What Applied Physical Chemistry Can Contribute to Understanding Cancer: Toward the Next Generation of Breakthroughs
Abstract
1. Introduction
2. Re-Visiting a Few Basic Chemical Principles in a Cellular Context
2.1. Concentration
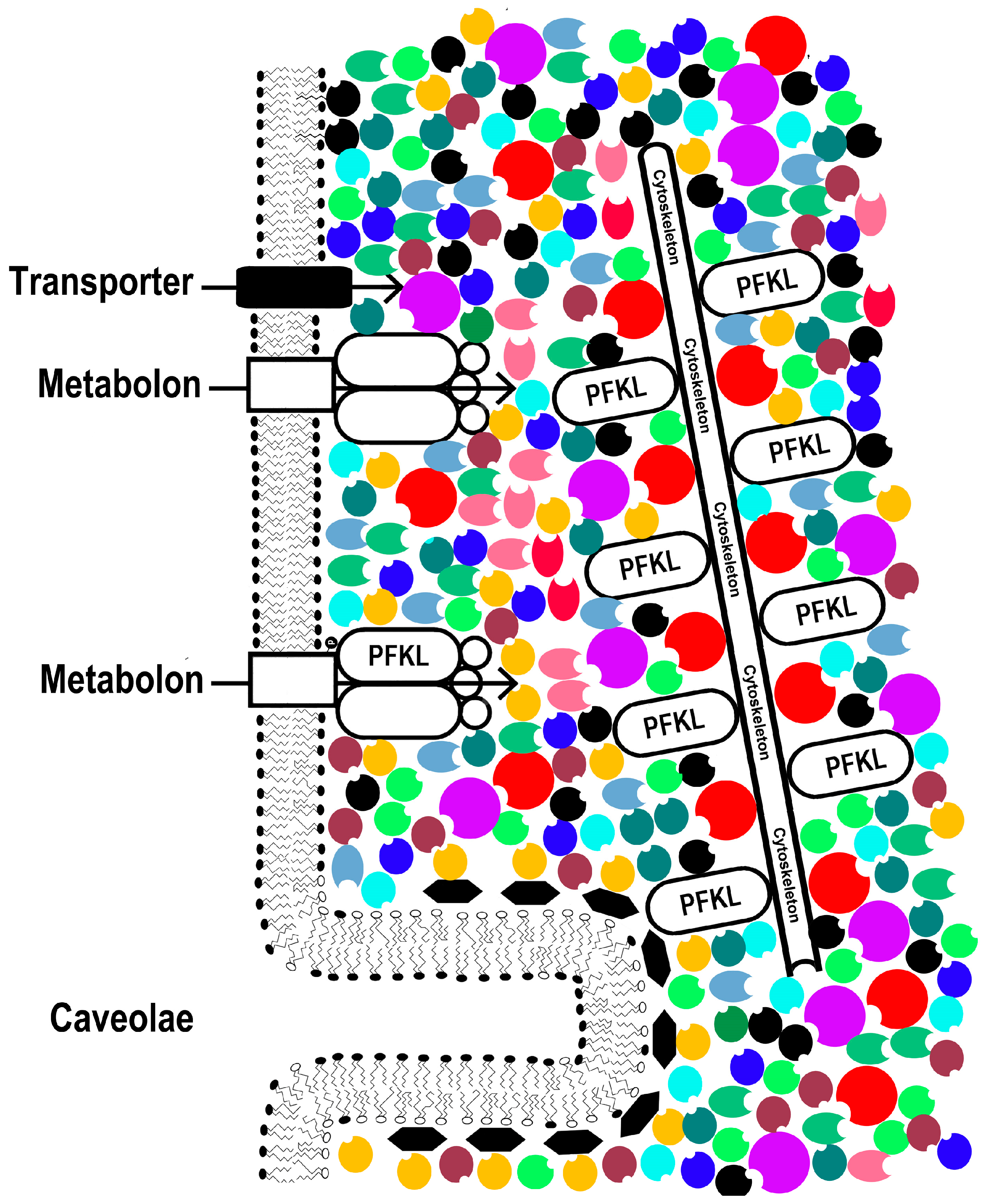
2.2. Intracellular Conditions: Concentration, Solvation, and Proximity
2.3. Phase Separations in Two and Three Dimensions
2.4. Redox Conditions
3. Carcinogenicity
4. Chemical Engineering Meets Cancer Biology: Enzyme Agglomeration in Cancer Recurrences
5. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Crick, F. Of Molecules and Men; Washington University Press: Seattle, WA, USA, 1966; p. 10. [Google Scholar]
- Ledford, H. How to solve the world’s biggest problems. Nature 2015, 525, 308–311. [Google Scholar] [CrossRef] [PubMed]
- Stone, F. Deconstructing silos and supporting collaboration. Employ. Relat. Today 2009, 31, 11–18. [Google Scholar] [CrossRef]
- Andrés, C.M.C.; Pérez de la Lastra, J.M.; Andrés Juan, C.; Plou, F.J.; Pérez-Lebeña, E. Superoxide anion chemistry-its role at the core of the innate immunity. Int. J. Mol. Sci. 2023, 24, 1841. [Google Scholar] [CrossRef] [PubMed]
- Missirlis, F.; Hu, J.; Kirby, K.; Hilliker, A.J.; Rouault, T.A.; Phillips, J.P. Compartment-specific protection of iron-sulfur proteins by superoxide dismutase. J. Biol. Chem. 2003, 278, 47365–47369. [Google Scholar] [CrossRef]
- Aikens, J.; Dix, T.A. Perhydroxyl radical (HOO·) initiated lipid peroxidation. The role of fatty acid hydroperoxides. J. Biol. Chem. 1991, 266, 15091–15098. [Google Scholar] [CrossRef]
- Bharara, M.S.; Atwood, D.A. Oxygen: Inorganic chemistry. In Encyclopedia of Inorganic Chemistry; King, R.B., Crabtree, R.H., Lukehart, C.M., Atwood, D.A., Scott, R.A., Eds.; Wiley Online Library: Hoboken, NJ, USA, 2006. [Google Scholar] [CrossRef]
- Li, D.; Wang, Y.; Liu, Z.-P. Academic background of Nobel prize laureates reveals the importance of multidisciplinary education in medicine. Soc. Sci. Hum. Open 2021, 3, 100114. [Google Scholar] [CrossRef]
- Traut, T.W. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 1994, 140, 1–22. [Google Scholar] [CrossRef]
- Betel, I. The endogenous substrate for nuclear oxidative phosphorylation. Arch. Biochem. Biophys. 1969, 134, 271–274. [Google Scholar] [CrossRef]
- Miller, D.S.; Horowitz, S.B. Intracellular compartmentalization of adenosine triphosphate. J. Biol. Chem. 1986, 261, 13911–13915. [Google Scholar] [CrossRef]
- Zimmerman, J.J.; von Saint André-von Arnim, A.; McLaughlin, J. Chapter 74—Cellular Respiration. In Pediatric Critical Care, 4th ed.; Fuhrman, B.P., Zimmerman, J.J., Eds.; Mosby: Maryland Heights, MO, USA, 2011; pp. 1058–1072. [Google Scholar]
- Liu, L.; Liang, X.; Li, Z.; Zhang, M.; Gao, M. Detection of ATP in cancer cells with a label-free fluorescent aptasensor. Nanomedicine 2022, 17, 765–774. [Google Scholar] [CrossRef]
- Imamura, H.; Nhat, K.P.; Togawa, H.; Saito, K.; Iino, R.; Kato-Yamada, Y.; Nagai, T.; Noji, H. Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. Proc. Natl. Acad. Sci. USA 2009, 106, 15651–15656. [Google Scholar] [CrossRef] [PubMed]
- Tamima, U.; Sarkar, S.; Islam, M.R.; Shil, A.; Kim, K.H.; Reo, Y.J.; Jun, Y.W.; Banna, H.; Lee, S.; Ahn, K.H. A small-molecule fluorescence probe for nuclear ATP. Angew. Chem. Int. Ed. 2023, 62, e202300580. [Google Scholar] [CrossRef] [PubMed]
- Boukouris, A.E.; Zervopoulos, S.D.; Michelakis, E.D. Metabolic enzymes moonlighting in the nucleus: Metabolic regulation of gene transcription. Trends Biochem. Sci. 2016, 41, 712–730. [Google Scholar] [CrossRef] [PubMed]
