Light and Shadow in Near-Infrared Spectroscopy: A Powerful Tool for Cannabis sativa L. Analysis
Abstract
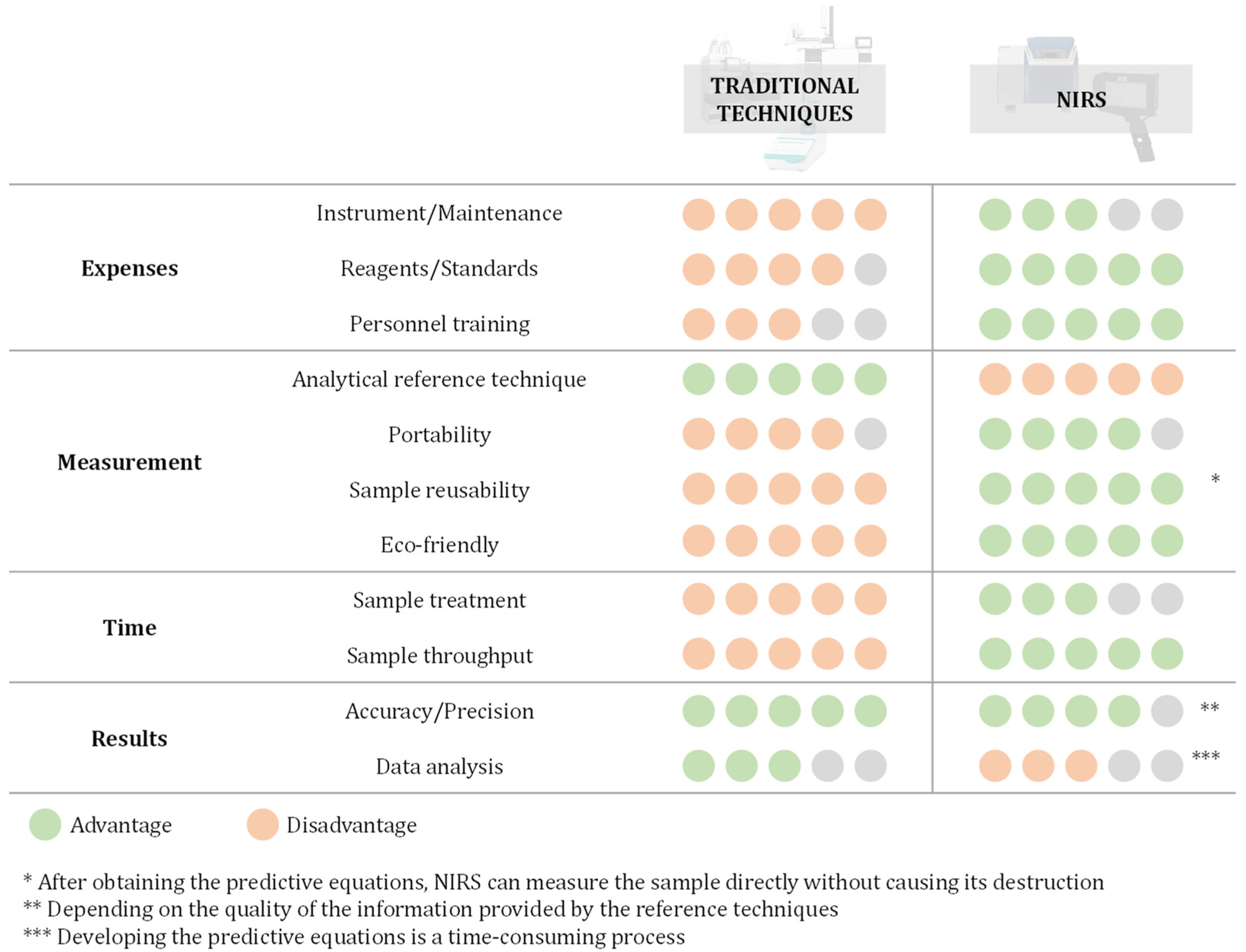
:1. Introduction
2. Qualitative Methods
2.1. Unsupervised Methods
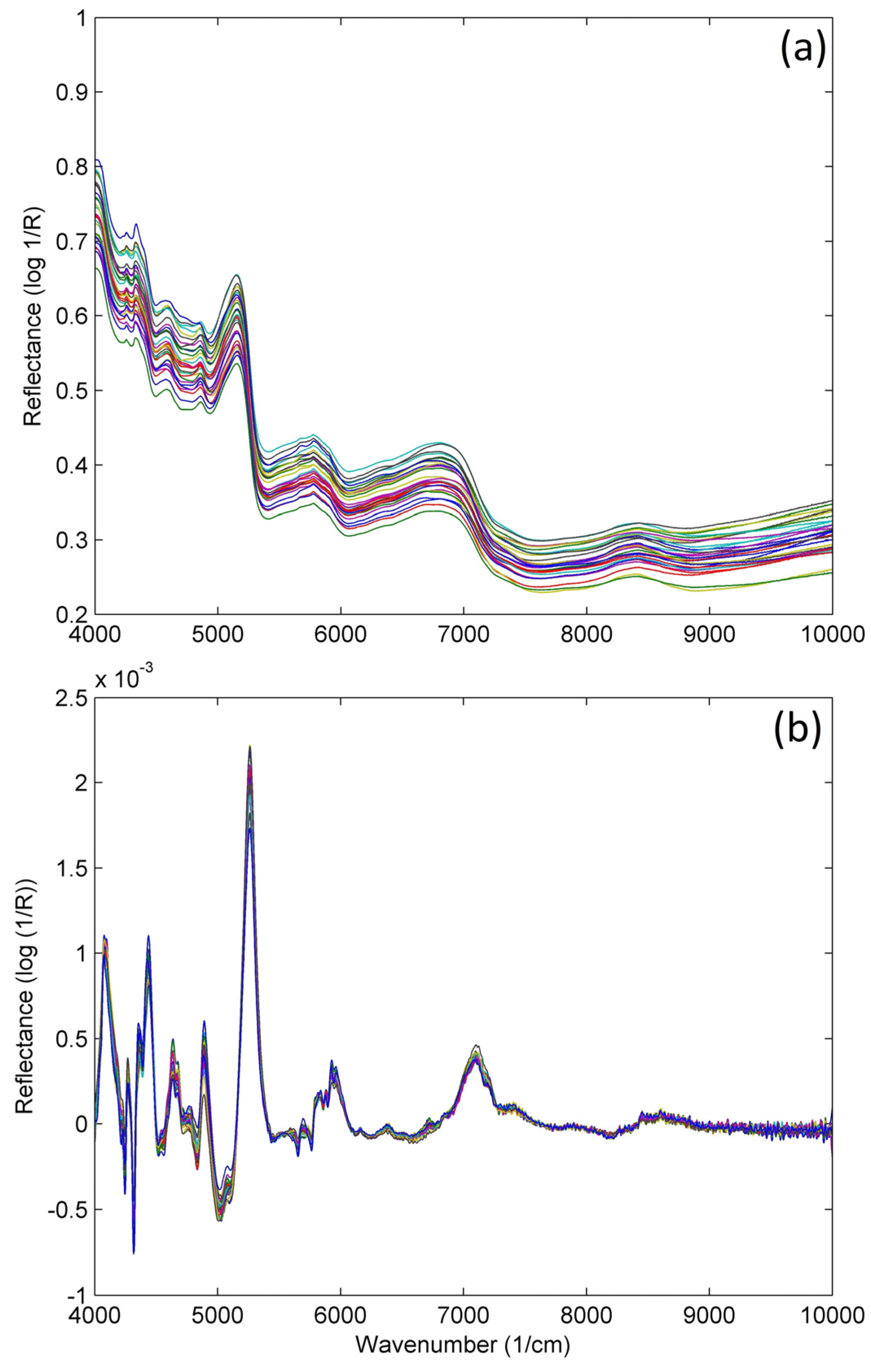
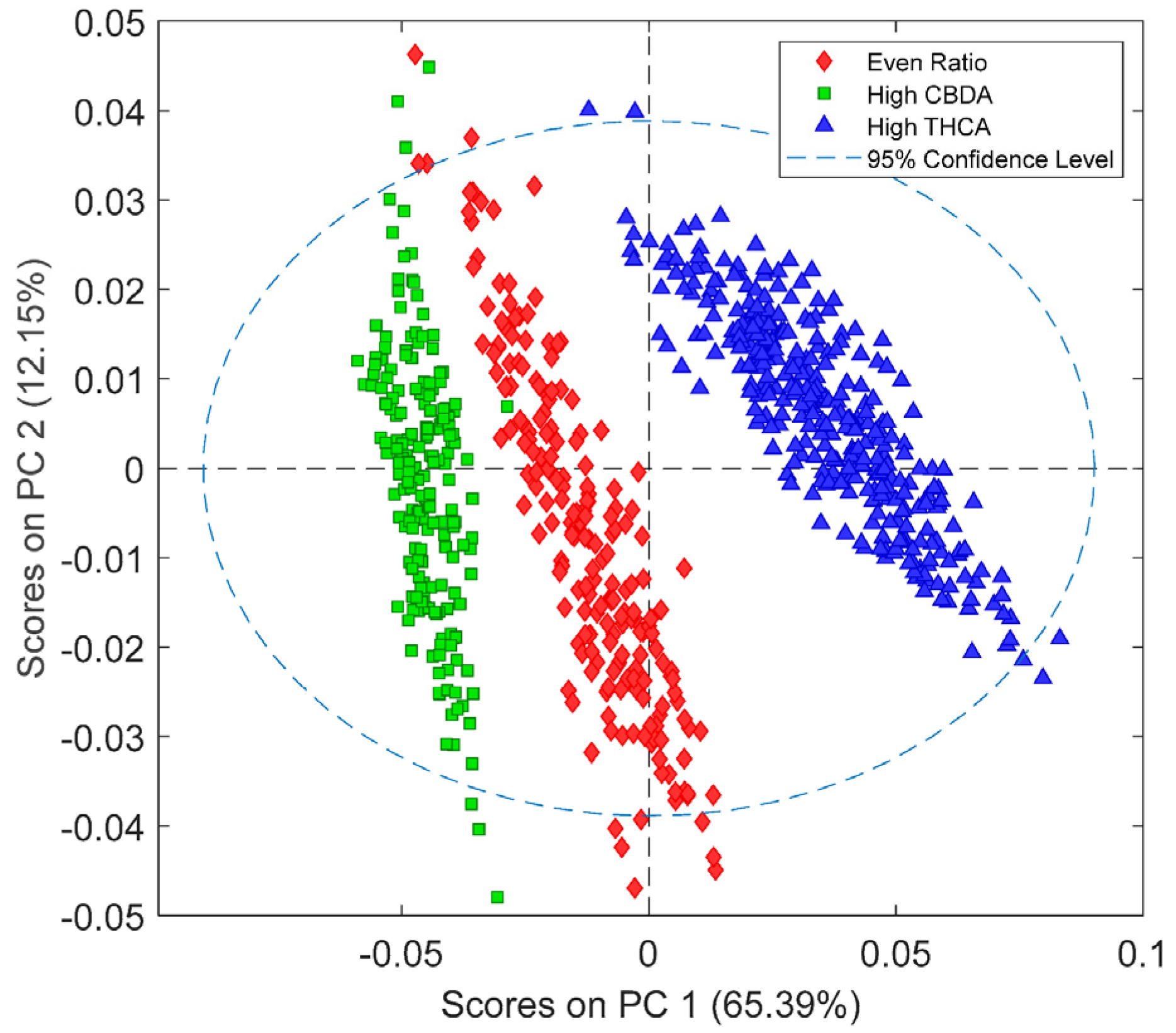
2.1.1. Principal Component Analysis
2.1.2. Hierarchical Clustering Analysis
2.1.3. Non-Hierarchical Clustering Analysis
2.2. Supervised Methods
2.2.1. Soft Independent Modelling of Class Analogy
2.2.2. Partial Least Squares Discriminant Analysis
| Instrument | Spectral Range (nm) | Samples | Parameter | Spectra Pretreatment | Chemometric Method | Ref. |
|---|---|---|---|---|---|---|
| FOSS NIRSystem 6500 (benchtop) | 1100–2500 | Inflorescences and leaves | Δ9-THC | - | PCA | [61] |
| PerkinElmer™ Frontier MIR/NIR (benchtop) Thermo Scientific MicroPHAZIR RX (handheld) | 1000–2500 1600–2500 | Inflorescences | Δ9-THC | SNV and Savitzky–Golay filters First derivative and SNV | PCA, HCA, SIMCA, k-NN, and PLS-DA | [52] |
| PerkinElmer 400 IR | 1000–2500 | Aerial parts | Growth stage | Savitzky–Golay filters, MSC, and mean centering | HCA, PCA, PLS-DA, and SVM-DA | [51] |
| Bruker MPA II FT-NIR (benchtop) Viavi MicroNIR Onsite-W (handheld) | 870–2500 950–1650 | Dried inflorescences | 14 cannabinoids | Detrend, SNV, and normalization | PCA and PLS-DA | [42] |
| Specim, SisuChema (handheld) | - | Leaves | Detection and classification of Cannabis | SNV, MSC, Savitzky–Golay filters | PCA and SIMCA | [50] |
| ThermoFisher, Antaris II FT-NIR (benchtop) | 1000–2500 | Inflorescences | CBDA, CBGA and THCA | Savitzky–Golay filters, SNV, MSC, and mean centering | PLS-DA | [20] |
| VIAVI, microNIR (portable) | 900–1700 | Hemp flours | CBD, Δ9-THC and CBG | SNV, MSC, mean centering, normalization, Savitzky–Golay filters | PCA | [69] |
| VIAVI, microNIR (portable) | 900–1700 | Oral fluids | Δ9-THC | SNV, MSC, normalization, and Savitzky–Golay filters | PLS-DA | [70] |
| Perten DA7250 | 950–1650 | Ground and whole hemp | Δ9-THC | Mean centering | LDA | [36] |
2.2.3. Parametric and Non-Parametric Methods
3. Quantitative Methods
3.1. Linear Regression Multivariate Statistical Techniques
3.1.1. Partial Least Squares Regression
3.1.2. Principal Component Regression
| Instrument | Spectral Range (nm) | Samples | Parameter | Regression Model | n | rv2 | RMSEv (%) | RMSEP (%) | SEP (%) | RPD | Ref. | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FT-NIR (handheld) | 1350–2560 | Dry hemp | CBD, total CBD, Δ9-THC, and total THC | PLS-R | 67–72 | 0.9100–0.9500 | - | - | 0.02–0.61 | - | [22] | |||||||||||
| VIAVI, microNIR (portable) | 900–1700 | Hemp flours | CBD, Δ9-THC, and CBG | PLS-R | 10 | 0.9741–0.9980 | 0.005 | 0.005–0.007 | - | - | [69] | |||||||||||
| Acousto-Optic Tunable Filter NIR | 1200–2200 | Dry hemp | Δ9-THC and CBD | PLS-R | 91–103 | 0.77 | 0.0140–0.4310 | - | - | 2.04–2.07 | [55] | |||||||||||
| Bruker MPA II FT-NIR (benchtop) | Viavi MicroNIR Onsite-W (handheld) | 870–2500 | 950–1650 | Dried inflorescences | 14 cannabinoids | PLS-R | 734 | 730 | 0.2500–0.9800 | 0.2100–0.9800 | 0.0800–7.000 | 0.0800–6.530 | 0.0600–5.5100 | 0.0800–6.2300 | - | - | [42] | |||||
| VIAVI, microNIR (portable) | 900–1700 | Oral fluids | Δ9-THC | PLS-R | 50 | 0.989 | 1.1 | 1.3 | - | - | [70] | |||||||||||
| Resonon Pika XC2 hyperspectral camera | 400–1000 | Fresh flowers | Fresh leaves | CBD, Δ9-THC, CBG, CBDA, THCA, and CBGA | PLS-R | 100 | 0.5100–0.8500 | 0.4200–0.71 | 0.9000–20.6700 | 0.1600–3.7600 | - | - | 1.43–2.62 | 1.32–1.88 | [44] | |||||||
| Perten DA7250 | 950–1650 | Ground hemp | MC | 5 cannabinoids | PLS-R | 115 | 0.91 | 0.0300–0.8500 | - | 1.28 | 0.02–0.92 | - | [36] | |||||||||
