1. Introduction
Bacterial infections are a serious cause of illness in cattle. Over the decades, much time and money has been spent on understanding the lifecycle and virulence factors of bacterial species that are known to cause serious disease. However, there are also rare bacteria responsible for chronic, latent disease that are more challenging to detect and therefore are in need of further study. Furthermore, within the context of latent infection, there is a confounding effect from the possibility of multiple concurrent infections with different species or multiple strains of a single species. A better understanding of such organisms and their complex relationships can lead to better detection and treatment, to allow for increased production outputs of mammalian hosts, as well as animal welfare outcomes.
One such under-studied bacterial pathogen is
Mycoplasma wenyonii, a species of hemotrophic
Mycoplasma that survives parasitically attached to the erythrocytes of cattle [
1]. In healthy animals, the infection often causes few to no outward signs. Severe disease is possible, however, with reports of hemolytic anemia, pyrexia, and internal abscess, often resulting in natural death or euthanasia [
2].
Due to the latent nature of the infection, little is known about how this bacterium survives, infects its host’s cells, and is transferred between individuals, as well as the true prevalence rate of infections. Latent hemoplasma infection has been established as a possible etiology of decreased milk production [
3].
M. wenyonii infection is also associated with swollen teats, rough coats, decreased milk production, and reduced reproductive productivity in first-calf dairy heifers [
4]. Male animals have been found to be aspermic during active infection [
5].
Currently, two distinct strains of
M. wenyonii have been described. The Massachusetts strain has been sequenced in full [
6]. More recently, another strain dubbed INIFAP02 was discovered in cattle from Chihuahua, Mexico, and its genomic draft has been published [
7]. A recent study has shown that older detection protocols based on the Massachusetts strain may not be successful at detecting the presence of the Mexican strain [
8]. It is likely that other strains exist with unquantified pathogenicity that may or may not be detectable using current methods.
Additionally, there is the possibility of interaction with other pathogenic bacterial species. This includes the closely related
Candidatus Mycoplasma haemobos, a more recently discovered hemotrophic
Mycoplasma that is being shown to be similarly prevalent or even more abundant than
M. wenyonii [
9]. Other bloodborne pathogens can also present an infection risk for cattle, such as those belonging to genus
Anaplasma, which are thought to be transferred by ticks [
8]. Another recent study has shown members of
Rickettsia,
Babesia, and
Borrelia to be present in cattle located in temperate regions [
10].
Bartonella has also been widely reported in cattle herds, showing a preference for pregnant females [
11]. These other genera of hemotrophic bacteria have confounded the reliability of
Mycoplasma detection tools, and since most cannot currently be cultured in the laboratory, scientists and veterinarians must rely heavily on DNA-based screening tools.
There is room for greater exploration of the bovine blood microbiome. Recent studies in humans have shown that there may be a substantial endogenous blood microbiome, contradictory to the previously accepted belief that blood must be sterile, only experiencing transient invasion from migratory microbial visitors from other flora-rich sites in the body [
12]. It has definitively been shown that there is a robust bovine-specific microbiome in the feces, ruminal fluid, and milk of cattle, but little is known concerning the microbial populations that may reside in blood. This is likely due to the inability to culture many bloodborne microbes.
For many decades, the standard method for discovering more about a bacterium has been by studying the species in culture. However, to date
M. wenyonii and any of the related hemotrophic
Mycoplasma species have not been successfully cultured [
13]. For this reason, it is necessary to utilize alternative methods to investigate such species. One of the most effective ways to conduct a survey of a bacterial population is through the process of 16S rRNA gene sequencing. 16S rRNA sequencing involves sequencing the ribosomal RNA gene that codes for a variable region of the 16S ribosomal subunit. Within the 16S gene, there exist multiple regions of greater sequence variability, known as Highly Variable Regions, including the frequently targeted V4 region [
14]. Upon PCR amplification and subsequent sequencing, 16S sequence data is used to group similar sequences together to form Operational Taxonomic Units (OTU), or alternatively Amplicon Sequence Variant (ASV) tables, which are then compared to established reference databases [
15]. The level of similarity between the sequences indicates how the organisms that are present may be related taxonomically. This technology allows for a quick and efficient survey of the relative differences in microbial populations within and between different samples. However, there are limitations to the inferences that can be made about absolute numbers of microbes present. Also, the presence of 16S microbial DNA does not directly correlate to a living microbial presence in the sample tissue, because the DNA may remain in samples for some time following microbial cell death. An RNA-based approach is necessary to confirm that living microbes are present in samples, or methods that degrade DNA from non-living cells are required [
16].
