vitro Responses to Dacomitinib and Afatinib in Bladder Cancer. Bl. Cancer 2018, 4, 77–90. [Google Scholar] [CrossRef] [Green Version]
- Donnelly, S.K.; Cabrera, R.; Mao, S.P.; Christin, J.R.; Wu, B.; Guo, W.; Bravo-Cordero, J.J.; Condeelis, J.S.; Segall, J.E.; Hodgson, L. Rac3 regulates breast cancer invasion and metastasis by controlling adhesion and matrix degradation. J. Cell Biol. 2017, 216, 4331–4349. [Google Scholar] [CrossRef] [PubMed]
- Kavarthapu, R.; Anbazhagan, R.; Dufau, M.L. Crosstalk between PRLR and EGFR/HER2 Signaling Pathways in Breast Cancer. Cancers 2021, 13, 4685. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; McGee, J.; Chen, X.; Doman, T.N.; Gong, X.; Zhang, Y.; Hamm, N.; Ma, X.; Higgs, R.E.; Bhagwat, S.V.; et al. Identification of Druggable Cancer Driver Genes Amplified across TCGA Datasets. PLoS ONE 2014, 9, e98293. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.-S.; Yuan, L.-T.; Kuei, C.-H.; Lin, H.-Y.; Chen, Y.-L.; Chiu, H.-W.; Lin, Y.-F. RGL2 Drives the Metastatic Progression of Colorectal Cancer via Preventing the Protein Degradation of β-Catenin and KRAS. Cancers 2021, 13, 1763. [Google Scholar] [CrossRef]
- Murakami, Y.; Sonoda, K.; Abe, H.; Watari, K.; Kusakabe, D.; Azuma, K.; Kawahara, A.; Akiba, J.; Oneyama, C.; Pachter, J.A.; et al. The activation of SRC family kinases and focal adhesion kinase with the loss of the amplified, mutated EGFR gene contributes to the resistance to afatinib, erlotinib and osimertinib in human lung cancer cells. Oncotarget 2017, 8, 70736–70751. [Google Scholar] [CrossRef] [Green Version]
- Stanley, A.; Ashrafi, G.H.; Seddon, A.M.; Modjtahedi, H. Synergistic effects of various Her inhibitors in combination with IGF-1R, C-MET and Src targeting agents in breast cancer cell lines. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef]
- Canonici, A.; Browne, A.L.; Ibrahim, M.F.K.; Fanning, K.P.; Roche, S.; Conlon, N.T.; O’Neill, F.; Meiller, J.; Cremona, M.; Morgan, C.; et al. Combined targeting EGFR and SRC as a potential novel therapeutic approach for the treatment of triple negative breast cancer. Ther. Adv. Med. Oncol. 2020, 12, 1758835919897546. [Google Scholar] [CrossRef]
- Conlon, N.; Canonici, A.; Morgan, C.; Cremona, M.; Hennessey, B.; Eustace, A.; O’Brien, N.; Slamon, D.; Crown, J.; O’Donovan, N. Targeting Src Kinase Blocks Development of Afatinib Resistance in HER2-Positive Breast Cancer. In Proceedings of the 2017 San Antonio Breast Cancer Symposium, San Antonio, TX; Philadelphia, PA, USA, 5–9 December 2017. [Google Scholar] [CrossRef]
- Manoharan, N.; Choi, J.; Chordas, C.; Zimmerman, M.A.; Scully, J.; Clymer, J.; Filbin, M.; Ullrich, N.J.; Bandopadhayay, P.; Chi, S.N.; et al. Trametinib for the treatment of recurrent/progressive pediatric low-grade glioma. J. Neuro-Oncol. 2020, 149, 253–262. [Google Scholar] [CrossRef]
- Grossauer, S.; Koeck, K.; Murphy, N.E.; Meyers, I.D.; Daynac, M.; Truffaux, N.; Truong, A.; Nicolaides, T.P.; McMahon, M.; Berger, M.S.; et al. Concurrent MEK targeted therapy prevents MAPK pathway reactivation during BRAFV600E targeted inhibition in a novel syngeneic murine glioma model. Oncotarget 2016, 7, 75839–75853. [Google Scholar] [CrossRef] [Green Version]
