Spectroscopic Investigations of Diethanolamine-Modified Nucleic Acids
Abstract
1. Introduction
2. Materials and Methods
2.1. General
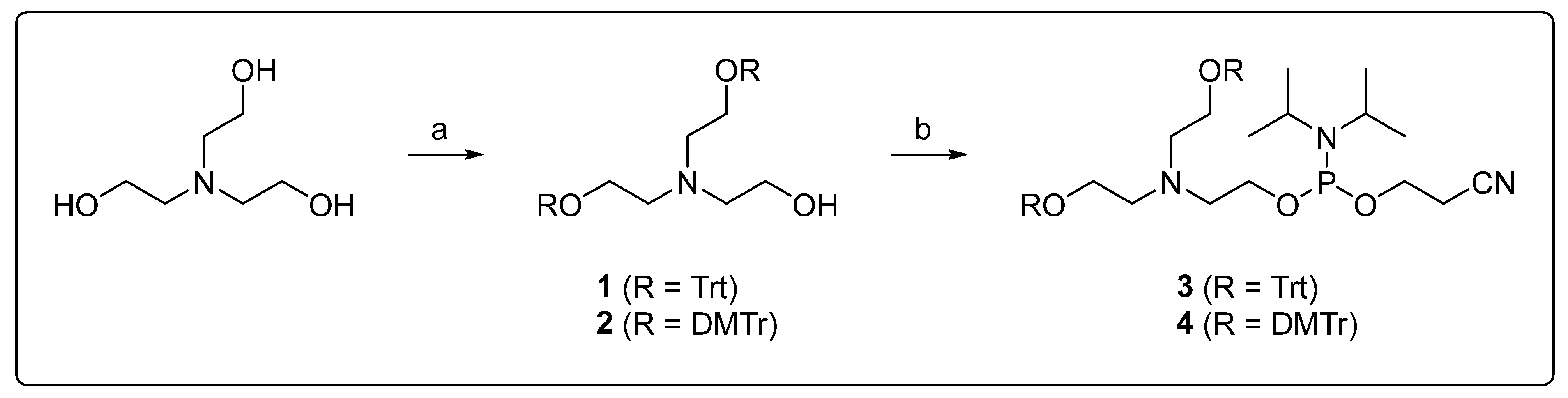
2.2. Synthesis of DMT-Protected Triethanolamine
2.3. Synthesis of the Trityl-Protected Phosphoramidite 3
2.4. Synthesis of the DMT-Protected Phosphoramidite 4
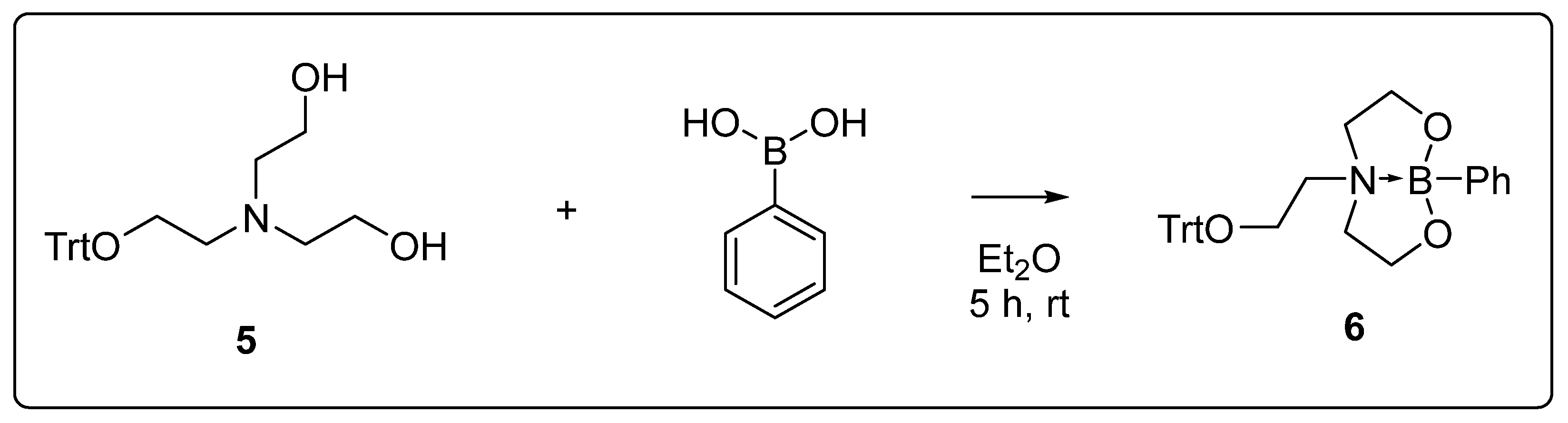
2.5. Synthesis of Dioxazaborocane 6
2.6. UV/vis Spectroscopy and DNA Melting Point Determination
2.7. CD Spectroscopy
2.8. DNA Synthesis
2.9. DNA Concentration
2.10. Single Crystal X-Ray Diffractometry
3. Results
3.1. Synthesis
3.2. Spectroscopic Characterisation of Nucleic Acids
3.3. Synthesis and Characterisation of Dioxazaborocane-Based Modification
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CD | circular dichroism |
| CEDIP-Cl | 2-cyanoethyl N,N-diisopropylchlorophosphoramidite |
| CHES | N-cyclohexyl-2-aminoethanesulfonic acid |
| DCM | Dichloromethane |
| DIPEA | N,N-diisopropylethylamine |
| DMSO | dimethyl sulfoxide |
| DMTr- | 4,4′-dimethoxytrityl- |
| MES | 2-(N-morpholino)ethanesulfonic acid |
| MOPS | 3-(N-morpholino)propanesulfonic acid |
| ODN | oligodeoxyribonucleotide |
| Trt | trityl |
| UV/vis | ultraviolet–visible |
References
- Bonner, G.; Klibanov, A.M. Structural stability of DNA in nonaqueous solvents. Biotechnol. Bioeng. 2000, 68, 339–344. [Google Scholar] [CrossRef]
- Manalo, M.N.; Kong, X.; LiWang, A. Sensitivity of hydrogen bonds of DNA and RNA to hydration, as gauged by 1JNH measurements in ethanol-water mixtures. J. Biomol. NMR 2007, 37, 257–263. [Google Scholar] [CrossRef]
- Nakano, S.I.; Sugimoto, N. The structural stability and catalytic activity of DNA and RNA oligonucleotides in the presence of organic solvents. Biophys. Rev. 2016, 8, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Hackl, E.V.; Blagoi, Y.P. Effect of ethanol on structural transitions of DNA and polyphosphates under Ca2+ ions action in mixed solutions. Acta Biochim. Pol. 2000, 47, 103–112. [Google Scholar] [CrossRef]
- Minier, M.A.S.; Farah; Lee, A.; Xue, L. Determination of the effect of water-miscible organic solvents on the stability of DNA duplexes via UV thermal denaturation and circular dichroism. J. Undergrad. Chem. Res. 2011, 10, 145–152. [Google Scholar]
- Beneventi, S.; Onori, G. Effect of ethanol on the thermal stability of tRNA molecules. Biophys. Chem. 1986, 25, 181–190. [Google Scholar] [CrossRef]