- Huangyang, P.; Simon, M.C. Hidden features: Exploring the non-canonical functions of metabolic enzymes. Dis. Model. Mech. 2018, 11, dmm033365. [Google Scholar] [CrossRef]
- Wright, R.H.; Lioutas, A.; Le Dily, F.; Soronellas, D.; Pohl, A.; Bonet, J.; Nacht, A.S.; Samino, S.; Font-Mateu, J.; Vicent, G.P.; et al. ADP-ribose-derived nuclear ATP synthesis by NUDIX5 is required for chromatin remodeling. Science 2016, 352, 1221–1225. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, L.Q.; Yang, C.C.; Li, J.; Tian, X.Y.; Li, D.N.; Cui, J.; Cai, J.P. The high expression of NUDT5 indicates poor prognosis of breast cancer by modulating AKT/Cyclin D signaling. PLoS ONE 2021, 16, e0245876. [Google Scholar] [CrossRef]
- Tan, D.; Zhang, Y. Silencing of Nudix type 5 represses proliferation and invasion and enhances chemosensitivity of gastric carcinoma cells by affecting the AKT/GSK-3β/β-catenin pathway. Toxicol. Appl. Pharmacol. 2022, 441, 115968. [Google Scholar] [CrossRef]
- Held, C.; Cameretti, L.F.; Sadowski, G. Measuring and modeling activity coefficients in aqueous amino acid solutions. Ind. Eng. Chem. Res. 2011, 50, 131–141. [Google Scholar] [CrossRef]
- Patel, A.; Malinovska, L.; Saha, S.; Wang, J.; Alberti, S.; Krishnan, Y.; Hyman, A.A. ATP as a biological hydrotrope. Science 2017, 356, 753–756. [Google Scholar] [CrossRef]
- Bilan, D.S.; Shokhina, A.G.; Lukyanov, S.A.; Belousov, V.V. Main cellular redox couples. Russ. J. Bioorganic Chem. 2015, 41, 341–356. [Google Scholar] [CrossRef]
- Martinovich, G.G.; Cherenkevich, S.N.; Sauer, H. Intracellular redox state: Towards quantitative description. Eur. Biophys. J. 2005, 34, 937–942. [Google Scholar] [CrossRef] [PubMed]
- Aprelev, A.; Weng, W.; Zakharov, M.; Rotter, M.; Yosmanovich, D.; Kwong, S.; Briehl, R.W.; Ferrone, F.A. Metastable polymerization of sickle hemoglobin in droplets. J. Mol. Biol. 2007, 369, 1170–1174. [Google Scholar] [CrossRef] [PubMed]
- Canterino, J.E.; Galkin, O.; Vekilov, P.G.; Hirsch, R.E. Phase separation and crystallization of hemoglobin C in transgenic mouse and human erythrocytes. Biophys. J. 2008, 95, 4025–4033. [Google Scholar] [CrossRef] [PubMed]
- Ciryam, P.; Kundra, R.; Morimoto, R.I.; Dobson, C.M.; Vendruscolo, M. Supersaturation is a major driving force for protein aggregation in neurodegenerative diseases. Trends Pharmacol. Sci. 2015, 36, 72–77. [Google Scholar] [CrossRef]
- Ellis, R.J. Macromolecular crowding: An important but neglected aspect of the intracellular environment. Curr. Opin. Struct. Biol. 2001, 11, 114–119. [Google Scholar] [CrossRef]
- Bañuelos, M.; Gancedo, C. In situ study of the glycolytic pathway in Saccharomyces cerevisiae. Arch. Microbiol. 1978, 117, 197–201. [Google Scholar] [CrossRef]
- Aragón, J.J.; Felíu, J.E.; Frenkel, R.A.; Sols, A. Permeabilization of animal cells for kinetic studies of intracellular enzymes: In situ behavior of the glycolytic enzymes of erythrocytes. Proc. Natl. Acad. Sci. USA 1980, 77, 6324–6328. [Google Scholar] [CrossRef]
- Clegg, J.S.; Jackson, S.A. Glycolysis in permeabilized L-929 cells. Biochem. J. 1988, 255, 335–344. [Google Scholar]
- Walev, I.; Bhakdi, S.C.; Hofmann, F.; Djonder, N.; Valeva, A.; Aktories, K.; Bhakdi, S. Delivery of proteins into living cells by reversible membrane permeabilization with streptolysin-O. Proc. Natl. Acad. Sci. USA 2001, 98, 3185–3190. [Google Scholar] [CrossRef]
- Dunaway, G.A. A review of animal phosphofructokinase isozymes with an emphasis on their physiological role. Mol. Cell. Biochem. 1983, 52, 75–91. [Google Scholar] [CrossRef]
