Fine-Scale Patterns in Bacterial Communities on a Gulf Coast Beach
Abstract
1. Introduction
2. Materials and Methods
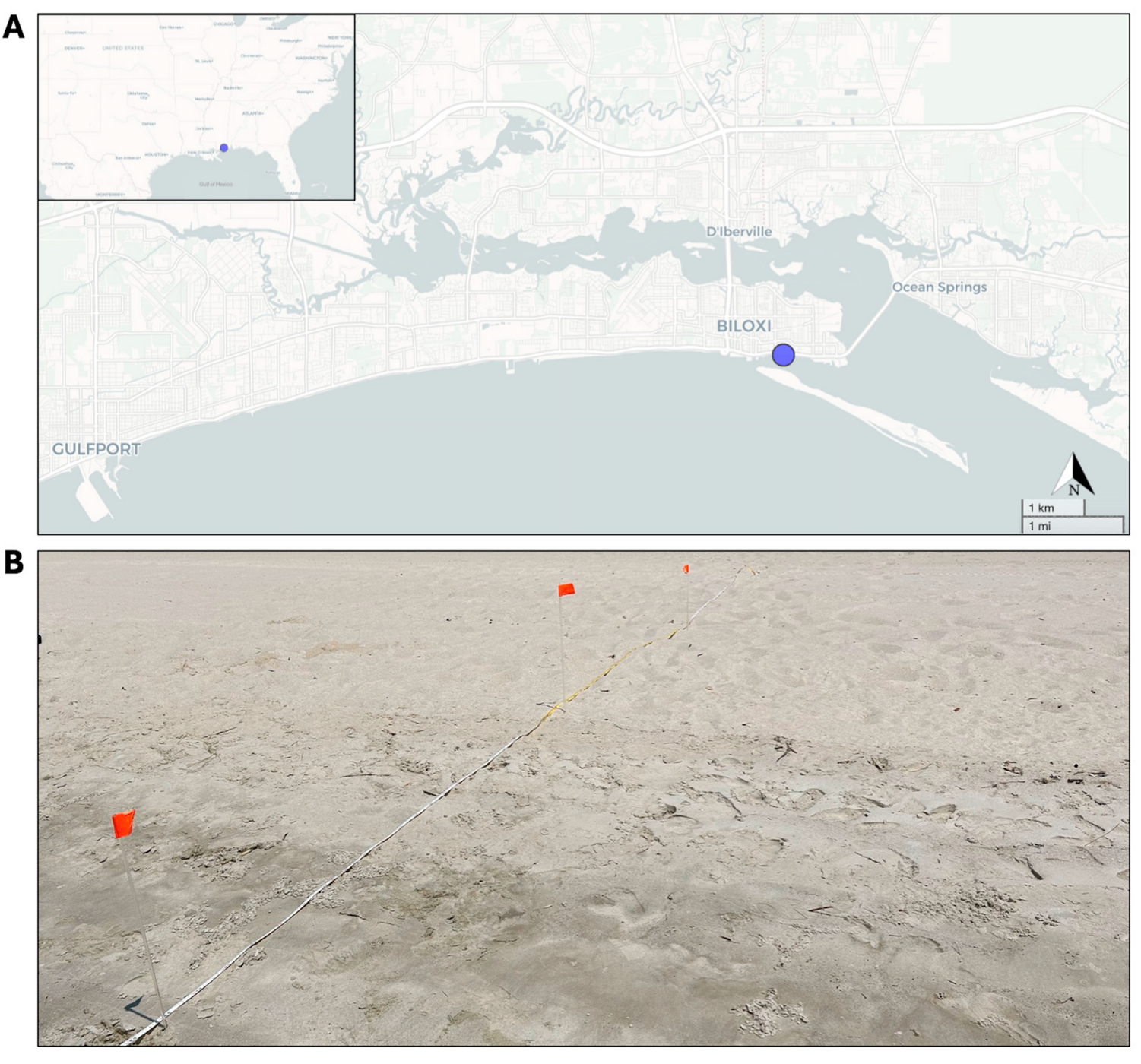
2.1. Sampling and Processing of Beach Sediments
2.2. Assays of Microbial Extracellular Enzyme Activity
2.3. DNA Extraction, Amplification, and Sequencing
2.4. Statistical Analyses
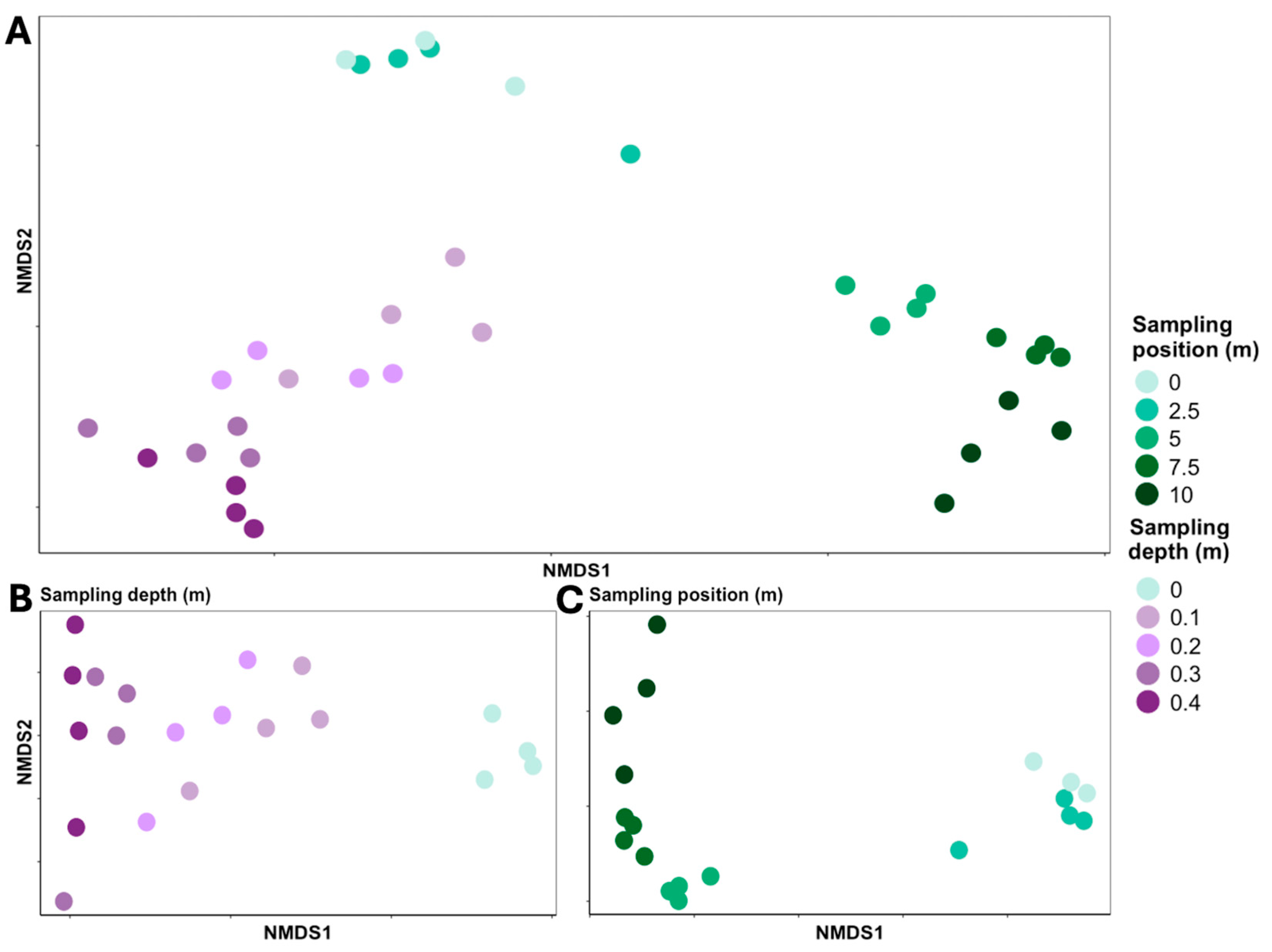
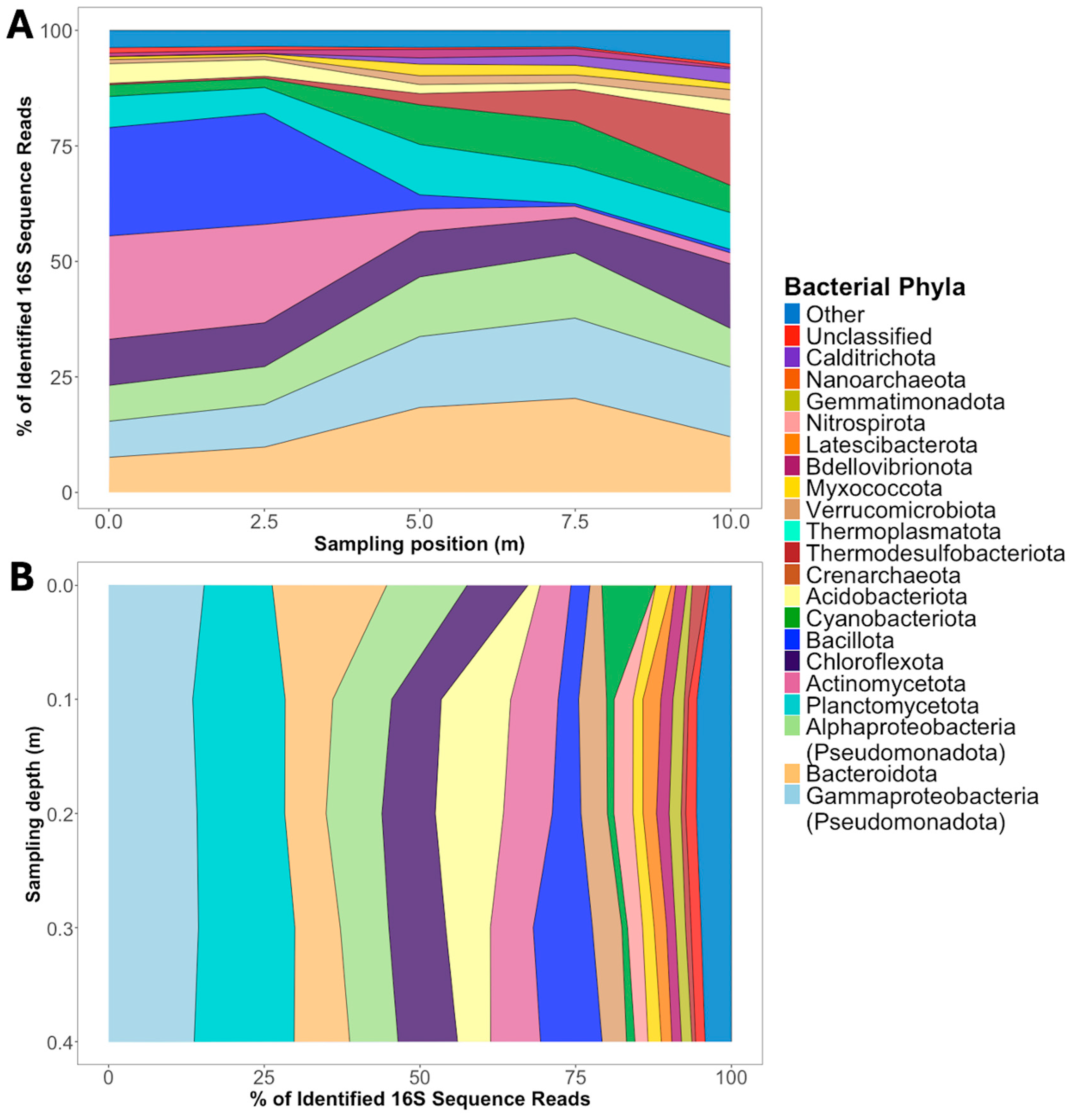
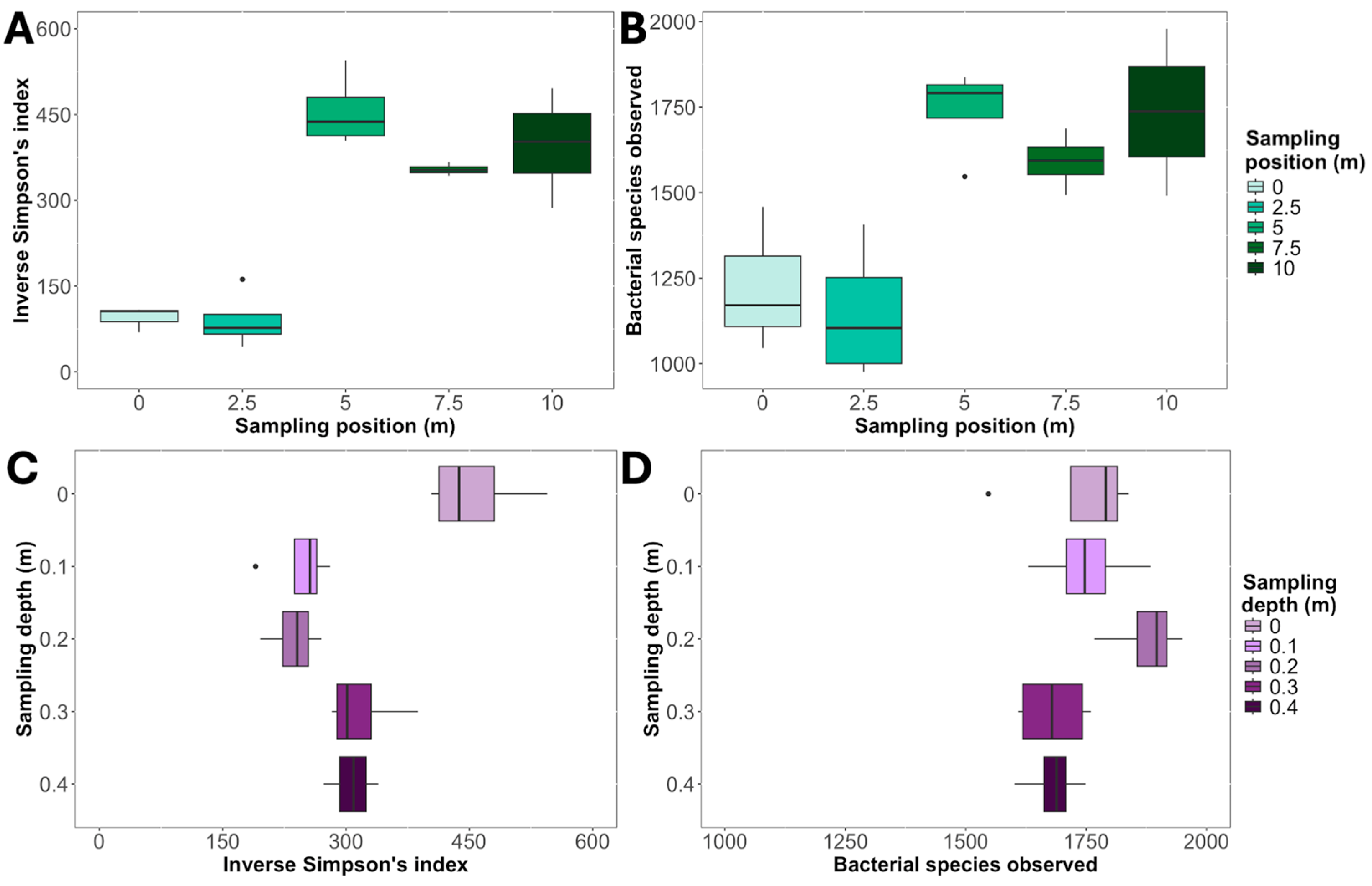
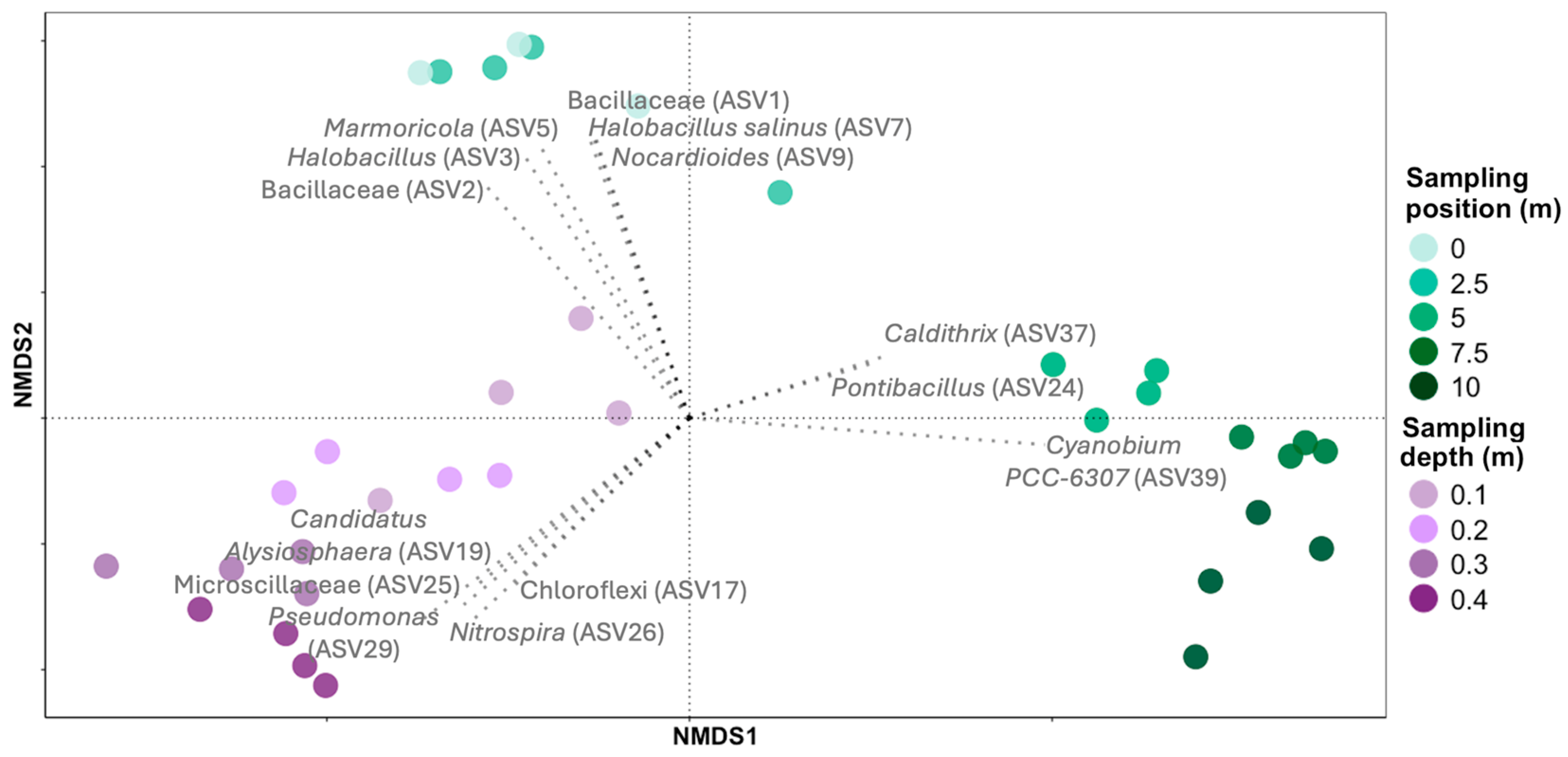
3. Results
4. Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Brick, S.; Niggemann, J.; Reckhardt, A.; Könneke, M.; Engelen, B. Interstitial microbial communities of coastal sediments are dominated by Nanoarchaeota. Front. Microbiol. 2025, 16, 1532193. [Google Scholar] [CrossRef]
- Luijendijk, A.; Hagenaars, G.; Ranasinghe, R.; Baart, F.; Donchyts, G.; Aarninkhof, S. The state of the World’s beaches. Sci. Rep. 2018, 8, 6641. [Google Scholar] [CrossRef]
- McLachlan, A.; Defeo, O. The Ecology of Sandy Shores, 3rd ed.; Academic Press: London, UK, 2017; pp. 1–4. [Google Scholar]
- Whitman, R.L.; Harwood, V.J.; Edge, T.A.; Nevers, M.B.; Byappanahalli, M.; Vijayavel, K.; Brandão, J.; Sadowsky, M.J.; Alm, E.W.; Crowe, A.; et al. Microbes in beach sands: Integrating environment, ecology and public health. Rev. Environ. Sci. Biotechnol. 2014, 13, 329–368. [Google Scholar] [CrossRef]