| Whole hemp | MC | 5 cannabinoids | 194 | 0.94 | 0.03–0.89 | 1.24 | 0.01–0.60 | |||||||||||||||
| Tellspec NIR-S-G1 (handheld) | 900–1700 | Resins | Δ9-THC | PLS-R | - | 0.02 | 5.19 | 3.87 | - | 1.51 | [43] | |||||||||||
| Viavi Solutions MicroNIR (handheld) | 950–1650 | 0.67 | 2.5 | 1.46 | 2.26 | |||||||||||||||||
| ThermoFisher Antaris II FT-NIR | 1000–2500 | Dried inflorescences | 10 cannabinoids | 9 terpenes | PLS-R | 47–237 | 84–218 | 0.6250–0.9900 | 0.7000–0.8870 | 0.0100–1.0080 | 0.0032–0.0400 | 0.0110–1.2750 | 0.0037–0.0416 | - | 1.87–10.87 | 1.78–3.00 | [20] | |||||
| FOSS NIR Systems 6500 (benchtop) | Bruker FT-NIR (portable) | 400–2498 | 800–2500 | Dried leaves and inflorescences | 8 cannabinoids | PLS-R | 189 | 0.5400–0.9800 | 0.7800–0.9900 | - | - | 0.03–1.72 | 0.04–1.79 | 1.25–6.03 | 1.52–6.00 | [48] | ||||||
| PerkinElmer Spectrum Two FT-NIR | 1000–2500 | Dried flowers | CBD and Δ9-THC | PCR | 302 | 0.9700–0.9800 | - | - | 0.73–0.92 | - | [91] | |||||||||||
| Hone HL-EVT9-Neo NIR (portable) | 1250–2500 | Dried flowers | 12 cannabinoids | ANN | 249 | 0.0300–1.0000 | 0.0010–0.5600 | - | - | - | [47] | |||||||||||
| Control development NIR spectrophotometer | 904–1699 | Hemp extracts | TDS, EY, TPC, and AC | ANN (SLE) | ANN (MAE) | - | 0.5925–0.9547 | 0.6459–0.9434 | 0.0140–305.5601 | 0.0320–21.8810 | - | - | - | [76] | ||||||||
| Bruker Matrix-F FT-NIR | 800–2500 | Hemp oil | CBD and total CBD | SOSVEN | sPLS-R | 20 | 0.9828–0.9864 | 0.9810–0.9844 | 6.4000–6.6000 | 6.8700–7.0000 | - | - | - | [57] | ||||||||
3.2. Non-Linear Regression Multivariate Statistical Techniques
3.2.1. Artificial Neural Networks
3.2.2. Support Vector Machine
3.3. Near-Infrared Hyperspectral Imaging
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zeine, R.R.; Teasdale, B.W. Medical Cannabis and the Effects of Cannabinoids on Fighting Cancer, Multiple Sclerosis, Epilepsy, Parkinson’s, and Other Neurodegenerative Diseases; IGI Global: Hershey, PA, USA, 2023; ISBN 1668456532. [Google Scholar]
- EMCDDA. EMCDDA Cannabis Laws in Europe. Questions and Answers for Policymaking; EMCDDA: Lisbon, Portugal, 2023. [Google Scholar]
- Sgrò, S.; Lavezzi, B.; Caprari, C.; Polito, M.; D’Elia, M.; Lago, G.; Furlan, G.; Girotti, S.; Ferri, E.N. Delta9-THC Determination by the EU Official Method: Evaluation of Measurement Uncertainty and Compliance Assessment of Hemp Samples. Anal. Bioanal. Chem. 2021, 413, 3399–3410. [Google Scholar] [CrossRef]
- Hädener, M.; König, S.; Weinmann, W. Quantitative Determination of CBD and THC and Their Acid Precursors in Confiscated Cannabis Samples by HPLC-DAD. Forensic Sci. Int. 2019, 299, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Micalizzi, G.; Vento, F.; Alibrando, F.; Donnarumma, D.; Dugo, P.; Mondello, L. Cannabis sativa L.: A Comprehensive Review on the Analytical Methodologies for Cannabinoids and Terpenes Characterization. J. Chromatogr. A 2021, 1637, 461864. [Google Scholar] [CrossRef] [PubMed]
- Izzo, L.; Castaldo, L.; Narváez, A.; Graziani, G.; Gaspari, A.; Rodríguez-Carrasco, Y.; Ritieni, A. Analysis of Phenolic Compounds in Commercial Cannabis sativa L. Inflorescences Using UHPLC-Q-Orbitrap HRMS. Molecules 2020, 25, 631. [Google Scholar] [CrossRef] [PubMed]
- Radwan, M.M.; Chandra, S.; Gul, S.; ElSohly, M.A. Cannabinoids, Phenolics, Terpenes and Alkaloids of Cannabis. Molecules 2021, 26, 2774. [Google Scholar] [CrossRef] [PubMed]
- Macherone, A. Chapter One-Comprehensive Analytical Testing of Cannabis and Hemp. In Analysis of Cannabis; Ferrer, I., Thurman, E.M., Eds.; Comprehensive Analytical Chemistry; Elsevier: Amsterdam, The Netherlands, 2020; Volume 90, pp. 3–29. [Google Scholar]
