Insights into Ecological Features of Microbial Dark Matter Within the Symbiotic Community During Alexandrium pacificum Bloom: Co-Occurrence Interactions and Assembly Processes
Abstract
1. Introduction
2. Materials and Methods
2.1. Dinoflagellate Strain, Cultivation and Sampling
2.2. DNA Extraction and Data Processing
2.3. Bioinformatics
3. Results
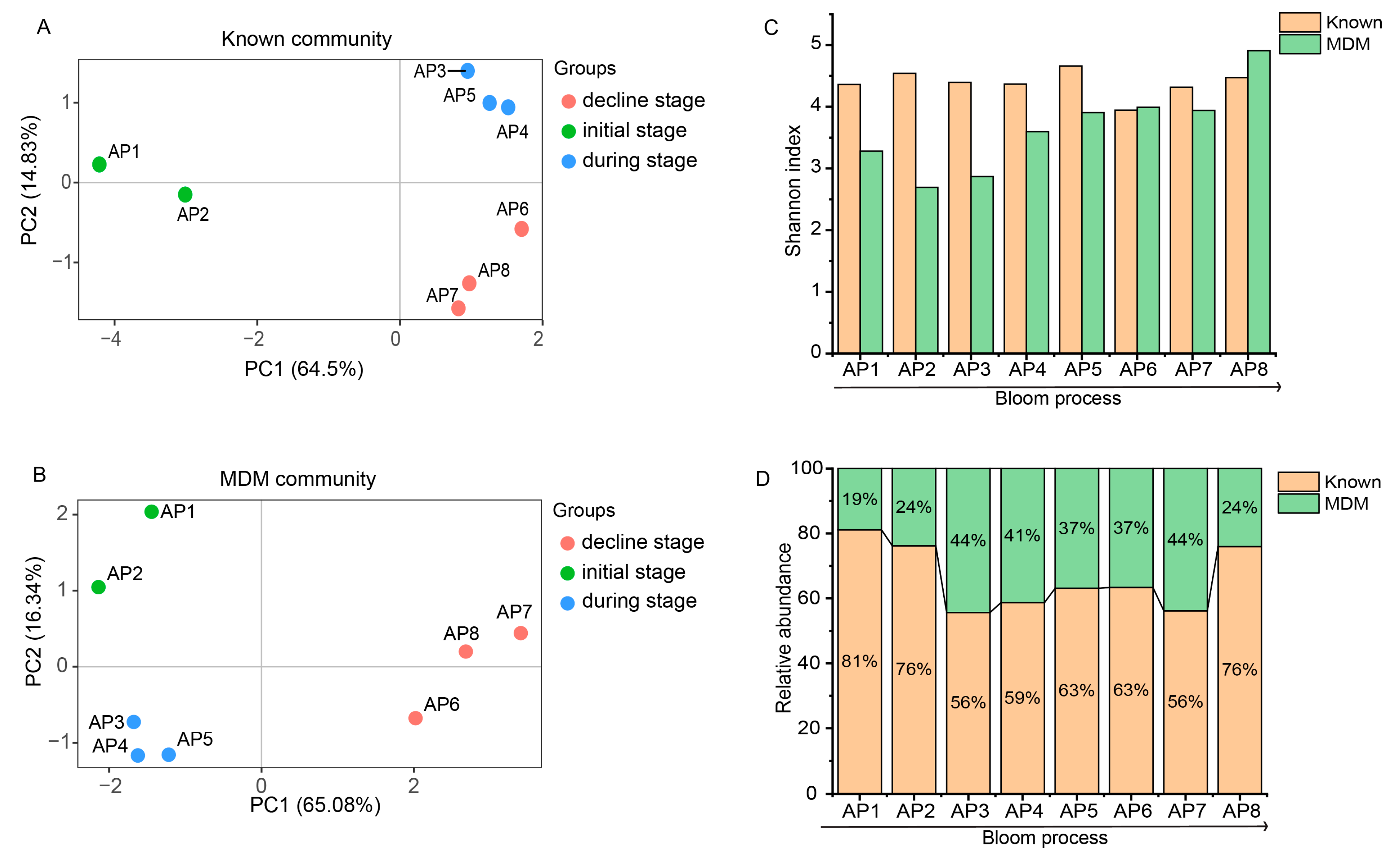
3.1. Diversity and Composition of Known Taxa and MDM
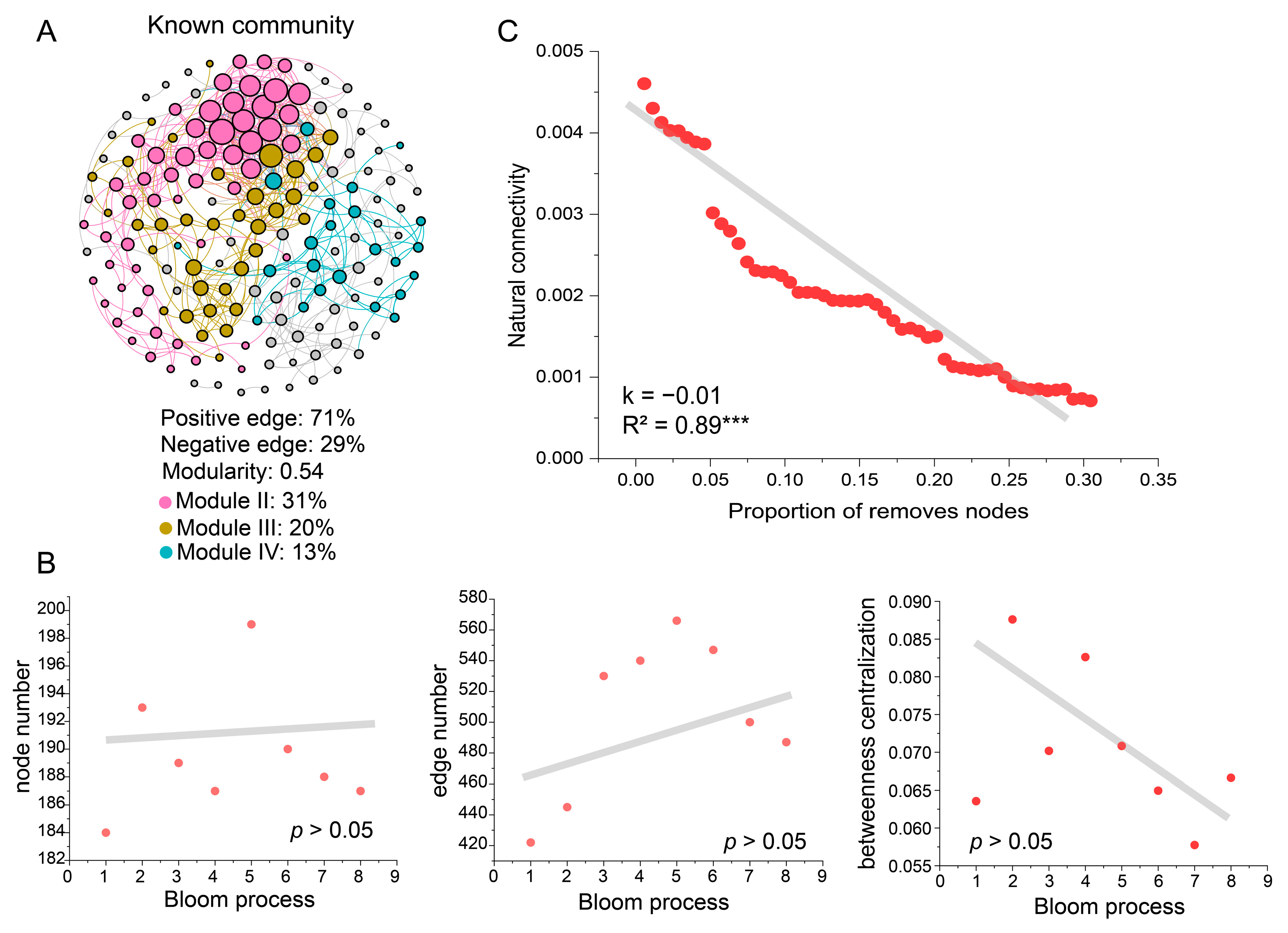
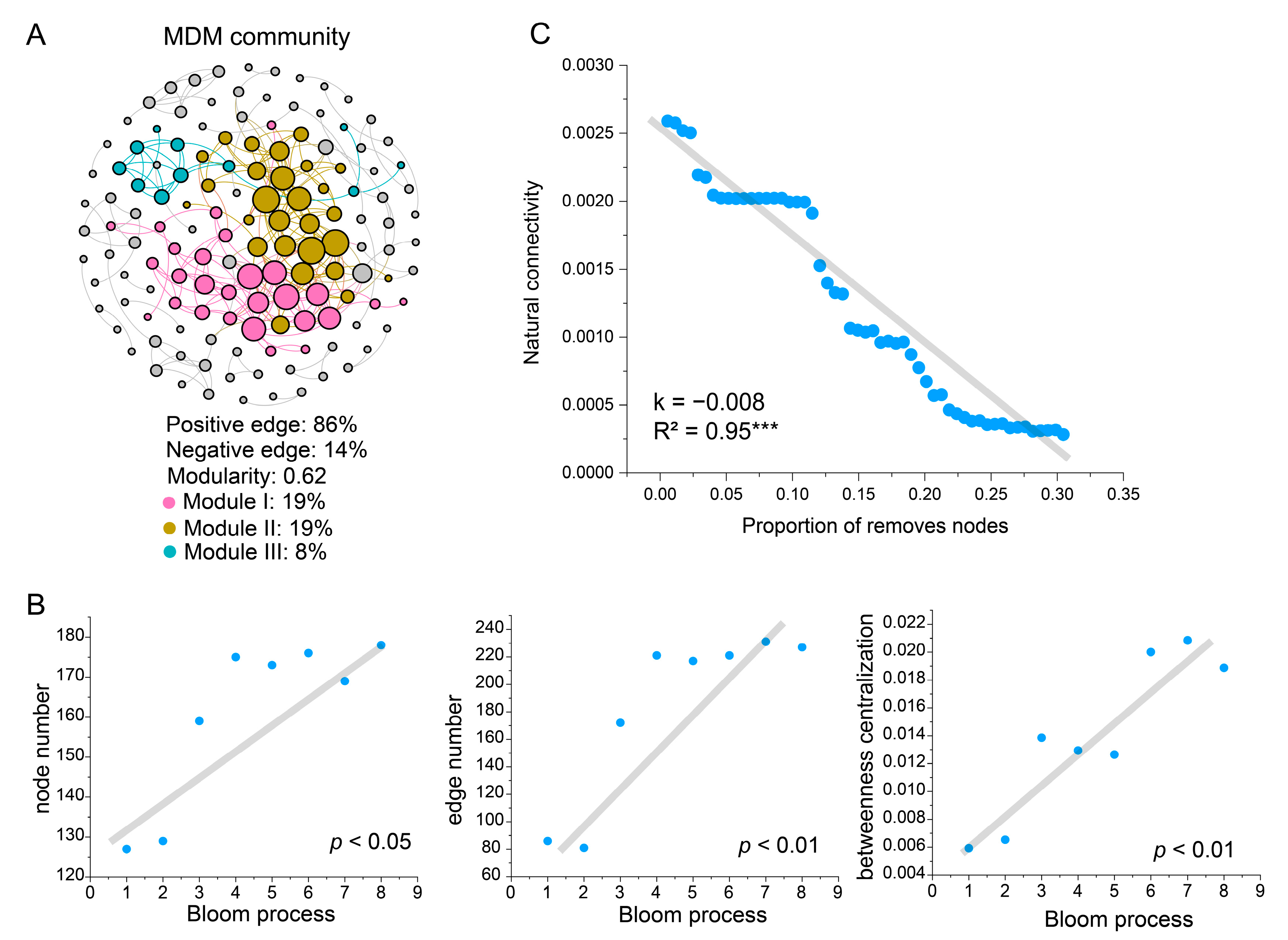
3.2. Ecological Networks of Known Taxa and MDM
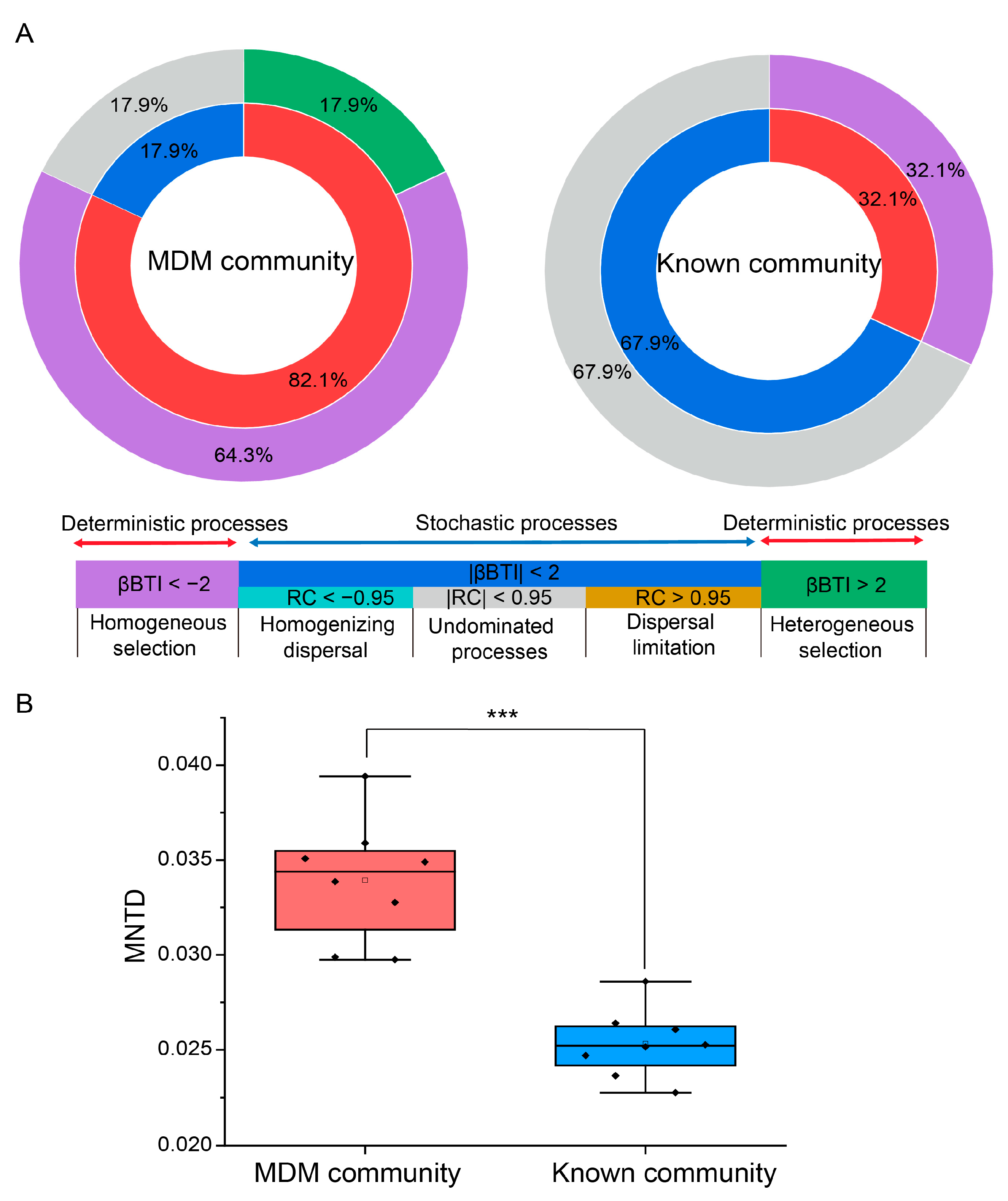
3.3. Assembly Process of Known Taxa and MDM
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MDM | Microbial dark matter |
| MNTD | Mean nearest taxon distance |
References
- Falkowski, P.G.; Fenchel, T.; Delong, E.F. The microbial engines that drive Earth’s biogeochemical cycles. Science 2008, 320, 1034–1039. [Google Scholar] [CrossRef]
- Lloyd, K.G.; Steen, A.D.; Ladau, J.; Yin, J.; Crosby, L. Phylogenetically novel uncultured microbial cells dominate earth microbiomes. MSystems 2018, 3, 10–1128. [Google Scholar] [CrossRef]
- Marcy, Y.; Ouverney, C.; Bik, E.M.; Lösekann, T.; Ivanova, N.; Martin, H.G.; Szeto, E.; Platt, D.; Hugenholtz, P.; Relman, D.A.; et al. Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated tm7 microbes from the human mouth. Proc. Natl. Acad. Sci. USA 2007, 104, 11889–11894. [Google Scholar] [CrossRef] [PubMed]