- Piskur, J.; Rupprecht, A. Aggregated DNA in ethanol solution. FEBS Lett. 1995, 375, 174–178. [Google Scholar] [CrossRef]
- Schultz, J.; Rupprecht, A.; Song, Z.; Piskur, J.; Nordenskiold, L.; Lahajnar, G. A mechanochemical study of MgDNA fibers in ethanol-water solutions. Biophys. J. 1994, 66, 810–819. [Google Scholar] [CrossRef][Green Version]
- Darzynkiewicz, Z.; Traganos, F.; Sharpless, T.; Melamed, M.R. DNA denaturation in situ. Effect of divalent cations and alcohols. J. Cell Biol. 1976, 68, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Rupprecht, A.; Piškur, J.; Schultz, J.; Nordenskiöld, L.; Song, Z.; Lahajnar, G. Mechanochemical study of conformational transitions and melting of Li-, Na-, K-, and CsDNA fibers in ethanol–water solutions. Biopolymers 1994, 34, 897–920. [Google Scholar] [CrossRef]
- Song, Z.; Rupprecht, A.; Fritzsche, H. Mechanochemical study of NaDNA and NaDNA-netropsin fibers in ethanol-water and trifluoroethanol-water solutions. Biophys. J. 1995, 68, 1050–1062. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J. Precipitation of DNA with Ethanol. Cold Spring Harb. Protoc. 2016, 2016, 1116–1120. [Google Scholar] [CrossRef]
- He, S.; Cao, B.; Yi, Y.; Huang, S.; Chen, X.; Luo, S.; Mou, X.; Guo, T.; Wang, Y.; Wang, Y.; et al. DNA precipitation revisited: A quantitative analysis. Nano Select 2021, 3, 617–626. [Google Scholar] [CrossRef]
- Del Vecchio, P.; Esposito, D.; Ricchi, L.; Barone, G. The effects of polyols on the thermal stability of calf thymus DNA. Int. J. Biol. Macromol. 1999, 24, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Duggan, E.L. Deformation of DNA. III. The effect of glycol and glycerol on the ultraviolet absorbances of DNA. Renaturation by dilution. Biochem. Biophys. Res. Commun. 1961, 6, 93–99. [Google Scholar] [CrossRef]
- Lee, J.; Vogt, C.E.; McBrairty, M.; Al-Hashimi, H.M. Influence of dimethylsulfoxide on RNA structure and ligand binding. Anal. Chem. 2013, 85, 9692–9698. [Google Scholar] [CrossRef]
- Stockx, J. The influence of strong solutions of uerea and poly alcohols on the spectroscopic behavior of ribonucleic acid and nucleotides. Biochim. Biophys. Acta 1963, 68, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Sen, A.; Nielsen, P.E. On the stability of peptide nucleic acid duplexes in the presence of organic solvents. Nucleic Acids Res. 2007, 35, 3367–3374. [Google Scholar] [CrossRef][Green Version]