In this study, we have employed a 16S rRNA V4 sequencing approach, in conjunction with both digital polymerase chain reaction (dPCR), and quantitative PCR (qPCR), to survey the microbial population present in bovine blood and examine the variability of the M. wenyonii population therein. We hypothesized that bovine blood would contain diverse microbial populations, similar to the blood microbiomes found in other mammalian species, including pathogenic species, commensals, and obligate cellular parasites. We also hypothesized that multiple strains of M. wenyonii may be present within the samples. Third, we expected that cow, calf, and bull categories may have different levels of species abundance and different dominant strains of Mycoplasma. Finally, it is hypothesized that the INFAP01 strain of M. wenyonii from Mexico will be present in this Texas herd in differing proportions.
2. Materials and Methods
2.1. Sampling
Prior to the start of the study, blood was collected by a local veterinarian and veterinary staff from a single herd of beef cattle located in Erath County, Texas, during the course of routine veterinary care. Jugular arterial blood was taken from 61 adult females, 55 of their calves, and 4 adult bulls. Samples were collected in 10 mL ethylenediaminetetraacetic acid (EDTA) tubes to prevent coagulation. Upon receipt by the university, blood was separated into 200 µL aliquots, stored in 1.5 mL flip-cap tubes, and frozen at −20 °C until DNA extraction.
2.2. Extraction
DNA extraction was performed utilizing a QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany) according to manufacturer instructions. The final 100 µL elution was stored frozen at −20 °C for later use.
2.3. 16S Library Preparation
16S rRNA V4 sequencing amplicons were created with Illumina-tagged universal prokaryotic primers 515F-Y 5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGGTGYCAGCMGCCGCGGTAA-3′ [
17] and 806RB 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGGACTACNVGGGTWTCTAAT-3′ [
18] according to the Illumina 16S Metagenomic Sequencing Library Preparation Guide and subsequently underwent index PCR for the application of identifying barcodes (Illumina, San Diego, CA, USA). To ensure the quality and quantity of the dsDNA, a Qubit (ThermoFisher Scientific, Waltham, MA, USA) fluorometric quantification dsDNA high sensitivity assay was performed before pooling the samples for sequencing. For size selection of amplicon products, a Pippin (Sage Science, Beverly, MA, USA) preparation protocol was performed to select for appropriately sized fragments in the 300–600 bp range. The samples were sent to the Texas A&M AgriLife Genomics and Bioinformatics Service (College Station, TX, USA) for 300 × 2 bp paired-end MiSeq sequencing using an Illumina v3 600-cycle sequencing kit (Illumina, San Diego, CA, USA).
2.4. Quantitative Real-Time PCR (qPCR)
The first qPCR target utilized the following primer and probe sequences, as reported by Meli and others [
19], to target the 16S gene of
M. wenyonii: forward 5′-CCACGTGAACGATGAAGGTCTT-3′, reverse 5′-GGCACATAGTTAGCTGTCACTTATTCAA-3′, and finally the probe 5′-Cy5-AGTACCATCAAGGCGCGCTCATTTCCTAG-TQ5-3′ [
19]. For the second target, forward and reverse primers, as well as a TaqMan probe, were synthesized according to the template outlined by Persson Waller and others in 2023 [
8] to specifically amplify targets similar to the strain of
M. wenyonii recently reported in Mexico, INFIAP02, via the polC gene. The probe and primer sequences used were as follows: forward 5′-ATTTGAGCTTACCTCCGCCT-3′, reverse 5′-GAGGATGTCTTTTCCCGCCTAT-3′, and probe 5′-FAM-ACCTTAGAGGAAATTCAAGGCCT-BHQ1-3′ [
8].
Standard curves were generated from plasmids for absolute quantification, and gradient PCR was performed to optimize the primer annealing temperature. Serial dilutions of the plasmids listed in
Supplemental Table S1 were created to function as positive controls for both targets. The copy number for each plasmid was estimated by running the serial dilution set on both the qPCR instrument and the dPCR instrument. Absolute quantification values for the standards were generated through a dPCR run, allowing comparison of Cq values from the qPCR instrument to estimate the copy number present in each dilution.
Primer concentrations were 400 nM each, forward and reverse, with probes at a 200 nM concentration. Kapa Robust 5X Master Mix (Roche, Basel, Switzerland) was used in a 10 µL reaction with 1 µL template DNA. A Bio-Rad (Hercules, CA, USA) CFX384 Touch real-time PCR detection instrument was used for amplification and fluorescence detection. The PCR protocol included an initial 3 min denaturation step at 95 °C, followed by 40 cycles of 10 s at 95 °C for denaturation and 30 s at 55 °C for annealing. After PCR, the data were transferred to Microsoft Excel 365 MSO Version 2504 (Redmond, WA, USA) Excel for analysis.