- AL Ubeed, H.M.S.; Bhuyan, D.J.; Alsherbiny, M.A.; Basu, A.; Vuong, Q.V. A Comprehensive Review on the Techniques for Extraction of Bioactive Compounds from Medicinal Cannabis. Molecules 2022, 27, 604. [Google Scholar] [CrossRef]
- Atapattu, S.N.; Johnson, K.R.D. Pesticide Analysis in Cannabis Products. J. Chromatogr. A 2020, 1612, 460656. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.K.; Xia, Z.; Bulloch, P.; Idowu, I.; Francisco, O.; Stetefeld, J.; Stout, J.; Zimmer, J.; Marvin, C.; Letcher, R.J.; et al. Validated Quantitative Cannabis Profiling for Canadian Regulatory Compliance-Cannabinoids, Aflatoxins, and Terpenes. Anal. Chim. Acta 2019, 1088, 79–88. [Google Scholar] [CrossRef]
- Buchicchio, L.; Asselborn, L.; Schneider, S.; van Nieuwenhuyse, A.; Moris, G.; Schummer, C. Investigation of Aflatoxin and Ochratoxin A Contamination of Seized Cannabis and Cannabis Resin Samples. Mycotoxin Res. 2022, 38, 71–78. [Google Scholar] [CrossRef]
- Wilcox, J.; Pazdanska, M.; Milligan, C.; Chan, D.; MacDonald, S.J.; Donnelly, C. Analysis of Aflatoxins and Ochratoxin A in Cannabis and Cannabis Products by LC–Fluorescence Detection Using Cleanup with Either Multiantibody Immunoaffinity Columns or an Automated System with In-Line Reusable Immunoaffinity Cartridges. J. AOAC Int. 2020, 103, 494–503. [Google Scholar] [CrossRef]
- Cardenia, V.; Gallina Toschi, T.; Scappini, S.; Rubino, R.C.; Rodriguez-Estrada, M.T. Development and Validation of a Fast Gas Chromatography/Mass Spectrometry Method for the Determination of Cannabinoids in Cannabis sativa L. J. Food Drug Anal. 2018, 26, 1283–1292. [Google Scholar] [CrossRef]
- Pellegrini, M.; Marchei, E.; Pacifici, R.; Pichini, S. A Rapid and Simple Procedure for the Determination of Cannabinoids in Hemp Food Products by Gas Chromatography-Mass Spectrometry. J. Pharm. Biomed. Anal. 2005, 36, 939–946. [Google Scholar] [CrossRef] [PubMed]
- Lachenmeier, D.W.; Kroener, L.; Musshoff, F.; Madea, B. Determination of Cannabinoids in Hemp Food Products by Use of Headspace Solid-Phase Microextraction and Gas Chromatography? Mass Spectrometry. Anal. Bioanal. Chem. 2004, 378, 183–189. [Google Scholar] [CrossRef]
- Gul, W.; Gul, S.W.; Radwan, M.M.; Wanas, A.S.; Mehmedic, Z.; Khan, I.I.; Sharaf, M.H.M.; ElSohly, M.A. Determination of 11 Cannabinoids in Biomass and Extracts of Different Varieties of Cannabis Using High-Performance Liquid Chromatography. J. AOAC Int. 2015, 98, 1523–1528. [Google Scholar] [CrossRef] [PubMed]
- McRae, G.; Melanson, J.E. Quantitative Determination and Validation of 17 Cannabinoids in Cannabis and Hemp Using Liquid Chromatography-Tandem Mass Spectrometry. Anal. Bioanal. Chem. 2020, 412, 7381–7393. [Google Scholar] [CrossRef] [PubMed]
- Stolker, A.A.M.; van Schoonhoven, J.; de Vries, A.J.; Bobeldijk-Pastorova, I.; Vaes, W.H.J.; van den Berg, R. Determination of Cannabinoids in Cannabis Products Using Liquid Chromatography–Ion Trap Mass Spectrometry. J. Chromatogr. A 2004, 1058, 143–151. [Google Scholar] [CrossRef]
- Birenboim, M.; Kengisbuch, D.; Chalupowicz, D.; Maurer, D.; Barel, S.; Chen, Y.; Fallik, E.; Paz-Kagan, T.; Shimshoni, J.A. Use of Near-Infrared Spectroscopy for the Classification of Medicinal Cannabis Cultivars and the Prediction of Their Cannabinoid and Terpene Contents. Phytochemistry 2022, 204, 113445. [Google Scholar] [CrossRef]
- Pourseyed Lazarjani, M.; Torres, S.; Hooker, T.; Fowlie, C.; Young, O.; Seyfoddin, A. Methods for Quantification of Cannabinoids: A Narrative Review. J. Cannabis Res. 2020, 2, 35. [Google Scholar] [CrossRef] [PubMed]
- Yao, S.; Ball, C.; Miyagusuku-Cruzado, G.; Giusti, M.M.; Aykas, D.P.; Rodriguez-Saona, L.E. A Novel Handheld FT-NIR Spectroscopic Approach for Real-Time Screening of Major Cannabinoids Content in Hemp. Talanta 2022, 247, 123559. [Google Scholar] [CrossRef] [PubMed]