- Srivastava, D.K.; Bernhard, S.A. Biophysical chemistry of metabolic reaction sequences in concentrated enzyme solution and in the cell. Annu. Rev. Biophys. Biophys. Chem. 1987, 16, 175–204. [Google Scholar] [CrossRef] [PubMed]
- Aragón, J.J.; Sols, A. Regulation of enzyme activity in the cell: Effect of enzyme concentration. FASEB J. 1991, 5, 2945–2950. [Google Scholar] [CrossRef] [PubMed]
- Boscá, L.; Aragón, J.J.; Sols, A. Modulation of muscle phosphofructokinase at physiological concentration of enzyme. J. Biol. Chem. 1985, 260, 2100–2107. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Costa, O.H.; Estévez, A.M.; Sánchez, V.; Aragón, J.J. Purification and properties of phosphofructokinase from Dictyostelium discoideum. Eur. J. Biochem. 1994, 226, 1007–1017. [Google Scholar] [CrossRef]
- Petty, H.R. Enzyme trafficking and co-clustering precede and accurately predict human breast cancer recurrences: An interdisciplinary review. Am. J. Physiol. Cell Physiol. 2022, 322, C991–C1010. [Google Scholar] [CrossRef]
- Castellana, M.; Wilson, M.Z.; Xu, Y.; Joshi, P.; Cristea, I.M.; Rabinowitz, J.D.; Gitai, Z.; Wingreen, N.S. Enzyme clustering accelerates processing of intermediates through metabolic channeling. Nat. Biotechnol. 2014, 32, 1011–1018. [Google Scholar] [CrossRef] [PubMed]
- Bar-Even, A.; Noor, E.; Savir, Y.; Liebermeister, W.; Davidi, D.; Tawfik, D.S.; Milo, R. The moderately efficient enzyme: Evolutionary and physicochemical trends shaping enzyme parameters. Biochemistry 2011, 50, 4402–4410. [Google Scholar] [CrossRef]
- Doigneaux, C.; Pedley, A.M.; Mistry, I.N.; Papayova, M.; Benkovic, S.J.; Tavassoli, A. Hypoxia drives the assembly of the multienzyme purinosome complex. J. Biol. Chem. 2020, 295, 9551–9566. [Google Scholar] [CrossRef]
- An, S.; Kumar, R.; Sheets, E.D.; Benkovic, S.J. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science 2008, 320, 103–106. [Google Scholar] [CrossRef]
- French, J.B.; Jones, S.A.; Deng, H.; Pedley, A.M.; Kim, D.; Chan, C.Y.; Hu, H.; Pugh, R.J.; Zhao, H.; Zhang, Y.; et al. Spatial colocalization and functional link of purinosomes with mitochondria. Science 2016, 351, 733–737. [Google Scholar] [CrossRef]
- Chan, C.Y.; Pedley, A.M.; Kim, D.; Xia, C.; Zhuang, X.; Benkovic, S.J. Microtubule-directed transport of purine metabolons drives their cytosolic transit to mitochondria. Proc. Natl Acad. Sci. USA 2018, 115, 13009–13014. [Google Scholar] [CrossRef] [PubMed]
- Pareek, V.; Tian, H.; Winograd, N.; Benkovic, S.J. Metabolomics and mass spectrometry imaging reveal channeled de novo purine synthesis in cells. Science 2020, 368, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Pedley, A.M.; Benkovic, S.J. A new view into the regulation of purine metabolism: The Purinosome. Trends Biochem. Sci. 2017, 42, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Bar-Peled, L.; Kory, N. Principles and functions of metabolic compartmentalization. Nat. Metab. 2022, 4, 1232–1244. [Google Scholar] [CrossRef] [PubMed]
- Haluska, C.K.; Baptista, M.S.; Fernandes, A.U.; Schroder, A.P.; Marques, C.M.; Itri, R. Photo-activated phase separation in giant vesicles made from different lipid mixtures. Biochim. Biophys. Acta 2012, 1818, 666–672. [Google Scholar] [CrossRef]
- Baoukina, S.; Tieleman, D.P. Computer simulations of phase separation in lipid bilayers and monolayers. Methods Membr. Lipids 2015, 1232, 307–322. [Google Scholar]
- Williams, W.P. Cold-induced lipid phase transitions. Trans. R. Soc. Lond. B Biol. Sci. 1990, 326, 555–567. [Google Scholar]