2.5. Digital Polymerase Chain Reaction (dPCR)
For dPCR, samples were thawed immediately before preparation and then vortexed and centrifuged briefly to ensure that all contents were adequately mixed. A master solution was then created for each sample reaction that included 23.5 µL molecular water, 7 µL Roche (Roche, Basel, Switzerland) Digital LightCycler® Master Mix 5×, 3.5 µL primer/probe working solution, and finally 1 µL template DNA for a 35 µL total reaction volume, resulting in a final concentration of 100 nM for the probe and 250 nM for each primer. The reagents were mixed in 1.5 mL centrifuge tubes that remained on ice during preparation. All 35 µL reaction mixes were loaded into eight-well Roche Digital LightCycler® Universal Nanowell Plates. The dPCR protocol consisted of incubation for 5 min at 50 °C, initial denaturation for 3 min at 95 °C, and finally 40 cycles of 10 s at 95 °C and 30 s at 60 °C. The initial results were then visualized with Roche Digital LightCycler® Development Software version 1.1 to identify positive nanowell clustering and perform absolute quantification.
2.6. Sequence Pipeline and Statistical Analysis
The raw fastq sequence file was converted into an ASV table using a 64-bit version of Usearch 11.0.6 [
20]. Based on average read quality, raw reads were length-trimmed using the fastx_truncate command to 220 bp for the forward reads and 110 bp for the reverse reads before merging with the fastq_mergepairs command. Amplicons lacking both forward and reverse primers were discarded, and primers were trimmed from amplicons using the search_pcr2 command. The resulting full-length primer-trimmed amplicons were quality-filtered to remove any sequences with one or more Ns and any sequences with more than 0.5 expected errors using the fastq_filter command. Quality-filtered sequences were dereplicated with the fastx_uniques command. ASVs were picked as unique zero-radius OTUs, and chimeras were filtered with the unoise3 command. An ASV table was created using the otutab command, and taxonomy was assigned using the sintax command with the Greengenes 13.8 database [
21]. Data was rarified to 1000 sequences per sample prior to community analyses, and the taxonomy was edited with a Linux stream editor (sed) command in Ubuntu version 22.04.5 before import into MicrobiomeAnalyst Version 2.0.
MicrobiomeAnalyst Version 2.0 was used to create histograms representing prokaryotic alpha diversity, a
Mycoplasma ASV heatmap, and a correlation network among the top three blood pathogen-associated genera [
22]. To illustrate alpha diversity, histograms were generated using both Chao1 and Shannon indices, with a Mann–Whitney test performed to determine significance between adult and juvenile populations, and Kruskal–Wallis ANOVA was conducted to determine significance between groups of cows, calves, and bulls. A heatmap was generated to show abundance differences only between
Mycoplasma-associated ASVs, with all other ASVs dropped for that purpose. A correlation network was created at the ASV level for genera known for blood infection, with a Spearman ranked correlation test having a correlation threshold of 0.40, a significance threshold < 0.05, and 100 permutations.
To visualize differences in the blood parasite microbiome, PCOa plots were created using PRIMER7 software (Primer-e, Quest Research Limited, Auckland, NZ, USA). Plots used a Bray–Curtis similarity matrix as input created from the rarefied, square root-transformed ASV sequence counts from only the families Anaplasmataceae, Bartonellaceae, and Mycoplasmataceae (other ASV sequences were removed from the rarefied ASV tables). PRIMER7 version 7.0.17 was also used to conduct PERMANOVAs comparing both bull-cow-calf groups and male-female groups of samples.
Mycoplasma ASVs were aligned with the MegAlign Pro module in DNASTAR software version 17.6.2.9 (DNASTAR, Madison, WI, USA) to identify SNP locations within the amplicons. An NCBI Nucleotide BLAST+ Version 2.16.0 search against the non-redundant database was used to confirm likely identification of Mycoplasma ASVs.