- Rinke, C.; Schwientek, P.; Sczyrba, A.; Ivanova, N.N.; Anderson, I.J.; Cheng, J.-F.; Darling, A.; Malfatti, S.; Swan, B.K.; Gies, E.A.; et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature 2013, 499, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Zamkovaya, T.; Foster, J.S.; de Crécy-Lagard, V.; Conesa, A. A network approach to elucidate and prioritize microbial dark matter in microbial communities. ISME J. 2021, 15, 228–244. [Google Scholar] [CrossRef]
- Anderson, D.M.; Fensin, E.; Gobler, C.J.; Hoeglund, A.E.; Hubbard, K.A.; Kulis, D.M.; Landsberg, J.H.; Lefebvre, K.A.; Provoost, P.; Richlen, M.L.; et al. Marine harmful algal blooms (HABs) in the United States: History, current status and future trends. Harmful Algae 2021, 102, 101975. [Google Scholar] [CrossRef]
- Wong, B.Y.K.; Chen, Y.H.; Cui, K.H.; Zhou, H.C.; Li, F.L.; Tam, N.F.Y.; Lee, F.W.F.; Xu, S.J.L. Differential allelopathic effects of mangrove plants Kandelia obovata and Aegiceras corniculatum on harmful algal species: Potential applications in algal bloom control. Mar. Pollut. Bull. 2024, 207, 116874. [Google Scholar] [CrossRef]
- Liu, Y.; Hu, Z.; Deng, Y.; Shang, L.; Gobler, C.J.; Tang, Y.Z. Dependence of genome size and copy number of rRNA gene on cell volume in dinoflagellates. Harmful Algae 2021, 109, 102108. [Google Scholar] [CrossRef]
- Zhou, J.; Richlen, M.L.; Sehein, T.R.; Kulis, D.M.; Anderson, D.M.; Cai, Z. Microbial community structure and associations during a marine dinoflagellate bloom. Front. Microbiol. 2018, 9, 1201. [Google Scholar] [CrossRef]