- Staley, C.; Sadowsky, M.J. Regional similarities and consistent patterns of local variation in beach sand bacterial communities throughout the Northern Hemisphere. Appl. Environ. Microbiol. 2016, 82, 2751–2762. [Google Scholar] [CrossRef] [PubMed]
- Taylor, H.B.; Kurtz, H.D. Microbial community structure shows differing levels of temporal stability in intertidal beach sands of the grand strand region of South Carolina. PLoS ONE 2020, 15, e0229387. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, N.; Keeling, P.J.; Leander, B.S.; Tai, V. Microbial communities in sandy beaches from the three domains of life differ by microhabitat and intertidal location. Mol. Ecol. 2022, 31, 3210–3227. [Google Scholar] [CrossRef]
- Cui, H.; Yang, K.; Pagaling, E.; Yan, T. Spatial and temporal variation in enterococcal abundance and its relationship to the microbial community in Hawaii beach sand and water. Appl. Environ. Microbiol. 2013, 79, 3601–3609. [Google Scholar] [CrossRef]
- Fierer, N.; Schimel, J.P.; Holden, P.A. Variations in microbial community composition through two soil depth profiles. Soil Biol. Biochem. 2003, 35, 167–176. [Google Scholar] [CrossRef]
- Halliday, E.; McLellan, S.L.; Amaral-Zettler, L.A.; Sogin, M.L.; Gast, R.J. Comparison of bacterial communities in sands and water at beaches with bacterial water quality violations. PLoS ONE 2014, 9, e90815. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.R.; Liew, K.C.; Yule, C.M. Structural and functional changes with depth in microbial communities in a tropical Malaysian peat swamp forest. Microb. Ecol. 2019, 57, 402–412. [Google Scholar] [CrossRef]
- Stone, M.M.; DeForest, J.L.; Plante, A.F. Changes in extracellular enzyme activity and microbial community structure with soil depth at the Luquillo Critical Zone Observatory. Soil Biol. Biochem. 2014, 75, 237–247. [Google Scholar] [CrossRef]
- Strom, S.L. Microbial ecology of ocean biogeochemistry: A community perspective. Science 2008, 320, 1043–1045. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, Q.; Chen, B.; Yu, Y.; Wang, T.; Xu, N.; Fan, X.; Penuelas, J.; Fu, Z.; Deng, Y.; et al. Global biogeography of microbes driving ocean ecological status under climate change. Nat. Commun. 2024, 15, 4657. [Google Scholar] [CrossRef]
- Turner, R.E.; Rabalais, N.N. The Gulf of Mexico. In World Seas: An Environmental Evaluation, 2nd ed.; Sheppard, C., Ed.; Academic Press: London, UK, 2019; Volume 1, pp. 445–464. [Google Scholar]