- García-Valverde, M.T.; Sánchez-Carnerero Callado, C.; Díaz-Liñán, M.C.; Sánchez de Medina, V.; Hidalgo-García, J.; Nadal, X.; Hanuš, L.; Ferreiro-Vera, C. Effect of Temperature in the Degradation of Cannabinoids: From a Brief Residence in the Gas Chromatography Inlet Port to a Longer Period in Thermal Treatments. Front. Chem. 2022, 10, 1038729. [Google Scholar] [CrossRef]
- Vacek, J.; Vostalova, J.; Papouskova, B.; Skarupova, D.; Kos, M.; Kabelac, M.; Storch, J. Antioxidant Function of Phytocannabinoids: Molecular Basis of Their Stability and Cytoprotective Properties under UV-Irradiation. Free Radic. Biol. Med. 2021, 164, 258–270. [Google Scholar] [CrossRef]
- Yangsud, J.; Santasanasuwan, S.; Ahkkarachinoreh, P.; Maha, A.; Madaka, F.; Suksaeree, J.; Songsak, T.; Vutthipong, A.; Monton, C. Stability of Cannabidiol, ∆9-Tetrahydrocannabinol, and Cannabinol under Stress Conditions. Adv. Tradit. Med. 2021, 21, 475–484. [Google Scholar] [CrossRef]
- Golombek, P.; Müller, M.; Barthlott, I.; Sproll, C.; Lachenmeier, D.W. Conversion of Cannabidiol (CBD) into Psychotropic Cannabinoids Including Tetrahydrocannabinol (THC): A Controversy in the Scientific Literature. Toxics 2020, 8, 41. [Google Scholar] [CrossRef]
- Czégény, Z.; Nagy, G.; Babinszki, B.; Bajtel, Á.; Sebestyén, Z.; Kiss, T.; Csupor-Löffler, B.; Tóth, B.; Csupor, D. CBD, a Precursor of THC in e-Cigarettes. Sci. Rep. 2021, 11, 8951. [Google Scholar] [CrossRef] [PubMed]
- Hazekamp, A.; Peltenburg, A.; Verpoorte, R.; Giroud, C. Chromatographic and Spectroscopic Data of Cannabinoids from Cannabis sativa L. J. Liq. Chromatogr. Relat. Technol. 2005, 28, 2361–2382. [Google Scholar] [CrossRef]
- Mano-Sousa, B.J.; Maia, G.A.S.; Lima, P.L.; Campos, V.A.; Negri, G.; Chequer, F.M.D.; Duarte-Almeida, J.M. Color Determination Method and Evaluation of Methods for the Detection of Cannabinoids by Thin-layer Chromatography (TLC). J. Forensic Sci. 2021, 66, 854–865. [Google Scholar] [CrossRef]
- Leite, J.D.A.; de Oliveira, M.V.L.; Conti, R.; Borges, W.d.S.; Rosa, T.R.; Filgueiras, P.R.; Lacerda, V.; Romão, W.; Neto, Á.C. Extraction and Isolation of Cannabinoids from Marijuana Seizures and Characterization by 1H NMR Allied to Chemometric Tools. Sci. Justice 2018, 58, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Malík, M.; Praus, L.; Tlustoš, P. Comparison of Recirculation and Drain-to-Waste Hydroponic Systems in Relation to Medical Cannabis (Cannabis sativa L.). Plants. Ind. Crops Prod. 2023, 202, 117059. [Google Scholar] [CrossRef]
- Michaud, K. Using Flame Atomic Absorption Spectroscopy to Analyze for Heavy Metal Content in Cannabidiol Products. Ph.D. Thesis, University of Massachusetts Lowell, Lowell, MA, USA, 2021. [Google Scholar]
- Menezes, I.M.N.R.; Nascimento, P.D.A.; Yamamoto, C.I.; Oliveira, A. Evaluation of Trace Elements in Cannabis Products. J. Food Compos. Anal. 2022, 113, 104721. [Google Scholar] [CrossRef]
- Potin, F.; Lubbers, S.; Husson, F.; Saurel, R. Hemp (Cannabis sativa L.) Protein Extraction Conditions Affect Extraction Yield and Protein Quality. J. Food Sci. 2019, 84, 3682–3690. [Google Scholar] [CrossRef]
- Craven, C.B.; Wawryk, N.; Jiang, P.; Liu, Z.; Li, X.-F. Pesticides and Trace Elements in Cannabis: Analytical and Environmental Challenges and Opportunities. J. Environ. Sci. 2019, 85, 82–93. [Google Scholar] [CrossRef]
- Su, K.; Maghirang, E.; Tan, J.W.; Yoon, J.Y.; Armstrong, P.; Kachroo, P.; Hildebrand, D. NIR Spectroscopy for Rapid Measurement of Moisture and Cannabinoid Contents of Industrial Hemp (Cannabis sativa). Ind. Crops Prod. 2022, 184, 115007. [Google Scholar] [CrossRef]