- Zhou, J.; Chen, G.-F.; Ying, K.Z.; Jin, H.; Song, J.-T.; Cai, Z.H. Phycosphere microbial succession patterns and assembly mechanisms in a marine dinoflagellate bloom. Appl. Environ. Microbiol. 2019, 85, e00349-19. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, B.Y.; Yu, K.; Du, X.P.; Zhu, J.M.; Zeng, Y.H.; Cai, Z.H. Functional profiles of phycospheric microorganisms during a marine dinoflagellate bloom. Water Res. 2020, 173, 115554. [Google Scholar] [CrossRef]
- Deng, Y.; Wang, K.; Hu, Z.; Hu, Q.; Tang, Y.Z. Identification and implications of a core bacterial microbiome in 19 clonal cultures laboratory-reared for months to years of the cosmopolitan dinoflagellate Karlodinium veneficum. Front. Microbiol. 2022, 13, 967610. [Google Scholar] [CrossRef]
- Deng, Y.; Wang, K.; Hu, Z.; Hu, Q.; Tang, Y.Z. Toxic and non-toxic dinoflagellates host distinct bacterial communities in their phycospheres. Commun. Earth Environ. Microbiol. 2023, 4, 263. [Google Scholar] [CrossRef]
- Lawson, C.A.; Raina, J.B.; Kahlke, T.; Seymour, J.R.; Suggett, D.J. Defining the core microbiome of the symbiotic dinoflagellate, Symbiodinium. Environ. Microbiol. Rep. 2018, 10, 7–11. [Google Scholar] [CrossRef]
- Maire, J.; Girvan, S.K.; Barkla, S.E.; Perez-Gonzalez, A.; Suggett, D.J.; Blackall, L.L.; van Oppen, M.J.H. Intracellular bacteria are common and taxonomically diverse in cultured and in hospite algal endosymbionts of coral reefs. ISME J. 2021, 15, 2028–2042. [Google Scholar] [CrossRef] [PubMed]