- McKinney, L.D.; Shepherd, J.G.; Wilson, C.A.; Hogarth, W.T.; Chanton, J.; Murawski, S.A.; Sandifer, P.A.; Sutton, T.; Yoskowitz, D.; Wowk, K.; et al. The Gulf of Mexico: An overview. Oceanography 2021, 34, 30–43. [Google Scholar] [CrossRef]
- Michel, J.; Owens, E.H.; Zengel, S.; Graham, A.; Nixon, Z.; Allard, T.; Holton, W.; Reimer, P.D.; Lamarche, A.; White, M.; et al. Extent and degree of shoreline oiling: Deepwater Horizon Oil Spill, Gulf of Mexico, USA. PLoS ONE 2013, 8, e65087. [Google Scholar] [CrossRef] [PubMed]
- Kostka, J.E.; Prakash, O.; Overholt, W.A.; Green, S.J.; Freyer, G.; Canion, A.; Delgardio, J.; Norton, N.; Hazen, T.C.; Huettel, M. Hydrocarbon-degrading bacteria and the bacterial community response in Gulf of Mexico beach sands impacted by the deepwater horizon oil spill. Appl. Environ. Microbiol. 2011, 77, 7962–7974. [Google Scholar] [CrossRef]
- Joye, S.B.; Teske, A.P.; Kostka, J.E. Microbial dynamics following the Macondo oil well blowout across Gulf of Mexico environments. BioScience 2014, 64, 766–777. [Google Scholar] [CrossRef]
- Rodriguez-R, L.M.; Overholt, W.A.; Hagan, C.; Huettel, M.; Kostka, J.E.; Konstantinidis, K.T. Microbial community successional patterns in beach sands impacted by the Deepwater Horizon oil spill. ISME J. 2015, 9, 1928–1940. [Google Scholar] [CrossRef]