- Alonso-Esteban, J.I.; Pinela, J.; Ćirić, A.; Calhelha, R.C.; Soković, M.; Ferreira, I.C.F.R.; Barros, L.; Torija-Isasa, E.; de Sánchez-Mata, M.C. Chemical Composition and Biological Activities of Whole and Dehulled Hemp (Cannabis sativa L.). Seeds. Food Chem. 2022, 374, 131754. [Google Scholar] [CrossRef] [PubMed]
- Pattnaik, F.; Nanda, S.; Kumar, V.; Naik, S.; Dalai, A.K.; Mohanty, M.K. Extraction of Sugars and Cellulose Fibers from Cannabis Stems by Hydrolysis, Pulping, and Bleaching. Chem. Eng. Technol. 2022, 45, 962–970. [Google Scholar] [CrossRef]
- Nie, B.; Henion, J.; Ryona, I. The Role of Mass Spectrometry in the Cannabis Industry. J. Am. Soc. Mass Spectrom. 2019, 30, 719–730. [Google Scholar] [CrossRef] [PubMed]
- Nelson, J.; Jones, C.; Heckle, S.; Anderson, L. Determination of Heavy Metals in a Variety of Cannabis and Cannabis-Derived Products, First Action 2021 03. J. AOAC Int. 2022, 105, 1640–1651. [Google Scholar] [CrossRef]
- Laza, A.; Orozco, E.; Baldo, M.F.; Raba, J.; Aranda, P.R. Determination of Arsenic (V) in Cannabis Oil by Adsorption on Multiwall Carbon Nanotubes Thin Film Using XRF Technique. Microchem. J. 2020, 158, 105265. [Google Scholar] [CrossRef]
- Tran, J.; Vassiliadis, S.; Elkins, A.C.; Cogan, N.O.I.; Rochfort, S.J. Developing Prediction Models Using Near-Infrared Spectroscopy to Quantify Cannabinoid Content in Cannabis sativa. Sensors 2023, 23, 2607. [Google Scholar] [CrossRef] [PubMed]
- Deidda, R.; Coppey, F.; Damergi, D.; Schelling, C.; Coïc, L.; Veuthey, J.-L.; Sacré, P.-Y.; De Bleye, C.; Hubert, P.; Esseiva, P.; et al. New Perspective for the In-Field Analysis of Cannabis Samples Using Handheld near-Infrared Spectroscopy: A Case Study Focusing on the Determination of Δ9-Tetrahydrocannabinol. J. Pharm. Biomed. Anal. 2021, 202, 114150. [Google Scholar] [CrossRef]
- Lu, Y.; Li, X.; Young, S.; Li, X.; Linder, E.; Suchoff, D. Hyperspectral Imaging with Chemometrics for Non-Destructive Determination of Cannabinoids in Floral and Leaf Materials of Industrial Hemp (Cannabis sativa L.). Comput. Electron. Agric. 2022, 202, 107387. [Google Scholar] [CrossRef]
- Rossi, G.B.; Lozano, V.A. Simultaneous Determination of Quality Parameters in Yerba Mate (Ilex Paraguariensis) Samples by Application of near-Infrared (NIR) Spectroscopy and Partial Least Squares (PLS). LWT 2020, 126, 109290. [Google Scholar] [CrossRef]
- Sajid, M.; Płotka-Wasylka, J. Green Analytical Chemistry Metrics: A Review. Talanta 2022, 238, 123046. [Google Scholar] [CrossRef] [PubMed]
- Gloerfelt-Tarp, F.; Hewavitharana, A.K.; Mieog, J.; Palmer, W.M.; Fraser, F.; Ansari, O.; Kretzschmar, T. Using a Global Diversity Panel of Cannabis sativa L. to Develop a near InfraRed-Based Chemometric Application for Cannabinoid Quantification. Sci. Rep. 2023, 13, 2253. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Carnerero Callado, C.; Núñez-Sánchez, N.; Casano, S.; Ferreiro-Vera, C. The Potential of near Infrared Spectroscopy to Estimate the Content of Cannabinoids in Cannabis sativa L.: A Comparative Study. Talanta 2018, 190, 147–157. [Google Scholar] [CrossRef]
- Deidda, R.; Dispas, A.; De Bleye, C.; Hubert, P.; Ziemons, É. Critical Review on Recent Trends in Cannabinoid Determination on Cannabis Herbal Samples: From Chromatographic to Vibrational Spectroscopic Techniques. Anal. Chim. Acta 2022, 1209, 339184. [Google Scholar] [CrossRef] [PubMed]
- Pereira, J.F.Q.; Pimentel, M.F.; Amigo, J.M.; Honorato, R.S. Detection and Identification of Cannabis sativa L. Using near Infrared Hyperspectral Imaging and Machine Learning Methods. A Feasibility Study. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2020, 237, 118385. [Google Scholar] [CrossRef]