- Sofińska, K.; Lupa, D.; Chachaj-Brekiesz, A.; Czaja, M.; Kobierski, J.; Seweryn, S.; Skirlińska-Nosek, K.; Szymonski, M.; Wilkosz, N.; Wnętrzak, A.; et al. Revealing local molecular distribution, orientation, phase separation, and formation of domains in artificial lipid layers: Towards comprehensive characterization of biological membranes. Adv. Colloid Interface Sci. 2022, 301, 102614. [Google Scholar] [CrossRef]
- Gohrbandt, M.; Lipski, A.; Grimshaw, J.W.; Buttress, J.A.; Baig, Z.; Herkenhoff, B.; Walter, S.; Kurre, R.; Deckers-Hebestreit, G.; Strahl, H. Low membrane fluidity triggers lipid phase separation and protein segregation in living bacteria. EMBO J. 2022, 41, e109800. [Google Scholar] [CrossRef]
- Leveille, C.L.; Cornell, C.E.; Merz, A.J.; Keller, S.L. Yeast cells actively tune their membranes to phase separate at temperatures that scale with growth temperatures. Proc. Natl. Acad. Sci. USA 2022, 119, e2116007119. [Google Scholar] [CrossRef]
- Xue, S.; Zhou, F.; Zhao, T.; Zhao, H.; Wang, X.; Chen, L.; Li, J.P.; Luo, S.Z. Phase separation on cell surface facilitates bFGF signal transduction with heparan sulphate. Nat. Commun. 2022, 13, 1112. [Google Scholar] [CrossRef] [PubMed]
- Rayermann, S.P.; Rayermann, G.E.; Cornell, C.E.; Merz, A.J.; Keller, S.L. Hallmarks of reversible separation of living, unperturbed cell membranes into two liquid phases. Biophys. J. 2017, 113, 2425–2432. [Google Scholar] [CrossRef] [PubMed]
- Toulmay, A.; Prinz, W.A. Direct imaging reveals stable, micrometer-scale lipid domains that segregate proteins in live cells. J. Cell Biol. 2013, 202, 35–44. [Google Scholar] [CrossRef]
- Sohn, H.W.; Tolar, P.; Jin, T.; Pierce, S.K. Fluorescence resonance energy transfer in living cells reveals dynamic membrane changes in the initiation of B cell signaling. Proc. Natl. Acad. Sci. USA 2006, 103, 8143–8148. [Google Scholar] [CrossRef] [PubMed]
- Bálint, Š.; Dustin, M.L. Localizing order to boost signaling. Elife 2017, 6, e25375. [Google Scholar] [CrossRef]
- Stone, M.B.; Shelby, S.A.; Núñez, M.F.; Wisser, K.; Veatch, S.L. Protein sorting by lipid phase-like domains supports emergent signaling function in B lymphocyte plasma membranes. Elife 2017, 6, e19891. [Google Scholar] [CrossRef]
- Tong, X.; Tang, R.; Xu, J.; Wang, W.; Zhao, Y.; Yu, X.; Shi, S. Liquid-liquid phase separation in tumor biology. Signal Transduct. Target. Ther. 2022, 7, 221. [Google Scholar] [CrossRef]
- Wang, B.; Zhang, L.; Dai, T.; Qin, Z.; Lu, H.; Zhang, L.; Zhou, F. Liquid-liquid phase separation in human health and diseases. Signal Transduct. Target. Ther. 2021, 6, 290. [Google Scholar] [CrossRef]
- Wright, R.H.G.; Le Dily, F.; Beato, M. ATP, Mg2+, Nuclear phase separation, and genome accessibility. Trends Biochem. Sci. 2019, 44, 565–574. [Google Scholar] [CrossRef]
- Das, S.; Lin, Y.H.; Vernon, R.M.; Forman-Kay, J.D.; Chan, H.S. Comparative roles of charge, π, and hydrophobic interactions in sequence-dependent phase separation of intrinsically disordered proteins. Proc. Natl. Acad. Sci. USA 2020, 117, 28795–28805. [Google Scholar] [CrossRef]
- Vernon, R.M.; Chong, P.A.; Tsang, B.; Kim, T.H.; Bah, A.; Farber, P.; Lin, H.; Forman-Kay, J.D. Pi-Pi contacts are an overlooked protein feature relevant to phase separation. Elife 2018, 7, e31486. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.; Xuan, Z.; Lagoy, R.C.; Jawerth, L.M.; Gonzalez, I.J.; Singh, M.; Prashad, S.; Kim, H.S.; Patel, A.; Albrecht, D.R.; et al. Phosphofructokinase relocalizes into subcellular compartments with liquid-like properties in vivo. Biophys. J. 2021, 120, 1170–1186. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Narlikar, G.J.; Kutateladze, T.G. Enzymatic reactions inside biological condensates. J. Mol. Biol. 2021, 433, 166624. [Google Scholar] [CrossRef] [PubMed]