- Huettel, M.; Overholt, W.A.; Kostka, J.E.; Hagan, C.; Kaba, J.; Wells, W.B.; Dudley, S. Degradation of Deepwater Horizon oil buried in a Florida beach influenced by tidal pumping. Mar. Pollut. Bull. 2018, 126, 488–500. [Google Scholar] [CrossRef]
- Rietl, A.J.; Jackson, C.R. Effects of the ecological restoration practices of prescribed burning and mechanical thinning on soil microbial enzyme activities and leaf litter decomposition. Soil Biol. Biochem. 2012, 50, 47–57. [Google Scholar] [CrossRef]
- Rietl, A.J.; Overlander, M.E.; Nyman, A.J.; Jackson, C.R. Microbial community composition and extracellular enzyme activities associated with Juncus roemerianus and Spartina alterniflora vegetated sediments in Louisiana saltmarshes. Microb. Ecol. 2016, 71, 290–303. [Google Scholar] [CrossRef]
- Jackson, C.R.; Tyler, H.L.; Millar, J.J. Determination of microbial extracellular enzyme activity in waters, soils, and sediments using high throughput microplate assays. J. Vis. Exp. 2013, 80, 50399. [Google Scholar] [CrossRef]
- Jackson, E.F.; Echlin, H.L.; Jackson, C.R. Changes in the phyllosphere community of the resurrection fern, Polypodium polypodioides, associated with rainfall and wetting. FEMS Microbiol. Ecol. 2006, 58, 236–246. [Google Scholar] [CrossRef]