- Kimura, T.; Iwai, S.; Moritan, T.; Nam, K.; Mutsuo, S.; Yoshizawa, H.; Okada, M.; Furuzono, T.; Fujisato, T.; Kishida, A. Preparation of poly(vinyl alcohol)/DNA hydrogels via hydrogen bonds formed on ultra-high pressurization and controlled release of DNA from the hydrogels for gene delivery. J. Artif. Organs 2007, 10, 104–108. [Google Scholar] [CrossRef]
- Zhou, L.; Emenuga, M.; Kumar, S.; Lamantia, Z.; Figueiredo, M.; Emrick, T. Designing Synthetic Polymers for Nucleic Acid Complexation and Delivery: From Polyplexes to Micelleplexes to Triggered Degradation. Biomacromolecules 2022, 23, 4029–4040. [Google Scholar] [CrossRef]
- Kim, Y.H.; Lee, K.; Li, S. Nucleic Acids Based Polyelectrolyte Complexes: Their Complexation Mechanism, Morphology, and Stability. Chem. Mater. 2021, 33, 7923–7943. [Google Scholar] [CrossRef]
- Kurz, M. Compatible solute influence on nucleic acids: Many questions but few answers. Saline Syst. 2008, 4, 6. [Google Scholar] [CrossRef]
- Lenz, T.; Layh, M.; Hebenbrock, M. One trityl, two trityl, three trityl groups—Structural differences of differently substituted triethanolamines. Z. Kristallogr. Cryst. Mater. 2025, 240, 101–111. [Google Scholar] [CrossRef]
- Böttcher, A.; Kowerko, D.; Sigel, R.K.O. Explicit analytic equations for multimolecular thermal melting curves. Biophys. J. 2015, 202, 32–39. [Google Scholar] [CrossRef]
- Le, L.V.; Kim, T.J.; Kim, Y.D.; Aspnes, D.E. Decoding ‘Maximum Entropy’ Deconvolution. Entropy 2022, 24, 1238. [Google Scholar] [CrossRef]
- Cavaluzzi, M.J.; Borer, P.N. Revised UV extinction coefficients for nucleoside-5’-monophosphates and unpaired DNA and RNA. Nucleic Acids Res. 2004, 32, e13. [Google Scholar] [CrossRef]
- APEX5, Version 2023.9.2; Bruker AXS Inc.: Madison, WI, USA, 2023.
- SAINT, Version 8.40B; includes Xprep, SADABS and TWINABS; Bruker AXS Inc.: Madison, WI, USA, 2001.
- Sheldrick, G.M. SADABS. University of Göttingen: Göttingen, Germany, 1996. [Google Scholar]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Lubben, J.; Wandtke, C.M.; Hubschle, C.B.; Ruf, M.; Sheldrick, G.M.; Dittrich, B. Aspherical scattering factors for SHELXL—Model, implementation and application. Acta Crystallogr. A 2019, 75, 50–62. [Google Scholar] [CrossRef] [PubMed]
- Diamond—Crystal and Molecular Structure Visualization, Crystal Impact; Dr. H. Putz & Dr. K. Brandenburg GbR: Bonn, Germany.