- Behringer, G.; Ochsenkühn, M.A.; Fei, C.; Fanning, J.; Koester, J.A.; Amin, S.A. Bacterial communities of diatoms display strong conservation across strains and time. Front. Microbiol. 2018, 9, 659. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.W.; Kang, J.; Park, J.S.; Joo, H.M.; Suh, S.S.; Kang, D.; Lee, T.K.; Kim, H.J. Dynamic bacterial community response to Akashiwo sanguinea (Dinophyceae) bloom in indoor marine microcosms. Sci. Rep. 2021, 11, 6983. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.K.; Chun, S.J.; Kim, J.H.; Park, B.S.; Baek, S.H. Short-term response of pelagic planktonic communities after inoculation with the mass cultured dinoflagellate Alexandrium affine in a large-scale mesocosm experiment. J. Appl. Phycol. 2021, 33, 3123–3137. [Google Scholar] [CrossRef]
- Röttjers, L.; Faust, K. From hairballs to hypotheses-biological insights from microbial networks. FEMS Microbiol. Rev. 2018, 42, 761–780. [Google Scholar] [CrossRef]
- Wuchty, S.; Ravasz, E.; Barabási, A.L. The architecture of biological networks. In Complex Systems Science in Biomedicine; Deisboeck, T.S., Kresh, J.Y., Eds.; Springer: Boston, MA, USA, 2006; pp. 165–181. [Google Scholar]
- Zhou, J.; Ning, D. Stochastic Community Assembly: Does It Matter in Microbial Ecology? Microbiol. Mol. Biol. Rev. 2017, 81, e00002-17. [Google Scholar] [CrossRef]
- Zhang, H.; Hou, F.; Xie, W.; Wang, K.; Zhou, X.; Zhang, D.; Zhu, X. Interaction and assembly processes of abundant and rare microbial communities during a diatom bloom process. Environ. Microbiol. 2020, 22, 1707–1719. [Google Scholar] [CrossRef] [PubMed]
- Gu, H.; Zeng, N.; Liu, T.; Yang, W.; Müller, A.; Krock, B. Morphology, toxicity, and phylogeny of Alexandrium (Dinophyceae) species along the coast of China. Harmful Algae 2013, 27, 68–81. [Google Scholar] [CrossRef]