- Jackson, C.R.; Stone, B.W.; Tyler, H.L. Emerging perspectives on the natural microbiome of fresh produce vegetables. Agriculture 2015, 5, 170–187. [Google Scholar] [CrossRef]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Oren, A.; Garrity, G.M. Valid publication of the names of forty-two phyla of prokaryotes. Int. J. Syst. Evol. Microbiol. 2021, 71, 005056. [Google Scholar] [CrossRef] [PubMed]
- Oren, A.; Mareš, J.; Rippka, R. Validation of the names Cyanobacterium and Cyanobacterium stanieri, and proposal of Cyanobacteriota phyl. nov. Int. J. Syst. Evol. Microbiol. 2022, 72, 005528. [Google Scholar] [CrossRef]
- Oren, A.; Göker, M. Candidatus List. Lists of names of prokaryotic Candidatus phyla. Int. J. Syst. Evol. Microbiol. 2023, 73, 005821. [Google Scholar] [CrossRef] [PubMed]
- Mikryukov, V. metagMisc: Miscellaneous Functions for Metagenomic Analysis. Zenodo. 2017. Available online: https://zenodo.org/ (accessed on 1 May 2025).
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Martinez Arbizu, P. Pairwiseadonis: Pairwise Multilevel Comparison Using Adonis. R Package Version 0.4. 2020. Available online: https://github.com/pmartinezarbizu/pairwiseAdonis (accessed on 1 May 2025).
- Oksanen, J. Vegan: Community Ecology Package; R Package Version 2.6-2. 2022. Available online: https://cran.r-project.org/web/packages/vegan/vegan.pdf (accessed on 1 May 2025).