- Farrugia, L.J. WinGX and ORTEP for Windows: An update. J. Appl. Crystallogr. 2012, 45, 849–854. [Google Scholar] [CrossRef]
- Zgarbova, M.; Otyepka, M.; Sponer, J.; Lankas, F.; Jurecka, P. Base Pair Fraying in Molecular Dynamics Simulations of DNA and RNA. J. Chem. Theory Comput. 2014, 10, 3177–3189. [Google Scholar] [CrossRef] [PubMed]
- Andreatta, D.; Sen, S.; Perez Lustres, J.L.; Kovalenko, S.A.; Ernsting, N.P.; Murphy, C.J.; Coleman, R.S.; Berg, M.A. Ultrafast dynamics in DNA: “fraying” at the end of the helix. J. Am. Chem. Soc. 2006, 128, 6885–6892. [Google Scholar] [CrossRef]
- Lenz, T.; Hepp, A.; Layh, M.; Hebenbrock, M. Anionic Boraza-Crown Ethers. Eur. J. Inorg. Chem. 2023, 26, e202300401. [Google Scholar] [CrossRef]
- Lenz, T.; Hebenbrock, M. Phenylsilver—An unexpected one-dimensional coordination polymer of silver(I) tetrads. Dalton Trans. 2024, 53, 423–427. [Google Scholar] [CrossRef]
- Contreras, R.; Garcia, C.; Mancilla, T.; Wrackmeyer, B. The N-B coordination in hindered cyclic thexylboronic esters derived from diethanolamines. J. Organomet. Chem. 1983, 246, 213–217. [Google Scholar] [CrossRef]
- Takahashi, A.; Yamanishi, M.; Kameyama, A.; Otsuka, H. Reversible modulation of polymer chain mobility by selective cage opening and closing of pendant boratrane units. Polym. J. 2024, 57, 259–268. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, S.; Jiang, Z.; Li, Y.; Jing, X. Boronic Ester Based Vitrimers with Enhanced Stability via Internal Boron-Nitrogen Coordination. J. Am. Chem. Soc. 2020, 142, 21852–21860. [Google Scholar] [CrossRef]
- Durka, K.; Kaminski, R.; Lulinski, S.; Serwatowski, J.; Wozniak, K. On the nature of the B⋯N interaction and the conformational flexibility of arylboronic azaesters. Phys. Chem. Chem. Phys. 2010, 12, 13126–13136. [Google Scholar] [CrossRef]
- Martin, A.R.; Barvik, I.; Luvino, D.; Smietana, M.; Vasseur, J.J. Dynamic and programmable DNA-templated boronic ester formation. Angew. Chem. Int. Ed. 2011, 50, 4193–4196. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.R.; Vasseur, J.J.; Smietana, M. Boron and nucleic acid chemistries: Merging the best of both worlds. Chem. Soc. Rev. 2013, 42, 5684–5713. [Google Scholar] [CrossRef] [PubMed]
- Barbeyron, R.; Martin, A.R.; Jean-Jacques Vasseur, J.-J.V.; Michael Smietana, M.S. DNA-templated borononucleic acid self assembly: A study of minimal complexity. RSC Adv. 2015, 5, 105587–105591. [Google Scholar] [CrossRef]
- Debiais, M.; Gimenez Molina, A.; Muller, S.; Vasseur, J.J.; Barvik, I.; Baraguey, C.; Smietana, M. Design and NMR characterization of reversible head-to-tail boronate-linked macrocyclic nucleic acids. Org. Biomol. Chem. 2022, 20, 2889–2895. [Google Scholar] [CrossRef]
- Lelievre-Buttner, A.; Schnarr, T.; Debiais, M.; Smietana, M.; Muller, S. Boronic Acid Assisted Self-Assembly of Functional RNAs. Chem. Eur. J. 2023, 29, e202300196. [Google Scholar] [CrossRef]
- Debiais, M.; Lelievre, A.; Vasseur, J.-J.; Müller, S.; Smietana, M. Boronic Acid-Mediated Activity Control of Split 10–23 DNAzymes. Chem. Eur. J. 2021, 27, 1138–1144. [Google Scholar] [CrossRef]