- Guillard, R.R.L. Culture of phytoplankton for feeding marine invertebrates. In Culture of Marine Invertebrate Animals; Smith, W.L., Chanley, M.H., Eds.; Plenum Press: New York, NY, USA, 1975; pp. 22–60. [Google Scholar]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Yan, X.; Al, M.A.; Ren, K.; Rensing, C.; Hu, A.; Tsyganov, A.N.; Mazei, Y.; Smirnov, A.; Mazei, N.; et al. Ecological and evolutionary processes involved in sharping microbial habitat generalists and specialists in urban park ecosystems. MSystems 2024, 9, e00469-24. [Google Scholar] [CrossRef]
- Deng, Y.; Jiang, Y.-H.; Yang, Y.; He, Z.; Luo, F.; Zhou, J. Molecular ecological network analyses. BMC Bioinform. 2012, 13, 113. [Google Scholar] [CrossRef]
- Wan, W.; Gadd, G.; Yang, Y.; Yuan, W.; Gu, J.; Ye, L.; Liu, W. Environmental adaptation is stronger for abundant rather than rare microorganisms in wetland soils from the Qinghai-Tibet Plateau. Mol. Ecol. 2021, 30, 2390–2403. [Google Scholar] [CrossRef]
- Liberati, D.; Guidolotti, G.; de Dato, G.; De Angelis, P. Enhancement of ecosystem carbon uptake in a dry shrubland under moderate warming: The role of nitrogen-driven changes in plant morphology. Glob. Change Biol. 2021, 27, 5629–5642. [Google Scholar] [CrossRef]
- Sun, J.; Chang, E.B. Exploring gut microbes in human health and disease: Pushing the envelope. Genes Dis. 2014, 1, 132–139. [Google Scholar] [CrossRef]
- Brisbin, M.M.; Mitarai, S.; Saito, M.A.; Alexander, H. Microbiomes of bloom-forming Phaeocystis algae are stable and consistently recruited, with both symbiotic and opportunistic modes. ISME J. 2022, 16, 2255–2264. [Google Scholar] [CrossRef]