- Borille, B.T.; Marcelo, M.C.A.; Ortiz, R.S.; Mariotti, K.d.C.; Ferrão, M.F.; Limberger, R.P. Near Infrared Spectroscopy Combined with Chemometrics for Growth Stage Classification of Cannabis Cultivated in a Greenhouse from Seized Seeds. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2017, 173, 318–323. [Google Scholar] [CrossRef]
- Duchateau, C.; Kauffmann, J.; Canfyn, M.; Stévigny, C.; De Braekeleer, K.; Deconinck, E. Discrimination of Legal and Illegal Cannabis Spp. According to European Legislation Using near Infrared Spectroscopy and Chemometrics. Drug Test. Anal. 2020, 12, 1309–1319. [Google Scholar] [CrossRef]
- Singh, M.; Bowman, M.J.; Berhow, M.A.; Price, N.P.J.; Liu, S.X. Application of near Infrared Spectroscopy for Determination of Relationship between Crop Year, Maturity Group, Location, and Carbohydrate Composition in Soybeans. Crop Sci. 2021, 61, 2409–2422. [Google Scholar] [CrossRef]
- Beć, K.B.; Grabska, J.; Huck, C.W. NIR Spectroscopy of Natural Medicines Supported by Novel Instrumentation and Methods for Data Analysis and Interpretation. J. Pharm. Biomed. Anal. 2021, 193, 113686. [Google Scholar] [CrossRef]
- Jarén, C.; Zambrana, P.C.; Pérez-Roncal, C.; López-Maestresalas, A.; Ábrego, A.; Arazuri, S. Potential of NIRS Technology for the Determination of Cannabinoid Content in Industrial Hemp (Cannabis sativa L.). Agronomy 2022, 12, 938. [Google Scholar] [CrossRef]
- Jianhua Yi Yifei Sun, Z.Z.N.L.; Lu, J. Near-Infrared Reflectance Spectroscopy for the Prediction of Chemical Composition in Walnut Kernel. Int. J. Food Prop. 2017, 20, 1633–1642. [Google Scholar] [CrossRef]
- Chen, Z.; de Boves Harrington, P.; Griffin, V.; Griffin, T. In Situ Determination of Cannabidiol in Hemp Oil by Near-Infrared Spectroscopy. J. Nat. Prod. 2021, 84, 2851–2857. [Google Scholar] [CrossRef] [PubMed]
- Roggo, Y.; Chalus, P.; Maurer, L.; Lema-Martinez, C.; Edmond, A.; Jent, N. A Review of near Infrared Spectroscopy and Chemometrics in Pharmaceutical Technologies. J. Pharm. Biomed. Anal. 2007, 44, 683–700. [Google Scholar] [CrossRef]
- Jackson, J.E. Getting Started. In A User’s Guide to Principal Components; John Wiley & Sons: Hoboken, NJ, USA, 1991; pp. 4–25. [Google Scholar]
- Bro, R.; Smilde, A.K. Principal Component Analysis. Anal. Methods 2014, 6, 2812–2831. [Google Scholar] [CrossRef]
- Wilson, N.; Heinrich, M. The Use of Near Infrared Spectroscopy to Discriminate between THC-Rich and Hemp Forms of Cannabis. Planta Med. 2006, 72, P_260. [Google Scholar] [CrossRef]
- Brereton, R.G. Pattern Recognition. In Chemometrics: Data Analysis for the Laboratory and Chemical Plant; John Wiley & Sons, Ltd: Chichester, UK, 2003; pp. 183–269. [Google Scholar]
- Gülağız, F.K.; Şahin, S. Comparison of Hierarchical and Non-Hierarchical Clustering Algorithms. Int. J. Comput. Eng. Inf. Technol. 2017, 9, 6–14. [Google Scholar]
- Wold, S. Pattern Recognition by Means of Disjoint Principal Components Models. Pattern Recognit. 1976, 8, 127–139. [Google Scholar] [CrossRef]
- Wold, S.; Sjöström, M. SIMCA: A Method for Analyzing Chemical Data in Terms of Similarity and Analogy. In Chemometrics: Theory and Application; ACS Publications: Washington, DC, USA, 1977; pp. 243–282. [Google Scholar]
- Chen, Z.; de Boves Harrington, P. Automatic Soft Independent Modeling for Class Analogies. Anal. Chim. Acta 2019, 1090, 47–56. [Google Scholar] [CrossRef]
- Vitale, R.; Cocchi, M.; Biancolillo, A.; Ruckebusch, C.; Marini, F. Class Modelling by Soft Independent Modelling of Class Analogy: Why, When, How? A Tutorial. Anal. Chim. Acta 2023, 1270, 341304. [Google Scholar] [CrossRef]