- Debiais, M.; Vasseur, J.J.; Smietana, M. Applications of the Reversible Boronic Acids/Boronate Switch to Nucleic Acids. Chem. Rec. 2022, 22, e202200085. [Google Scholar] [CrossRef]
- Reverte, M.; Vasseur, J.J.; Smietana, M. Nuclease stability of boron-modified nucleic acids: Application to label-free mismatch detection. Org. Biomol. Chem. 2015, 13, 10604–10608. [Google Scholar] [CrossRef]
- Clave, G.; Reverte, M.; Vasseur, J.J.; Smietana, M. Modified internucleoside linkages for nuclease-resistant oligonucleotides. RSC Chem. Biol. 2021, 2, 94–150. [Google Scholar] [CrossRef] [PubMed]
- Barbeyron, R.; Vasseur, J.-J.; Baraguey, C.; Smietana, M. Synthesis of 3‘-deoxy-3‘-iminodiacetic acid and 3‘-deoxy-3‘-aminodiethanol thymidine analogues and NMR study of their complexation with boronic acids. Tetrahedron 2017, 73, 2468–2475. [Google Scholar] [CrossRef]
- Epple, S.; El-Sagheer, A.H.; Brown, T. Artificial nucleic acid backbones and their applications in therapeutics, synthetic biology and biotechnology. Emerg. Top. Life Sci. 2021, 5, 691–697. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, J.A.; Witzigmann, D.; Thomson, S.B.; Chen, S.; Leavitt, B.R.; Cullis, P.R.; van der Meel, R. The current landscape of nucleic acid therapeutics. Nat. Nanotechnol. 2021, 16, 630–643. [Google Scholar] [CrossRef]
- O’Donoghue, P.; Heinemann, I.U.; Fan, C. Editorial: Synthetic Nucleic Acids for Expanding Genetic Codes and Probing Living Cells. Front. Bioeng. Biotechnol. 2021, 9, 720534. [Google Scholar] [CrossRef]
- Roberts, T.C.; Langer, R.; Wood, M.J.A. Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov. 2020, 19, 673–694. [Google Scholar] [CrossRef]
- Curtis, E.A. Pushing the Limits of Nucleic Acid Function. Chem. Eur. J. 2022, 28, e202201737. [Google Scholar] [CrossRef]
- Springsteen, G.; Wang, B. A detailed examination of boronic acid–diol complexation. Tetrahedron 2002, 58, 5291–5300. [Google Scholar] [CrossRef]
- Pizer, R.; Tihal, C. Equilibria and reaction mechanism of the complexation of methylboronic acid with polyols. Inorg. Chem. 1992, 31, 3243–3247. [Google Scholar] [CrossRef]
- Lorand, J.P.; Edwards, J.O. Polyol Complexes and Structure of the Benzeneboronate Ion. J. Org. Chem. 1959, 24, 769–774. [Google Scholar] [CrossRef]
- Yan, J.; Springsteen, G.; Deeter, S.; Wang, B. The relationship among pKa, pH, and binding constants in the interactions between boronic acids and diols—It is not as simple as it appears. Tetrahedron 2004, 60, 11205–11209. [Google Scholar] [CrossRef]
- He, C.; Dong, J.; Xu, C.; Pan, X. N-Coordinated Organoboron in Polymer Synthesis and Material Science. ACS Polymers Au 2022, 3, 5–27. [Google Scholar] [CrossRef]
- Quinteros-Sedano, A.; Le Besnerais, B.; Van Zee, N.J.; Nicolaÿ, R. Exploiting Dioxazaborocane Chemistry for Preparing Elastomeric Vitrimers with Enhanced Processability and Mechanical Properties. Chem. Mater. 2025, 37, 2058–2070. [Google Scholar] [CrossRef]
- Ito, Y.; Aoki, D.; Otsuka, H. Functionalization of amine-cured epoxy resins by boronic acids based on dynamic dioxazaborocane formation. Polym. Chem. 2020, 11, 5356–5364. [Google Scholar] [CrossRef]


 N,
N,
 O,
O,
 B,
B,
 C.
C.
 N,
N,
 O,
O,
 B,
B,
 C.