- Nott, T.J.; Petsalaki, E.; Farber, P.; Jervis, D.; Fussner, E.; Plochowietz, A.; Craggs, T.D.; Bazett-Jones, D.P.; Pawson, T.; Forman-Kay, J.D.; et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell 2015, 57, 936–947. [Google Scholar] [CrossRef]
- Banani, S.F.; Lee, H.O.; Hyman, A.A.; Rosen, M.K. Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017, 18, 285–298. [Google Scholar] [CrossRef]
- Kuffner, A.M.; Prodan, M.; Zuccanni, R.; Capasso Palmiero, U.; Faltova, L.; Arosio, P. Acceleration of an enzymatic reaction in liquid phase separated compartments based on intrinsically disordered protein domains. ChemSystemsChem 2020, 2, e200001. [Google Scholar] [CrossRef]
- Alpha-Tocopherol, Beta Carotene Cancer Prevention Study Group. The effect of vitamin E and beta carotene on the incidence of lung cancer and other cancers in male smokers. N. Engl. J. Med. 1994, 330, 1029–1033. [Google Scholar] [CrossRef]
- Wondrak, G.T. Redox-directed cancer therapeutics: Molecular mechanisms and opportunities. Antioxid. Redox Signal. 2009, 11, 3013–3069. [Google Scholar] [CrossRef]
- Petty, H.R. Prognostic evaluation of ductal carcinoma in situ lesions using monoclonal antibodies and machine learning. In Handbook of Cancer and Immunology; Rezaei, N., Ed.; Springer-Nature: London, UK, 2022. [Google Scholar]
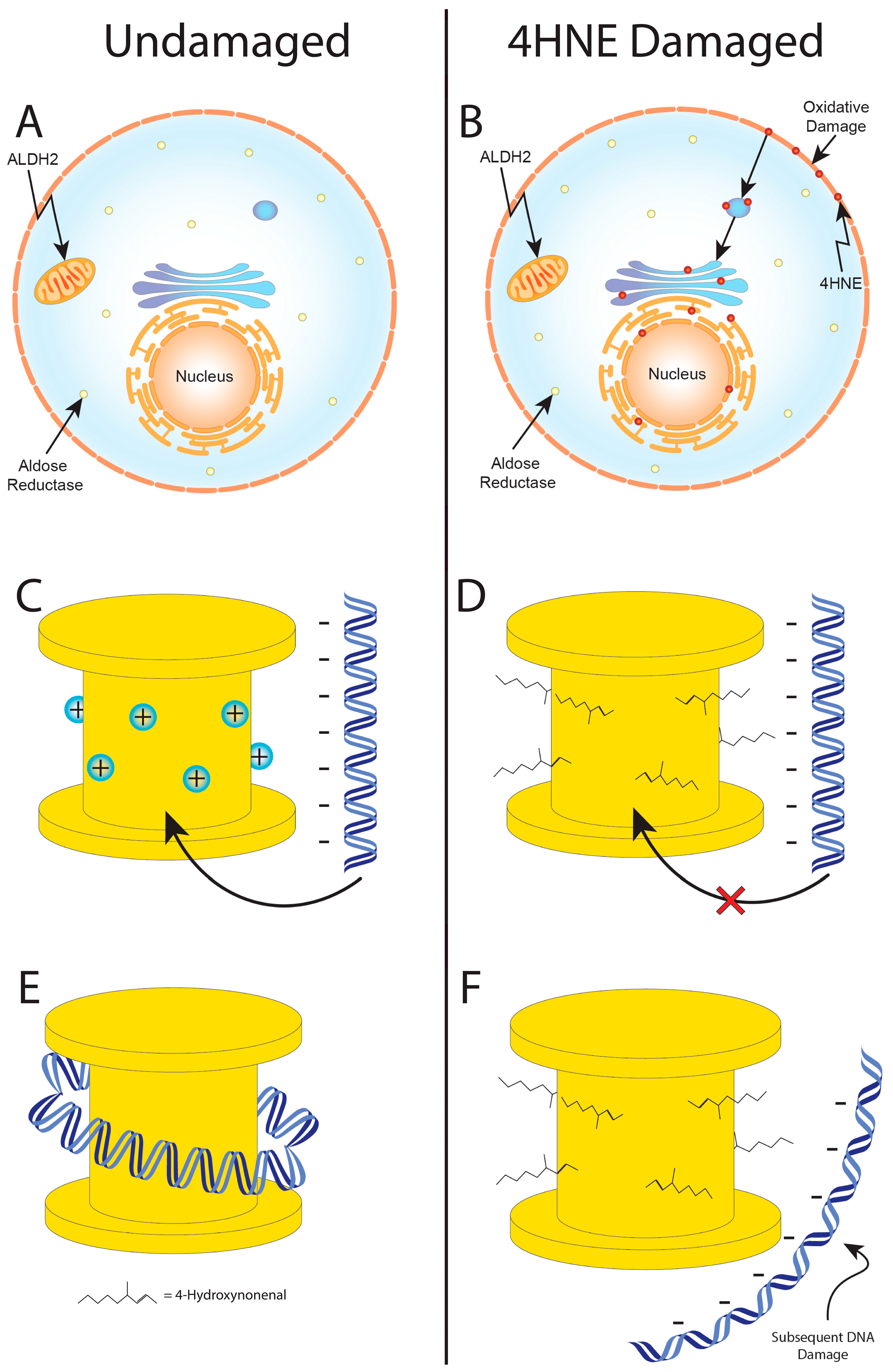
- Onufriev, A.V.; Schiessel, H. The nucleosome: From structure to function through physics. Curr. Opin. Struct. Biol. 2019, 56, 119–130. [Google Scholar] [CrossRef]
- Montanaro, L.; Treré, D.; Derenzini, M. Nucleolus, ribosomes, and cancer. Am. J. Pathol. 2008, 173, 301–310. [Google Scholar] [CrossRef]
- McAnena, P.; Brown, J.A.; Kerin, M.J. Circulating nucleosomes and nucleosome modifications as biomarkers in cancer. Cancers 2017, 9, 5. [Google Scholar] [CrossRef] [PubMed]
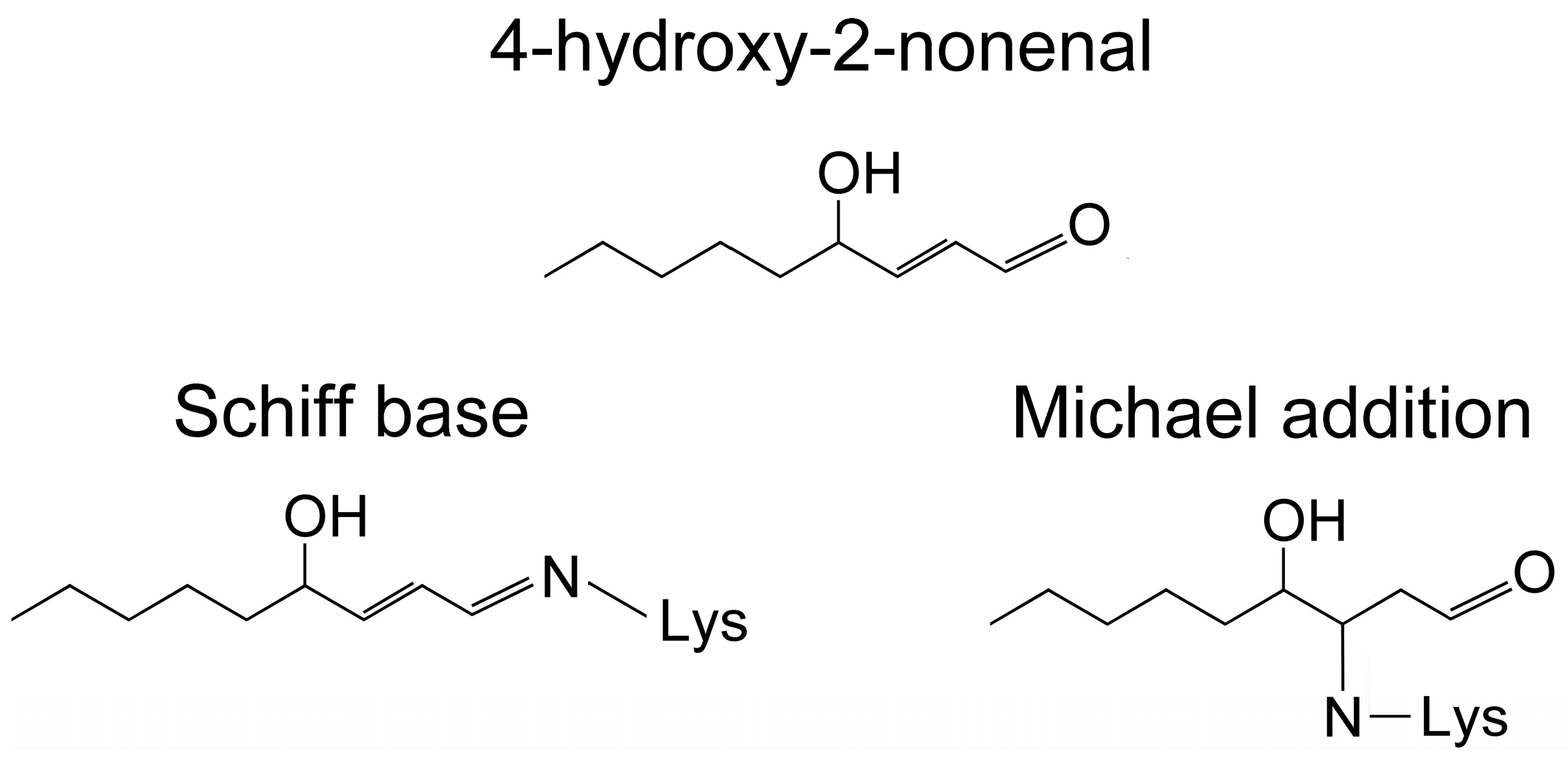
- Sayre, L.M.; Lin, D.; Yuan, Q.; Zhu, X.; Tang, X. Protein adducts generated from products of lipid oxidation: Focus on HNE and one. Drug Metab. Rev. 2006, 38, 651–675. [Google Scholar] [CrossRef] [PubMed]
- Rauniyar, N.; Prokai, L. Detection and identification of 4-hydroxy-2-nonenal Schiff-base adducts along with products of Michael addition using data-dependent neutral loss-driven MS3 acquisition: Method evaluation through an in vitro study on cytochrome c oxidase modifications. Proteomics 2009, 9, 5188–5193. [Google Scholar] [CrossRef] [PubMed]
- Vazdar, M.; Jurkiewicz, P.; Hof, M.; Jungwirth, P.; Cwiklik, L. Behavior of 4-hydroxynonenal in phospholipid membranes. J. Phys. Chem. B 2012, 116, 6411–6415. [Google Scholar] [CrossRef]
- Ou, X.; Lao, Y.; Xu, J.; Wutthinitikornkit, Y.; Shi, R.; Chen, X.; Li, J. ATP can efficiently stabilize protein through a unique mechanism. JACS Au 2021, 1, 1766–1777. [Google Scholar] [CrossRef]
- Galligan, J.J.; Rose, K.L.; Beavers, W.N.; Hill, S.; Tallman, K.A.; Tansey, W.P.; Marnett, L.J. Stable histone adduction by 4-oxo-2-nonenal: A potential link between oxidative stress and epigenetics. J. Am. Chem. Soc. 2014, 136, 11864–11866. [Google Scholar] [CrossRef]
- Galligan, J.J.; Marnett, L.J. Histone adduction and its functional impact on epigenetics. Chem. Res. Toxicol. 2017, 30, 376–387. [Google Scholar] [CrossRef]
- Schirmer, T.; Evans, P.R. Structural basis of the allosteric behaviour of phosphofructokinase. Nature 1990, 343, 140–145. [Google Scholar] [CrossRef]