- Lee, L.C.; Liong, C.-Y.; Jemain, A.A. Partial Least Squares-Discriminant Analysis (PLS-DA) for Classification of High-Dimensional (HD) Data: A Review of Contemporary Practice Strategies and Knowledge Gaps. Analyst 2018, 143, 3526–3539. [Google Scholar] [CrossRef]
- Risoluti, R.; Gullifa, G.; Battistini, A.; Materazzi, S. Monitoring of Cannabinoids in Hemp Flours by MicroNIR/Chemometrics. Talanta 2020, 211, 120672. [Google Scholar] [CrossRef]
- Risoluti, R.; Gullifa, G.; Battistini, A.; Materazzi, S. MicroNIR/Chemometrics: A New Analytical Platform for Fast and Accurate Detection of Δ9-Tetrahydrocannabinol (THC) in Oral Fluids. Drug Alcohol Depend. 2019, 205, 107578. [Google Scholar] [CrossRef]
- San Nicolas, M.; Villate, A.; Alvarez-Mora, I.; Olivares, M.; Aizpurua-Olaizola, O.; Amigo, J. NIR-Hyperspectral Imaging and Machine Learning for Non-Invasive Chemotype Classification in Cannabis sativa L.; Elsevier: Amsterdam, The Netherlands, 2023. [Google Scholar]
- Shaffer, R.E.; Rose-Pehrsson, S.L.; McGill, R.A. A Comparison Study of Chemical Sensor Array Pattern Recognition Algorithms. Anal. Chim. Acta 1999, 384, 305–317. [Google Scholar] [CrossRef]
- Wang, H.; Chen, D.; Chen, Y. The Integrated Strategy of Pattern Classification and Its Application in Chemistry. Chemom. Intell. Lab. Syst. 2004, 70, 23–31. [Google Scholar] [CrossRef]
- Specht, D.F. Probabilistic Neural Networks. Neural Netw. 1990, 3, 109–118. [Google Scholar] [CrossRef]
- Park, J.; Sandberg, I.W. Universal Approximation Using Radial-Basis-Function Networks. Neural Comput. 1991, 3, 246–257. [Google Scholar] [CrossRef] [PubMed]
- Valinger, D.; Jurina, T.; Šain, A.; Matešić, N.; Panić, M.; Benković, M.; Gajdoš Kljusurić, J.; Jurinjak Tušek, A. Development of ANN Models Based on Combined UV-vis-NIR Spectra for Rapid Quantification of Physical and Chemical Properties of Industrial Hemp Extracts. Phytochem. Anal. 2021, 32, 326–338. [Google Scholar] [CrossRef] [PubMed]
- Aasim, M.; Katırcı, R.; Akgur, O.; Yildirim, B.; Mustafa, Z.; Nadeem, M.A.; Baloch, F.S.; Karakoy, T.; Yılmaz, G. Machine Learning (ML) Algorithms and Artificial Neural Network for Optimizing in Vitro Germination and Growth Indices of Industrial Hemp (Cannabis sativa L.). Ind. Crops Prod. 2022, 181, 114801. [Google Scholar] [CrossRef]
- Coomans, D.; Massart, D.L. Alternative K-Nearest Neighbour Rules in Supervised Pattern Recognition. Anal. Chim. Acta 1982, 138, 153–165. [Google Scholar] [CrossRef]
- Fisher, R.A. The Use of Multiple Measurements in Taxonomic Problems. Ann. Eugen. 1936, 7, 179–188. [Google Scholar] [CrossRef]
- Xanthopoulos, P.; Pardalos, P.M.; Trafalis, T.B. Linear Discriminant Analysis. In Robust Data Mining; Pardalos, P.M., Pintér, J.D., Robinson, S.M., Terlaky, T., Thai, M.T., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 27–33. [Google Scholar]
- Massart, D.L.; Vandeginste, B.G.M.; Deming, S.N.; Michotte, Y.; Kaufman, L. Supervised Pattern Recognition. In Chemometrics: A Textbook; Vandeginste, B.G.M., Kaufman, L., Eds.; Elsevier: Amsterdam, The Netherlands, 2003; Volume 2, pp. 385–413. [Google Scholar]
- Borregaard, T.; Nielsen, H.; Nørgaard, L.; Have, H. Crop–Weed Discrimination by Line Imaging Spectroscopy. J. Agric. Eng. Res. 2000, 75, 389–400. [Google Scholar] [CrossRef]
- Geskovski, N.; Stefkov, G.; Gigopulu, O.; Stefov, S.; Huck, C.W.; Makreski, P. Mid-Infrared Spectroscopy as Process Analytical Technology Tool for Estimation of THC and CBD Content in Cannabis Flowers and Extracts. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2021, 251, 119422. [Google Scholar] [CrossRef] [PubMed]
- Food and Drug Administration. International Council for Harmonisation Validation of Analytical Procedures Q2(R2); Food and Drug Administration: Rockville, MD, USA, 2022.