- Trombetta, T.; Vidussi, F.; Roques, C.; Scotti, M.; Mostajir, B. Marine microbial food web networks during phytoplankton bloom and non-bloom periods: Warming favors smaller organism interactions and intensifies trophic cascade. Front. Microbiol. 2020, 11, 502336. [Google Scholar] [CrossRef]
- Myklestad, S.M. Dissolved organic carbon from phytoplankton. In Marine Chemistry; Springer: Boston, MA, USA, 2000; pp. 111–148. [Google Scholar]
- Azam, F.; Malfatti, F. Microbial structuring of marine ecosystems. Nat. Rev. Microbiol. 2007, 5, 782–791. [Google Scholar] [CrossRef] [PubMed]
- Faust, K.; Raes, J. Microbial interactions: From networks to models. Nat. Rev. Microbiol. 2012, 10, 538–550. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.; Xu, W.; Kong, L.; Shen, M.; Wang, S.; Sun, Y.; Gao, Y.; Jiang, Q.; Xue, J.; Cheng, D.; et al. Bacterial specialists playing crucial roles in maintaining system stability and governing microbial diversity in bioremediation of oil-polluted sediments under typical deep-sea condition. Bioresour. Technol. 2024, 413, 131498. [Google Scholar] [CrossRef] [PubMed]
- Chesson, P. Mechanisms of maintenance of species diversity. Annu. Rev. Ecol. Syst. 2000, 31, 343–366. [Google Scholar] [CrossRef]
- Chave, J. Neutral theory and community ecology. Ecol. Lett. 2004, 7, 241–253. [Google Scholar] [CrossRef]
- Wang, L.; Lian, C.; Wan, W.; Qiu, Z.; Luo, X.; Huang, Q.; Deng, Y.; Zhang, T.; Yu, K. Salinity-triggered homogeneous selection constrains the microbial function and stability in lakes. Appl. Microbiol. Biot. 2023, 107, 6591–6605. [Google Scholar] [CrossRef]