- Robinson, J.B., Jr.; Inman, L.; Sumegi, B.; Srere, P.A. Further characterization of the Krebs tricarboxylic acid cycle metabolon. J. Biol. Chem. 1987, 262, 1786–1790. [Google Scholar] [CrossRef]
- Islam, M.M.; Nautiyal, M.; Wynn, R.M.; Mobley, J.A.; Chuang, D.T.; Hutson, S.M. Branched-chain amino acid metabolon: Interaction of glutamate dehydrogenase with the mitochondrial branched-chain aminotransferase (BCATm). J. Biol. Chem. 2010, 285, 265–276. [Google Scholar] [CrossRef]
- Chu, H.; Low, P.S. Mapping of glycolytic enzyme-binding sites on human erythrocyte band 3. Biochem. J. 2006, 400, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Araiza-Olivera, D.; Chiquete-Felix, N.; Rosas-Lemus, M.; Sampedro, J.G.; Peña, A.; Mujica, A.; Uribe-Carvajal, S. A glycolytic metabolon in Saccharomyces cerevisiae is stabilized by F-actin. FEBS J. 2013, 280, 3887–3905. [Google Scholar] [CrossRef] [PubMed]
- Raikar, L.S.; Vallejo, J.; Lloyd, P.G.; Hardin, C.D. Overexpression of caveolin-1 results in increased plasma membrane targeting of glycolytic enzymes: The structural basis for a membrane associated metabolic compartment. J. Cell. Biochem. 2006, 98, 861–871. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.; Puchulu-Campanella, E.; Galan, J.A.; Tao, W.A.; Low, P.S.; Hoffman, J.F. Identification of cytoskeletal elements enclosing the ATP pools that fuel human red blood cell membrane cation pumps. Proc. Natl. Acad. Sci. USA 2012, 109, 12794–12799. [Google Scholar] [CrossRef] [PubMed]
- Tanner, L.B.; Goglia, A.G.; Wei, M.H.; Sehgal, T.; Parsons, L.R.; Park, J.O.; White, E.; Toettcher, J.E.; Rabinowitz, J.D. Four key steps control glycolytic flux in mammalian cells. Cell Syst. 2018, 7, 49–62. [Google Scholar] [CrossRef]
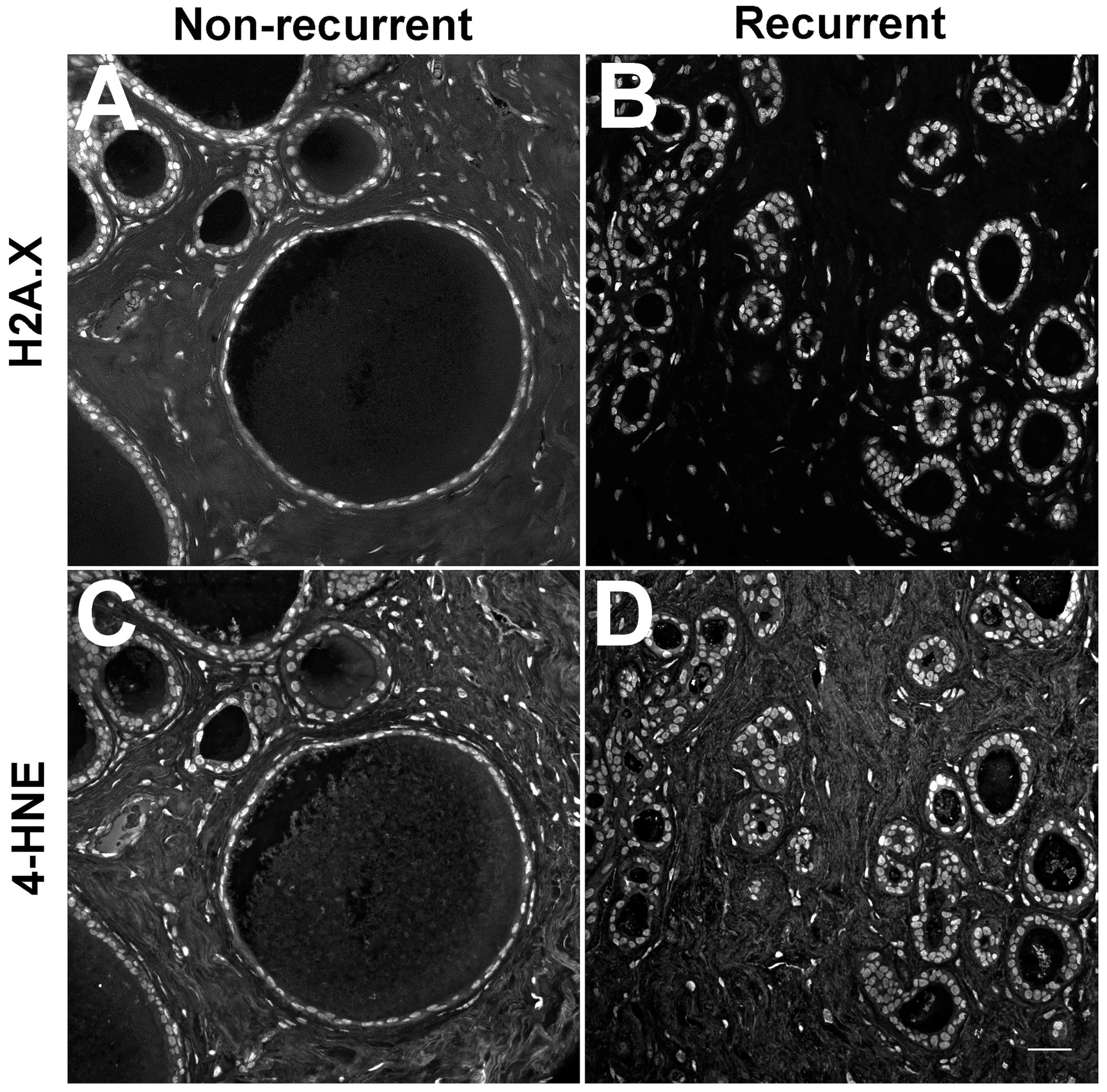
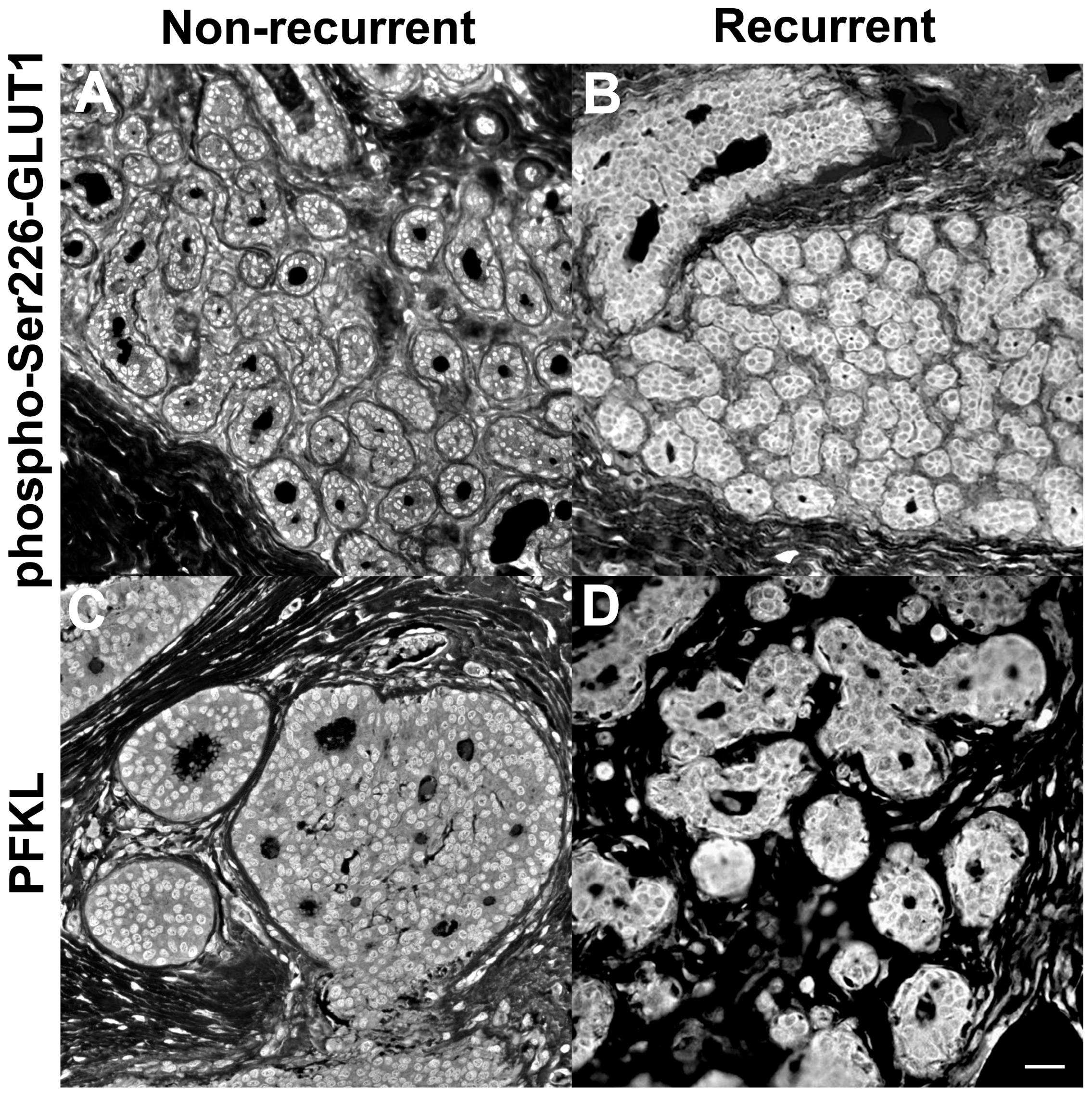
- Kraft, A.M.; Petty, H.R. Spatial locations of certain enzymes and transporters within preinvasive ductal epithelial cells predict human breast cancer recurrences. Am. J. Physiol. Cell Physiol. 2020, 319, C910–C921. [Google Scholar] [CrossRef]
- Cheung, R.A.; Kraft, A.M.; Petty, H.R. Relocation of phosphofructokinases within epithelial cells is a novel event preceding breast cancer recurrence that accurately predicts patient outcomes. Am. J. Physiol. Cell Physiol. 2021, 321, C654–C670. [Google Scholar] [CrossRef]
- Petty, H.R. Using machine vision of glycolytic elements to predict breast cancer recurrences: Design and implementation. Metabolites 2023, 3, 41. [Google Scholar] [CrossRef]
- Saatchi, Y.; Schanen, P.; Cheung, R.A.; Petty, H.R. Computer vision identifies recurrent and non-recurrent ductal carcinoma in situ lesions with special emphasis on African American women. Am. J. Pathol. 2023, in press. [Google Scholar] [CrossRef]
- Tomaiuolo, M.; Bertram, R.; Houle, D. Enzyme isoforms may increase phenotypic robustness. Evolution 2008, 62, 2884–2893. [Google Scholar] [CrossRef]
- Kahraman, A.; Karakulak, T.; Szklarczyk, D.; von Mering, C. Pathogenic impact of transcript isoform switching in 1,209 cancer samples covering 27 cancer types using an isoform-specific interaction network. Sci. Rep. 2020, 10, 14453. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.E.; Ma, J.; Sacharidou, A.; Mi, W.; Salato, V.K.; Nguyen, N.; Jiang, Y.; Pascual, J.M.; North, P.E.; Shaul, P.W.; et al. A protein kinase C phosphorylation motif in GLUT1 affects glucose transport and is mutated in GLUT1 deficiency syndrome. Mol. Cell 2015, 58, 845–853. [Google Scholar] [CrossRef] [PubMed]