- Mougi, A. pH adaptation stabilizes bacterial communities. NPJ Biodivers. 2024, 3, 32. [Google Scholar] [CrossRef]
- Dong, Y.; Zhang, J.; Chen, R.; Zhong, L.; Lin, X.; Feng, Y. Microbial community composition and activity in saline soils of coastal agroecosystems. Microorganisms 2022, 10, 835. [Google Scholar] [CrossRef]
- Weedon, J.T.; Bååth, E.; Rijkers, R.; Reischke, S.; Sigurdsson, B.D.; Oddsdottir, E.; van Hal, J.; Aerts, R.; Janssens, I.A.; van Bodegom, P.M. Community adaptation to temperature explains abrupt soil bacterial community shift along a geothermal gradient on Iceland. Soil Biol. Biochem. 2023, 177, 108914. [Google Scholar] [CrossRef]
- Craig, H.; Antwis, R.E.; Cordero, I.; Ashworth, D.; Robinson, C.H.; Osborne, T.Z.; Bardgett, R.D.; Rowntree, J.K.; Simpson, L.T. Nitrogen addition alters composition, diversity, and functioning of microbial communities in mangrove soils: An incubation experiment. Soil Biol. Biochem. 2021, 153, 108076. [Google Scholar] [CrossRef]
- Langenheder, S.; Prosser, J.I. Resource availability influences the diversity of a functional group of heterotrophic soil bacteria. Environ. Microbiol. 2008, 10, 2245–2256. [Google Scholar] [CrossRef] [PubMed]
- Franklin, R.B.; Blum, L.K.; McComb, A.C.; Mills, A.L. A geostatistical analysis of small-scale spatial variability in bacterial abundance and community structure in salt marsh creek bank sediments. FEMS Microbiol. Ecol. 2002, 42, 71–80. [Google Scholar] [CrossRef]
- Sinsabaugh, R.L.; Belnap, J.; Findlay, S.G.; Shah, J.J.F.; Hill, B.H.; Kuehn, K.A.; Kuske, C.R.; Litvak, M.E.; Martinez, N.G.; Moorhead, D.L.; et al. Extracellular enzyme kinetics scale with resource availability. Biogeochemistry 2014, 121, 287–304. [Google Scholar] [CrossRef]
- Dalai, R.C. Soil organic phosphorus. Adv. Agron. 1977, 29, 83–117. [Google Scholar] [CrossRef]
- Sinsabaugh, R.L.; Moorhead, D.L. Resource allocation to extracellular enzyme production: A model for nitrogen and phosphorus control of litter decomposition. Soil Biol. Biochem. 1994, 26, 1305–1311. [Google Scholar] [CrossRef]
- Sinsabaugh, R.L. Phenol oxidase, peroxidase and organic matter dynamics of soil. Soil Biol. Biochem. 2010, 42, 391–404. [Google Scholar] [CrossRef]
- Sinsabaugh, R.L.; Antibus, R.K.; Linkins, A.E.; McClaugherty, C.A.; Rayburn, L.; Repert, D.; Weiland, T. Wood decomposition over a first-order watershed: Mass loss as a function of lignocellulase activity. Soil Biol. Biochem. 1992, 24, 743–749. [Google Scholar] [CrossRef]
- Ljungdahl, L.G.; Eriksson, K.E. Ecology of microbial cellulose degradation. In Advances in Microbial Ecology; Marshall, K.C., Ed.; Springer: Boston, MA, USA, 1985; Volume 8, pp. 237–299. [Google Scholar] [CrossRef]
- Qu, Q.; Wang, Z.; Gan, Q.; Liu, R.; Xu, H. Impact of drought on soil microbial biomass and extracellular enzyme activity. Front. Plant Sci. 2023, 14, 1221288. [Google Scholar] [CrossRef]
- Bogati, K.; Walczak, M. The impact of drought stress on soil microbial community, enzyme activities, and plants. Agronomy 2022, 12, 189. [Google Scholar] [CrossRef]
- Gomez, E.J.; Delgado, J.A.; Gonzalez, J.M. Environmental factors affect the response of microbial extracellular enzyme activity in soils when determined as a function of water availability and temperature. Ecol. Evol. 2020, 10, 10105–10115. [Google Scholar] [CrossRef]
- Hao, J.; Chai, Y.N.; Lopes, L.D.; Ordóñez, R.A.; Wright, E.E.; Archontoulis, S.; Schachtman, D.P. The effects of soil depth on the structure of microbial communities in agricultural soils in Iowa, USA. Appl. Environ. Microbiol. 2021, 87, e02673-20. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Wan, X.; Che, M.; Li, L.; Liu, M. Soil depth exerts greater effect on bacterial community than spatial structure in Longmenshan fault zone. Appl. Environ. Microbiol. 2025, 91, e01161-24. [Google Scholar] [CrossRef] [PubMed]
- Arboleda-Baena, C.; Freilich, M.; Pareja, C.B.; Logares, R.; De la Iglesia, R.; Navarrete, S.A. Microbial community and network responses across strong environmental gradients: How do they compare with macroorganisms? FEMS Microbiol. Ecol. 2024, 100, fiae017. [Google Scholar] [CrossRef]