C.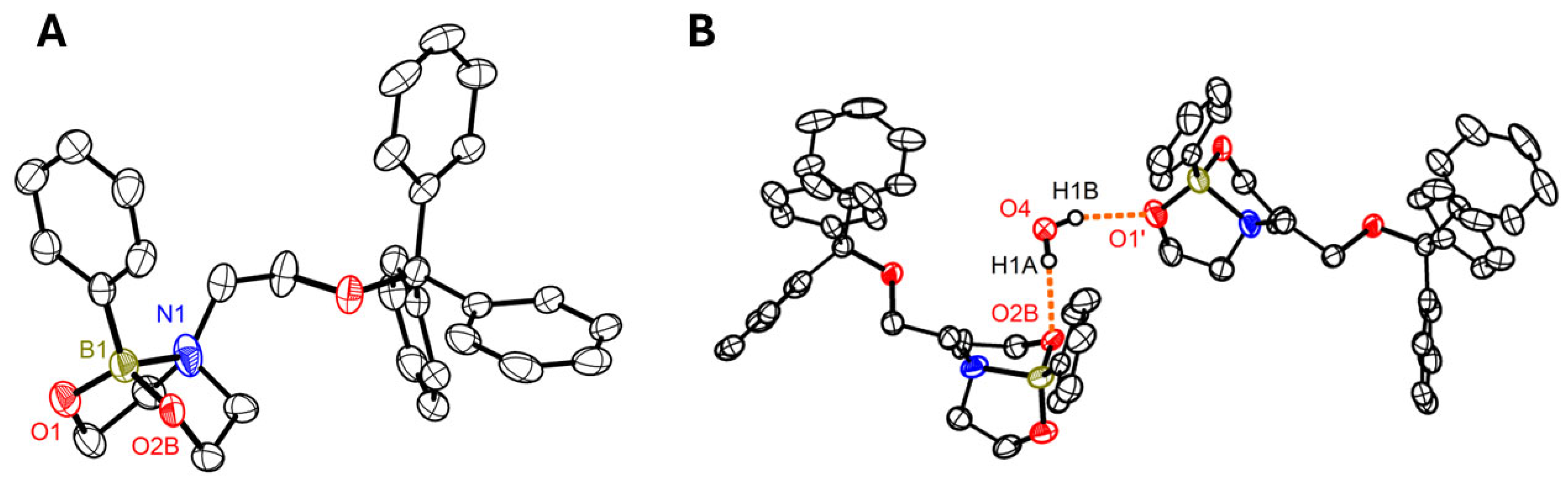

| Entry | Terminal Base | Sequence | Chemical Formula |
|---|---|---|---|
| ODN1 | N | 5′-d(NTA TAG GCC TAT A) | C124H163N45O75P12 |
| ODN2 | □ | 5′-d( TA TAG GCC TAT A) | C118H149N44O70P11 |
| ODN3 | N | 5′-d(NAA TAG GCC TAT T) | C124H163N45O75P12 |
| ODN4 | □ | 5′-d( AA TAG GCC TAT T) | C118H149N44O70P11 |
| ODN5 | N | 5′-d(NGA TAG GCC TAT C) | C123H162N46O75P12 |
| ODN6 | □ | 5′-d( GA TAG GCC TAT C) | C117H148N45O70P11 |
| ODN7 | N | 5′-d(NCA TAG GCC TAT G) | C123H162N46O75P12 |
| ODN8 | □ | 5′-d( CA TAG GCC TAT G) | C117H148N45O70P11 |
| ODN | Without Additive | With Phenylboronic Acid in DMSO | With DMSO |
|---|---|---|---|
| 1 | 37.9(1) | 37.7(1) | 38.0(1) |
| 2 | 40.6(1) | 40.9(1) | 40.7(1) |
| 3 | 38.5(1) | 40.2(2) | 40.5(1) |
| 4 | 40.2(1) | 40.1(1) | 40.5(1) |
| 5 | 41.8(1) | 42.7(1) | 42.8(1) |
| 6 | 41.5(2) | 42.3(1) | 42.2(1) |
| 7 | 43.9(2) | 43.4(3) | 44.7(1) |
| 8 | 45.1(1) | 46.1(1) | 45.5(1) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lenz, T.; Hebenbrock, M. Spectroscopic Investigations of Diethanolamine-Modified Nucleic Acids. AppliedChem 2025, 5, 40. https://doi.org/10.3390/appliedchem5040040
Lenz T, Hebenbrock M. Spectroscopic Investigations of Diethanolamine-Modified Nucleic Acids. AppliedChem. 2025; 5(4):40. https://doi.org/10.3390/appliedchem5040040
Chicago/Turabian StyleLenz, Tabea, and Marian Hebenbrock. 2025. "Spectroscopic Investigations of Diethanolamine-Modified Nucleic Acids" AppliedChem 5, no. 4: 40. https://doi.org/10.3390/appliedchem5040040
APA StyleLenz, T., & Hebenbrock, M. (2025). Spectroscopic Investigations of Diethanolamine-Modified Nucleic Acids. AppliedChem, 5(4), 40. https://doi.org/10.3390/appliedchem5040040