- Kondrat, S.; Karuss, U.; von Lieres, E. Enzyme co-localization: Mechanisms and benefits. Curr. Res. Chem. Biol. 2022, 2, 100031. [Google Scholar] [CrossRef]
- Giunta, G.; Tostevin, F.; Tănase-Nicola, S.; Gerland, U. Optimal spatial allocation of enzymes as an investment problem. Commun. Phys. 2022, 5, 319. [Google Scholar] [CrossRef]
- Hinzpeter, F.; Tostevin, F.; Buchner, A.; Gerland, U. Trade-offs and design principles in the spatial organization of catalytic particles. Nat. Phys. 2022, 18, 203–211. [Google Scholar] [CrossRef]
- Fink, T.; Stevović, B.; Verwaal, R.; Roubos, J.A.; Gaber, R.; Benčina, M.; Jerala, R.; Gradišar, H. Metabolic enzyme clustering by coiled coils improves the biosynthesis of resveratrol and mevalonate. AMB Express 2020, 10, 97. [Google Scholar] [CrossRef]
- Wheeldon, I.; Minteer, S.D.; Banta, S.; Barton, S.C.; Atanassov, P.; Sigman, M. Substrate channelling as an approach to cascade reactions. Nat. Chem. 2016, 8, 299–309. [Google Scholar] [CrossRef]
- Schmidt, S.; Castiglione, K.; Kourist, R. Overcoming the incompatibility challenge in chemoenzymatic and multi-catalytic cascade reactions. Chemistry 2018, 24, 1755–1768. [Google Scholar] [CrossRef]
- Qiu, X.Y.; Xie, S.S.; Min, L.; Wu, X.M.; Zhu, L.Y.; Zhu, L. Spatial organization of enzymes to enhance synthetic pathways in microbial chassis: A systematic review. Microb. Cell Factories 2018, 17, 120. [Google Scholar] [CrossRef]
- Gopich, I.V. Cluster channeling in cascade reactions. J. Phys. Chem. B 2021, 125, 2061–2073. [Google Scholar] [CrossRef]
- Stępiński, D. The nucleolus, an ally, and an enemy of cancer cells. Histochem. Cell Biol. 2018, 150, 607–629. [Google Scholar] [CrossRef] [PubMed]
- Helpap, B. Nucleolar grading of breast cancer. Comparative studies on frequency and localization of nucleoli and histology, stage, hormonal receptor status and lectin histochemistry. Virchows Arch. A Pathol. Anat. Histopathol. 1989, 415, 501–508. [Google Scholar] [CrossRef] [PubMed]
- Carotenuto, P.; Pecoraro, A.; Palma, G.; Russo, G.; Russo, A. Therapeutic approaches targeting nucleolus in cancer. Cells 2019, 8, 1090. [Google Scholar] [CrossRef] [PubMed]
- Spannl, S.; Tereshchenko, M.; Mastromarco, G.J.; Ihn, S.J.; Lee, H.O. Biomolecular condensates in neurodegeneration and cancer. Traffic 2019, 20, 890–911. [Google Scholar] [CrossRef]
- Glass-Marmor, L.; Beitner, R. Taxol (paclitaxel) induces a detachment of phosphofructokinase from cytoskeleton of melanoma cells and decreases the levels of glucose 1,6-bisphosphate, fructose 1,6-bisphosphate and ATP. Eur. J. Pharmacol. 1999, 370, 195–199. [Google Scholar] [CrossRef]
- Dhyani, P.; Quispe, C.; Sharma, E.; Bahukhandi, A.; Sati, P.; Attri, D.C.; Szopa, A.; Sharifi-Rad, J.; Docea, O.; Mardare, I.; et al. Anticancer potential of alkaloids: A key emphasis to colchicine, vinblastine, vincristine, vindesine, vinorelbine and vincamine. Cancer Cell Int. 2022, 22, 206. [Google Scholar] [CrossRef]
- Schwartz, D.; Beitner, R. Detachment of the glycolytic enzymes, phosphofructokinase, and aldolase, from cytoskeleton of melanoma cells, induced by local anesthetics. Mol. Genet. Metab. 2000, 69, 159–164. [Google Scholar] [CrossRef]
- Grandhi, R.K.; Perona, B. Mechanisms of action by which local anesthetics reduce cancer recurrence, a systematic review. Pain Med. 2020, 21, 401–414. [Google Scholar] [CrossRef]
- Tejeda-Muñoz, N.; Mei, K.C.; Sheladiya, P.; Monka, J. Targeting membrane trafficking as a strategy for cancer treatment. Vaccines 2022, 10, 790. [Google Scholar] [CrossRef]