- Urzì, C.; Salamone, P.; Schumann, P.; Stackebrandt, E. Marmoricola aurantiacus gen. nov., sp. nov., a coccoid member of the family Nocardioidaceae isolated from a marble statue. Int. J. Syst. Evol. Microbiol. 2000, 2, 529–536. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, J.; Liu, Y.; Wang, X.; Zhang, B.; Zhang, W.; Chen, T.; Liu, G.; Xue, L.; Cui, X. Nocardioides: “Specialists” for hard-to-degrade pollutants in the environment. Molecules 2023, 28, 7433. [Google Scholar] [CrossRef]
- Saadouli, I.; Marasco, R.; Mejri, L.; Hamden, H.; Guerfali, M.M.S.; Stathopoulou, P.; Daffonchio, D.; Cherif, A.; Ouzari, H.I.; Tsiamis, G.; et al. Diversity and adaptation properties of actinobacteria associated with Tunisian stone ruins. Front. Microbiol. 2022, 13, 997832. [Google Scholar] [CrossRef]
- Gurung, S.; Lee, C.M.; Weon, H.Y.; Han, S.R.; Oh, T.J. Comparative genome analysis of three Halobacillus strains isolated from saline environments reveal potential salt tolerance and algicidal mechanisms. Environ. Microbiol. Rep. 2025, 17, e70121. [Google Scholar] [CrossRef]
- Felföldi, T.; Somogyi, B.; Márialigeti, K.; Vörös, L. Notes on the biogeography of non-marine planktonic picocyanobacteria: Re-evaluating novelty. J. Plankton Res. 2011, 33, 1622–1626. [Google Scholar] [CrossRef]
- Flombaum, P.; Gallegos, J.L.; Gordillo, R.A.; Rincón, J.; Zabala, L.L.; Jiao, N.; Karl, D.M.; Li, W.K.; Lomas, M.W.; Veneziano, D.; et al. Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. Proc. Natl. Acad. Sci. USA 2013, 110, 9824–9829. [Google Scholar] [CrossRef] [PubMed]
- Callieri, C.; Cabello-Yeves, P.J.; Bertoni, F. The “Dark Side” of Picocyanobacteria: Life as we do not know it (yet). Microorganisms 2022, 10, 546. [Google Scholar] [CrossRef]
- Wang, J.; Wang, J.; Wu, S.; Zhang, Z.; Li, Y. Global geographic diversity and distribution of the myxobacteria. Microbiol. Spectr. 2021, 9, e00012-21. [Google Scholar] [CrossRef] [PubMed]
- Zou, D.; Zhang, C.; Liu, Y.; Li, M. Biogeographical distribution and community assembly of Myxococcota in mangrove sediments. Environ. Microbiome 2024, 19, 47. [Google Scholar] [CrossRef]
- Muñoz-Dorado, J.; Marcos-Torres, F.J.; García-Bravo, E.; Moraleda-Muñoz, A.; Pérez, J. Myxobacteria: Moving, killing, feeding, and surviving together. Front. Microbiol. 2016, 7, 781. [Google Scholar] [CrossRef] [PubMed]
- Fuerst, J.; Sagulenko, E. Beyond the bacterium: Planctomycetes challenge our concepts of microbial structure and function. Nat. Rev. Microbiol. 2011, 9, 403–413. [Google Scholar] [CrossRef]
- Wiegland, S.; Jogler, M.; Jogler, C. On the maverick Planctomycetes. FEMS Microbiol. Rev. 2018, 42, 739–760. [Google Scholar] [CrossRef]
- Torcello-Requena, A.; Murphy, A.R.J.; Lidbury, I.D.E.A.; Pitt, F.D.; Stark, R.; Millard, A.D.; Puxty, R.J.; Chen, Y.; Scanlan, D.J. A distinct, high-affinity, alkaline phosphatase facilitates occupation of P-depleted environments by marine picocyanobacteria. Proc. Natl. Acad. Sci. USA 2024, 121, e2312892121. [Google Scholar] [CrossRef] [PubMed]
- Palanisami, S.; Saha, S.K.; Lakshmanan, U. Laccase and polyphenol oxidase activities of marine cyanobacteria: A study with Poly R-478 decolorization. World J. Microbiol. Biotechnol. 2010, 26, 63–69. [Google Scholar] [CrossRef]
- Behl, K.; Devi, A.; Yadav, Y.; Jaiswal, P. Chapter 14-Cyanobacteria: A precious resource for bioremediation. In Cyanobacteria: Metabolisms to Molecules; Mishra, A.K., Singh, S.S., Eds.; Academic Press: Boston, MA, USA, 2023; pp. 341–382. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Basha, E.; Vaughn, S.N.; Pavlovsky, J.C.; Roth, H.; Jackson, C.R. Fine-Scale Patterns in Bacterial Communities on a Gulf Coast Beach. Coasts 2025, 5, 34. https://doi.org/10.3390/coasts5030034
Basha E, Vaughn SN, Pavlovsky JC, Roth H, Jackson CR. Fine-Scale Patterns in Bacterial Communities on a Gulf Coast Beach. Coasts. 2025; 5(3):34. https://doi.org/10.3390/coasts5030034
Chicago/Turabian StyleBasha, Elizabeth, Stephanie N. Vaughn, Jacqueline C. Pavlovsky, Hays Roth, and Colin R. Jackson. 2025. "Fine-Scale Patterns in Bacterial Communities on a Gulf Coast Beach" Coasts 5, no. 3: 34. https://doi.org/10.3390/coasts5030034
APA StyleBasha, E., Vaughn, S. N., Pavlovsky, J. C., Roth, H., & Jackson, C. R. (2025). Fine-Scale Patterns in Bacterial Communities on a Gulf Coast Beach. Coasts, 5(3), 34. https://doi.org/10.3390/coasts5030034