- Pudewell, S.; Wittich, C.; Jasemi, N.S.K.; Bazgir, F.; Ahmadian, M.R. Accessory proteins of the RAS-MAPK pathway: Moving from the side line to the front line. Commun. Biol. 2021, 4, 1–10. [Google Scholar] [CrossRef]
- Stegh, A.H.; Herrmann, H.; Lampel, S.; Weisenberger, D.; Andrä, K.; Seper, M.; Wiche, G.; Krammer, P.H.; Peter, M.E. Identification of the Cytolinker Plectin as a Major Early In Vivo Substrate for Caspase 8 during CD95- and Tumor Necrosis Factor Receptor-Mediated Apoptosis. Mol. Cell. Biol. 2000, 20, 5665–5679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nazarian, R.; Shi, H.; Wang, Q.; Kong, X.; Koya, R.C.; Lee, H.; Chen, Z.; Lee, M.-K.; Attar, N.; Sazegar, H.; et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature 2010, 468, 973–977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czyz, M. Fibroblast Growth Factor Receptor Signaling in Skin Cancers. Cells 2019, 8, 540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lito, P.; Pratilas, C.A.; Joseph, E.W.; Tadi, M.; Halilovic, E.; Zubrowski, M.; Huang, A.; Wong, W.L.; Callahan, M.K.; Merghoub, T.; et al. Relief of Profound Feedback Inhibition of Mitogenic Signaling by RAF Inhibitors Attenuates Their Activity in BRAFV600E Melanomas. Cancer Cell 2012, 22, 668–682. [Google Scholar] [CrossRef] [Green Version]
- Bowyer, S.; Lee, R.; Fusi, A.; Lorigan, P. Dabrafenib and its use in the treatment of metastatic melanoma. Melanoma Manag. 2015, 2, 199–208. [Google Scholar] [CrossRef]
- Vizirianakis, I.S. Clinical Translation of Genotyping and Haplotyping Data: Implementation of in Vivo Pharmacology Experience Leading Drug Prescription to Pharmacotyping. Clin. Pharmacokinet. 2007, 46, 807–824. [Google Scholar] [CrossRef]
- Pirmohamed, M. Personalized Pharmacogenomics: Predicting Efficacy and Adverse Drug Reactions. Annu. Rev. Genom. Hum. Genet. 2014, 15, 349–370. [Google Scholar] [CrossRef]
- Ahn, K.; Luo, J.; Berg, A.; Keefe, D.; Wu, R. Functional mapping of drug response with pharmacodynamic–pharmacokinetic principles. Trends Pharmacol. Sci. 2010, 31, 306–311. [Google Scholar] [CrossRef]
- Astras, G.; Papagiannopoulos, C.I.; Kyritsis, K.A.; Markitani, C.; Vizirianakis, I. Pharmacogenomic Testing to Guide Personalized Cancer Medicine Decisions in Private Oncology Practice: A Case Study. Front. Oncol. 2020, 10, 521. [Google Scholar] [CrossRef]
- Tafazoli, A.; Guchelaar, H.-J.; Miltyk, W.; Kretowski, A.J.; Swen, J.J. Applying Next-Generation Sequencing Platforms for Pharmacogenomic Testing in Clinical Practice. Front. Pharmacol. 2021, 12, 2025. [Google Scholar] [CrossRef]
- Kyriakidis, K.; Charalampidou, A.; Natsiavas, P.; Vizirianakis, I.S.; Malousi, A. Linking exome sequencing data with drug response aberrations. Stud. Health Technol. Inform. 2019, 264, 1845–1846. [Google Scholar] [CrossRef] [PubMed]