- Allal, C.; Favre, G.; Couderc, B.; Salicio, S.; Sixou, S.; Hamilton, A.D.; Sebti, S.M.; Lajoie-Mazenc, I.; Pradines, A. RhoA prenylation is required for promotion of cell growth and transformation and cytoskeleton organization but not for induction of serum response element transcription. J. Biol. Chem. 2000, 275, 31001–31008. [Google Scholar] [CrossRef]
- Wang, M.; Casey, P.J. Protein prenylation: Unique fats make their mark on biology. Nat. Rev. Mol. Cell Biol. 2016, 17, 110–122. [Google Scholar] [CrossRef] [PubMed]
- Harris, B.R.E.; Zhang, Y.; Tao, J.; Shen, R.; Zhao, X.; Cleary, M.P.; Wang, T.; Yang, D.Q. ATM inhibitor KU-55933 induces apoptosis and inhibits motility by blocking GLUT1-mediated glucose uptake in aggressive cancer cells with sustained activation of Akt. FASEB J. 2021, 35, e21264. [Google Scholar] [CrossRef] [PubMed]
- Csermely, P.; Agoston, V.; Pongor, S. The efficiency of multi-target drugs: The network approach might help drug design. Trends Pharmacol. Sci. 2005, 26, 178–182. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Kong, J.; Tucker-Burden, C.; Anand, M.; Rong, Y.; Rahman, F.; Moreno, C.S.; Van Meir, E.G.; Hadjipanayis, C.G.; Brat, D.J. Human Brat ortholog TRIM3 is a tumor suppressor that regulates asymmetric cell division in glioblastoma. Cancer Res. 2014, 74, 4536–4548. [Google Scholar] [CrossRef] [PubMed]
- Tiku, V.; Jain, C.; Raz, Y.; Nakamura, S.; Heestand, B.; Liu, W.; Späth, M.; Suchiman, H.E.D.; Müller, R.U.; Slagboom, P.E.; et al. Small nucleoli are a cellular hallmark of longevity. Nat. Commun. 2017, 8, 16083. [Google Scholar] [CrossRef]
| Influence Type | Selected Types and Examples 2 | |
|---|---|---|
| Local Environment | pH | PFK |
| Redox Condition | Oxidative, Reductive | |
| Ionic Condition | Potassium, Calcium | |
| Dielectric Constant | Active sites, LLPS | |
| Electrostatic | Nucleosomes and other binding events | |
| Chemical | Concentration | Activity coefficient, effective concentration |
| Isoform switching | PFK | |
| Phosphorylation | Signaling, enzyme, and transport molecules | |
| Solubility | Proteins, metabolites | |
| Low affinity binding sites | Asp-Asp-Thy-Glu-Asp sequences | |
| High affinity binding sites | Ligands, cofactors, ATP, GTP, Allosteric factors, ubiquitination, prenylation, etc. | |
| Emergent Behaviors | Hydrotropes | ATP, GTP, and other amphipathic molecules |
| Phase separations | Two and three dimensional | |
| Feedback | Positive and negative | |
| Translocation | Microfilaments and microtubules | |
| Assembly state | Monomer, oligomer, mixed oligomers, clusters, metabolons, metabolic platforms |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schanen, P.; Petty, H.R. What Applied Physical Chemistry Can Contribute to Understanding Cancer: Toward the Next Generation of Breakthroughs. AppliedChem 2023, 3, 378-399. https://doi.org/10.3390/appliedchem3030024
Schanen P, Petty HR. What Applied Physical Chemistry Can Contribute to Understanding Cancer: Toward the Next Generation of Breakthroughs. AppliedChem. 2023; 3(3):378-399. https://doi.org/10.3390/appliedchem3030024
Chicago/Turabian StyleSchanen, Parker, and Howard R. Petty. 2023. "What Applied Physical Chemistry Can Contribute to Understanding Cancer: Toward the Next Generation of Breakthroughs" AppliedChem 3, no. 3: 378-399. https://doi.org/10.3390/appliedchem3030024
APA StyleSchanen, P., & Petty, H. R. (2023). What Applied Physical Chemistry Can Contribute to Understanding Cancer: Toward the Next Generation of Breakthroughs. AppliedChem, 3(3), 378-399. https://doi.org/10.3390/appliedchem3030024