- Van Wyngaard, E.; Blancquaert, E.; Nieuwoudt, H.; Aleixandre-Tudo, J.L. Infrared Spectroscopy and Chemometric Applications for the Qualitative and Quantitative Investigation of Grapevine Organs. Front. Plant Sci. 2021, 12, 723247. [Google Scholar] [CrossRef] [PubMed]
- Helland, I. Partial Least Squares Regression. In Wiley StatsRef: Statistics Reference Online; Wiley: New York, NY, USA, 2014. [Google Scholar]
- Espel Grekopoulos, J. Construction and Validation of Quantification Methods for Determining the Cannabidiol Content in Liquid Pharma-Grade Formulations by Means of Near-Infrared Spectroscopy and Partial Least Squares Regression. Med. Cannabis Cannabinoids 2019, 2, 43–55. [Google Scholar] [CrossRef] [PubMed]
- Toonen, M.A.J.; Maliepaard, C.; Reijmers, T.H.; van der Voet, H.; Mastebroek, H.D.; van den Broeck, H.C.; Ebskamp, M.J.M.; Kessler, W.; Kessler, R.W. Predicting the Chemical Composition of Fibre and Core Fraction of Hemp (Cannabis sativa L.). Euphytica 2004, 140, 39–45. [Google Scholar] [CrossRef]
- Williams, P. The RPD Statistic: A Tutorial Note. NIR News 2014, 25, 22–26. [Google Scholar] [CrossRef]
- Jamwal, R.; Amit; Kumari, S.; Balan, B.; Kelly, S.; Cannavan, A.; Singh, D.K. Rapid and Non-Destructive Approach for the Detection of Fried Mustard Oil Adulteration in Pure Mustard Oil via ATR-FTIR Spectroscopy-Chemometrics. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2021, 244, 118822. [Google Scholar] [CrossRef]
- Townsend, D.; Eustis, I.; Lewis, M.; Rodgers, S.; Smith, K.; Bohman, A. The Determination of Total THC and CBD Content in Cannabis Flower by Fourier Transform Near Infrared Spectroscopy; PerkinElmer: Waltham, MA, USA, 2018; pp. 1–5. [Google Scholar]
- Marill, K.A. Advanced Statistics: Linear Regression, Part II: Multiple Linear Regression. Acad. Emerg. Med. 2004, 11, 94–102. [Google Scholar] [CrossRef]
- Zou, J.; Han, Y.; So, S.-S. Overview of Artificial Neural Networks. In Artificial Neural Networks—Methods and Applications; Humana Press: Totowa, NJ, USA, 2008; pp. 14–22. [Google Scholar]
- Mammone, A.; Turchi, M.; Cristianini, N. Support Vector Machines. WIREs Comput. Stat. 2009, 1, 283–289. [Google Scholar] [CrossRef]
- Pisner, D.A.; Schnyer, D.M. Support Vector Machine. In Machine Learning; Elsevier: Amsterdam, The Netherlands, 2020; pp. 101–121. [Google Scholar]
- Deidda, R.; Damergi, D.; Coppey, F.; De Bleye, C.; Coic, L.; Sacre, P.-Y.; Hubert, P.; Ziemons, E.; Esseiva, P.; Veuthey, J.-L. Handheld Near Infrared Spectroscopy for Cannabis Analysis: From the Analytical Problem to the Chemometric Solution. Chimiométrie 2020, 1, 2–3. Available online: https://orbi.uliege.be/handle/2268/244774 (accessed on 10 August 2023).
- Adesokan, M.; Alamu, E.O.; Otegbayo, B.; Maziya-Dixon, B. A Review of the Use of Near-Infrared Hyperspectral Imaging (NIR-HSI) Techniques for the Non-Destructive Quality Assessment of Root and Tuber Crops. Appl. Sci. 2023, 13, 5226. [Google Scholar] [CrossRef]
- Lu, B.; Dao, P.D.; Liu, J.; He, Y.; Shang, J. Recent Advances of Hyperspectral Imaging Technology and Applications in Agriculture. Remote Sens. 2020, 12, 2659. [Google Scholar] [CrossRef]
- Holmes, W.S.; Ooi, M.P.-L.; Abeysekera, S.; Kuang, Y.C.; Simpkin, R.; Caddie, M.; Nowak, J.; Demidenko, S. On Machine Learning Methods to Estimate Cannabidiolic Acid Content of Cannabis sativa L. from near-Infrared Hyperspectral Imaging. In Proceedings of the 2023 IEEE International Instrumentation and Measurement Technology Conference (I2MTC), Kuala Lumpur, Malaysia, 22–25 May 2023; IEEE: Piscataway, NJ, USA; pp. 1–6. [Google Scholar]
- Heil, K.; Schmidhalter, U. An Evaluation of Different NIR-Spectral Pre-Treatments to Derive the Soil Parameters C and N of a Humus-Clay-Rich Soil. Sensors 2021, 21, 1423. [Google Scholar] [CrossRef]
- Abeysekera, S.K.; Robinson, A.; Ooi, M.P.-L.; Kuang, Y.C.; Manley-Harris, M.; Holmes, W.; Hirst, E.; Nowak, J.; Caddie, M.; Steinhorn, G.; et al. Sparse Reproducible Machine Learning for near Infrared Hyperspectral Imaging: Estimating the Tetrahydrocannabinolic Acid Concentration in Cannabis sativa L. Ind. Crops Prod. 2023, 192, 116137. [Google Scholar] [CrossRef]
- Wang, C.; Liu, B.; Liu, L.; Zhu, Y.; Hou, J.; Liu, P.; Li, X. A Review of Deep Learning Used in the Hyperspectral Image Analysis for Agriculture. Artif. Intell. Rev. 2021, 54, 5205–5253. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Díaz-Liñán, M.d.C.; Sánchez de Medina, V.; Ferreiro-Vera, C.; García-Valverde, M.T. Light and Shadow in Near-Infrared Spectroscopy: A Powerful Tool for Cannabis sativa L. Analysis. AppliedChem 2023, 3, 526-545. https://doi.org/10.3390/appliedchem3040033
Díaz-Liñán MdC, Sánchez de Medina V, Ferreiro-Vera C, García-Valverde MT. Light and Shadow in Near-Infrared Spectroscopy: A Powerful Tool for Cannabis sativa L. Analysis. AppliedChem. 2023; 3(4):526-545. https://doi.org/10.3390/appliedchem3040033
Chicago/Turabian StyleDíaz-Liñán, María del Carmen, Verónica Sánchez de Medina, Carlos Ferreiro-Vera, and María Teresa García-Valverde. 2023. "Light and Shadow in Near-Infrared Spectroscopy: A Powerful Tool for Cannabis sativa L. Analysis" AppliedChem 3, no. 4: 526-545. https://doi.org/10.3390/appliedchem3040033
APA StyleDíaz-Liñán, M. d. C., Sánchez de Medina, V., Ferreiro-Vera, C., & García-Valverde, M. T. (2023). Light and Shadow in Near-Infrared Spectroscopy: A Powerful Tool for Cannabis sativa L. Analysis. AppliedChem, 3(4), 526-545. https://doi.org/10.3390/appliedchem3040033