- Kirouac, D.C.; Schaefer, G.; Chan, J.; Merchant, M.; Orr, C.; Huang, S.-M.A.; Moffat, J.; Liu, L.; Gadkar, K.; Ramanujan, S. Clinical responses to ERK inhibition in BRAF V600E-mutant colorectal cancer predicted using a computational model. NPJ Syst. Biol. Appl. 2017, 3, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Albeck, J.G.; Burke, J.M.; Aldridge, B.; Zhang, M.; Lauffenburger, D.A.; Sorger, P.K. Quantitative Analysis of Pathways Controlling Extrinsic Apoptosis in Single Cells. Mol. Cell 2008, 30, 11–25. [Google Scholar] [CrossRef] [Green Version]
- Paananen, J.; Fortino, V. An omics perspective on drug target discovery platforms. Brief. Bioinform. 2020, 21, 1937–1953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharifi-Noghabi, H.; Jahangiri-Tazehkand, S.; Smirnov, P.; Hon, C.; Mammoliti, A.; Nair, S.K.; Mer, A.S.; Ester, M.; Haibe-Kains, B. Drug sensitivity prediction from cell line-based pharmacogenomics data: Guidelines for developing machine learning models. Briefings Bioinform. 2021, 22, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.; Dang, C.C.; Ballester, P.J. Systematic Assessment of Multi-Gene Predictors of Pan-Cancer Cell Line Sensitivity to Drugs Exploiting Gene Expression Data. F1000Research 2017, 5, 2927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lloyd, J.P.; Soellner, M.B.; Merajver, S.D.; Li, J.Z. Impact of between-tissue differences on pan-cancer predictions of drug sensitivity. PLoS Comput. Biol. 2021, 17, e1008720. [Google Scholar] [CrossRef] [PubMed]
- Vizirianakis, I.S.; Chatzopoulou, M.; Bonovolias, I.D.; Nicolaou, I.; Demopoulos, V.J.; Tsiftsoglou, A.S. Toward the Development of Innovative Bifunctional Agents To Induce Differentiation and To Promote Apoptosis in Leukemia: Clinical Candidates and Perspectives. J. Med. Chem. 2010, 53, 6779–6810. [Google Scholar] [CrossRef]
- Noorbakhsh, J.; Vazquez, F.; McFarland, J.M. Bridging the gap between cancer cell line models and tumours using gene expression data. Br. J. Cancer 2021, 125, 311–312. [Google Scholar] [CrossRef]
- Lavacchi, D.; Roviello, G.; D’Angelo, A. Tumor-Agnostic Treatment for Cancer: When How is Better than Where. Clin. Drug Investig. 2020, 40, 519–527. [Google Scholar] [CrossRef]
- Adashek, J.J.; Subbiah, V.; Kurzrock, R. From Tissue-Agnostic to N-of-One Therapies: (R)Evolution of the Precision Paradigm. Trends Cancer 2021, 7, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Olsen, N.D.; Wagner, J.; McDaniel, J.; Stephens, S.H.; Westreich, S.T.; Prasanna, A.; Johanson, E.; Boja, E.; Maier, E.J.; Serang, O.; et al. PrecisionFDA Truth Challenge V2: Calling Variants from Short- and Long-Reads in Difficult-to-Map Regions. Biorxiv 2020, 11, 380741. [Google Scholar]
- Hsu, C.-H.; Tomiyasu, H.; Liao, C.-H.; Lin, C.-S. Genome-wide DNA methylation and RNA-seq analyses identify genes and pathways associated with doxorubicin resistance in a canine diffuse large B-cell lymphoma cell line. PLoS ONE 2021, 16, e0250013. [Google Scholar] [CrossRef] [PubMed]
- Krushkal, J.; Silvers, T.; Reinhold, W.C.; Sonkin, D.; Vural, S.; Connelly, J.; Varma, S.; Meltzer, P.S.; Kunkel, M.; Rapisarda, A.; et al. Epigenome-wide DNA methylation analysis of small cell lung cancer cell lines suggests potential chemotherapy targets. Clin. Epigenetics 2020, 12, 1–28. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).