- Whitacre, J.M.; Atamas, S.P. Degeneracy allows for both apparent homogeneity and diversification in populations. Biosystems 2012, 110, 34–42. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiao, Y.; Wang, S.; Wang, L.; Li, S.; Wang, F.; Wang, B.; Liu, Y. Insights into Ecological Features of Microbial Dark Matter Within the Symbiotic Community During Alexandrium pacificum Bloom: Co-Occurrence Interactions and Assembly Processes. Coasts 2025, 5, 31. https://doi.org/10.3390/coasts5030031
Qiao Y, Wang S, Wang L, Li S, Wang F, Wang B, Liu Y. Insights into Ecological Features of Microbial Dark Matter Within the Symbiotic Community During Alexandrium pacificum Bloom: Co-Occurrence Interactions and Assembly Processes. Coasts. 2025; 5(3):31. https://doi.org/10.3390/coasts5030031
Chicago/Turabian StyleQiao, Yanlu, Shuo Wang, Lingzhe Wang, Shijie Li, Feng Wang, Bo Wang, and Yuyang Liu. 2025. "Insights into Ecological Features of Microbial Dark Matter Within the Symbiotic Community During Alexandrium pacificum Bloom: Co-Occurrence Interactions and Assembly Processes" Coasts 5, no. 3: 31. https://doi.org/10.3390/coasts5030031
APA StyleQiao, Y., Wang, S., Wang, L., Li, S., Wang, F., Wang, B., & Liu, Y. (2025). Insights into Ecological Features of Microbial Dark Matter Within the Symbiotic Community During Alexandrium pacificum Bloom: Co-Occurrence Interactions and Assembly Processes. Coasts, 5(3), 31. https://doi.org/10.3390/coasts5030031