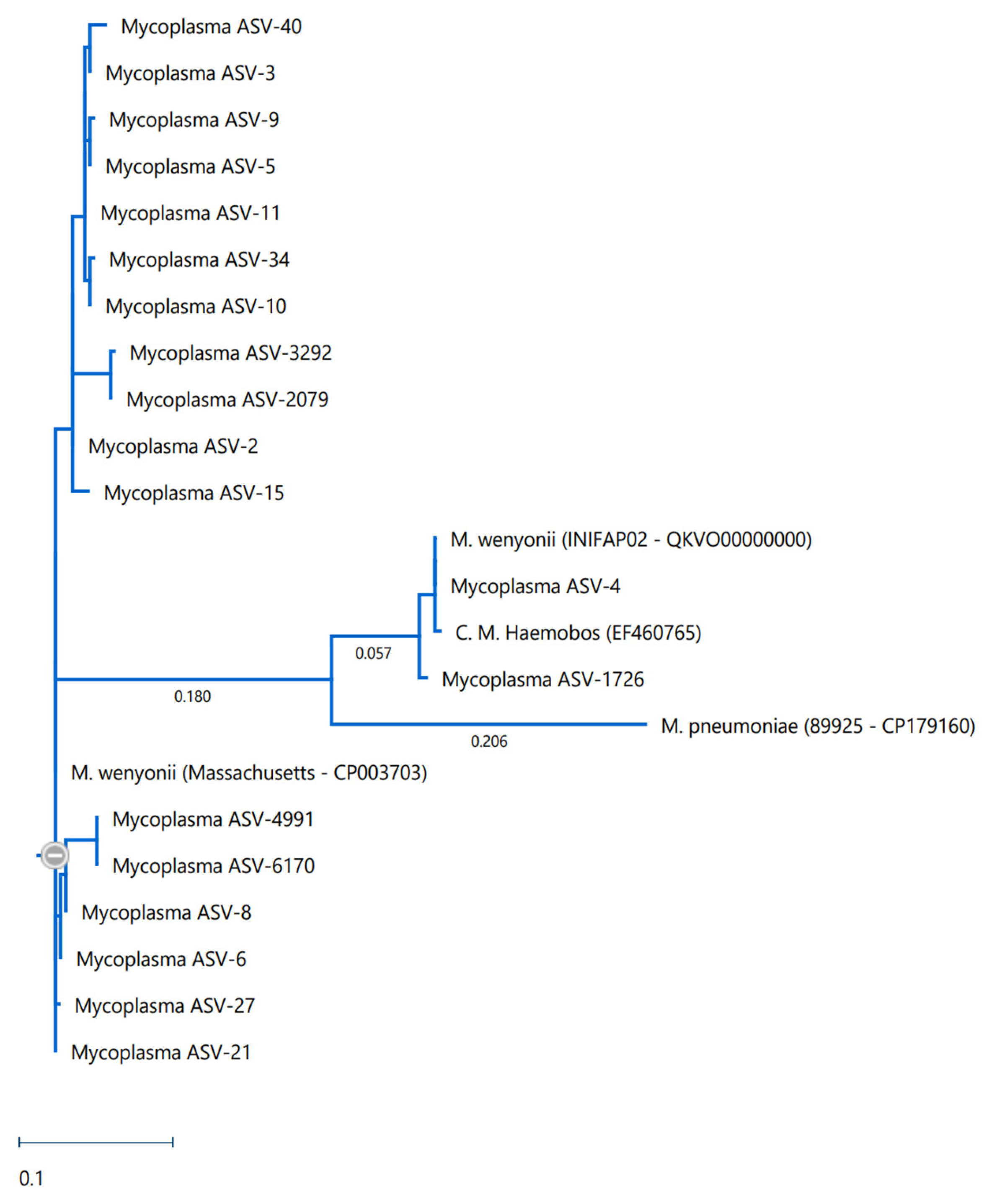
The phylogenetic tree was generated in DNASTAR MegAlign Pro using an alignment of 16S fragments assembled through Mauve, and the tree itself was compiled with RAxML maximum likelihood estimation and 100 bootstrap permutations. Published genomes from M. wenyonii Massachusetts (CP003703), M. wenyonii INIFAP02 (QKVO00000000), C. m. haemobos (EF460765), and M. pneumoniae 89,925 (CP179160) were used as comparison points to illustrate distances.
Differential abundance analysis was conducted using QIIME 1.9 [
23]. Prior to statistical comparisons, ASVs with fewer than 10 sequence counts were filtered from the raw ASV table using the filter_otus_from_otu_table.py script. The ASV table was then normalized by cumulative sum scaling using the normalize_table.py script [
24]. Differential abundance between groups of samples was then compared using the differential_abundance.py script using the DESeq2_nbinom flag [
25] to produce log fold change values between categorical groups. Relative abundance was visualized through the creation of graphs in Microsoft Excel (Microsoft, Remond, WA, USA).
Statistical analyses were performed in QIIME 1.9 and R Studio version 4.3.1. Differences in sample means for ordinal variables were generated through Kruskal–Wallis ANOVA and post hoc analysis by Wilcoxon test with Bonferroni adjustment for multiple tests. For categorical variable correlation analysis, bootstrapped Spearman rank correlation was utilized with 100 permutations. The results were visualized, and the images were generated, in MicrobiomeAnalyst software Version 2.0.
Frequency of infection across groups was statistically analyzed for both dPCR and qPCR data using a chi-square test. Mean cycles of quantification were analyzed using Kruskal–Wallis nonparametric ANOVA for relative differences across groups.
3. Results
The DNA sequencing run produced a total of 5,545,803 paired-end sequences. A total of 19 blood samples produced an insufficient number of DNA sequences, including 13 calves, 4 adult cows, and 1 bull, and were eliminated from analysis after rarefaction.
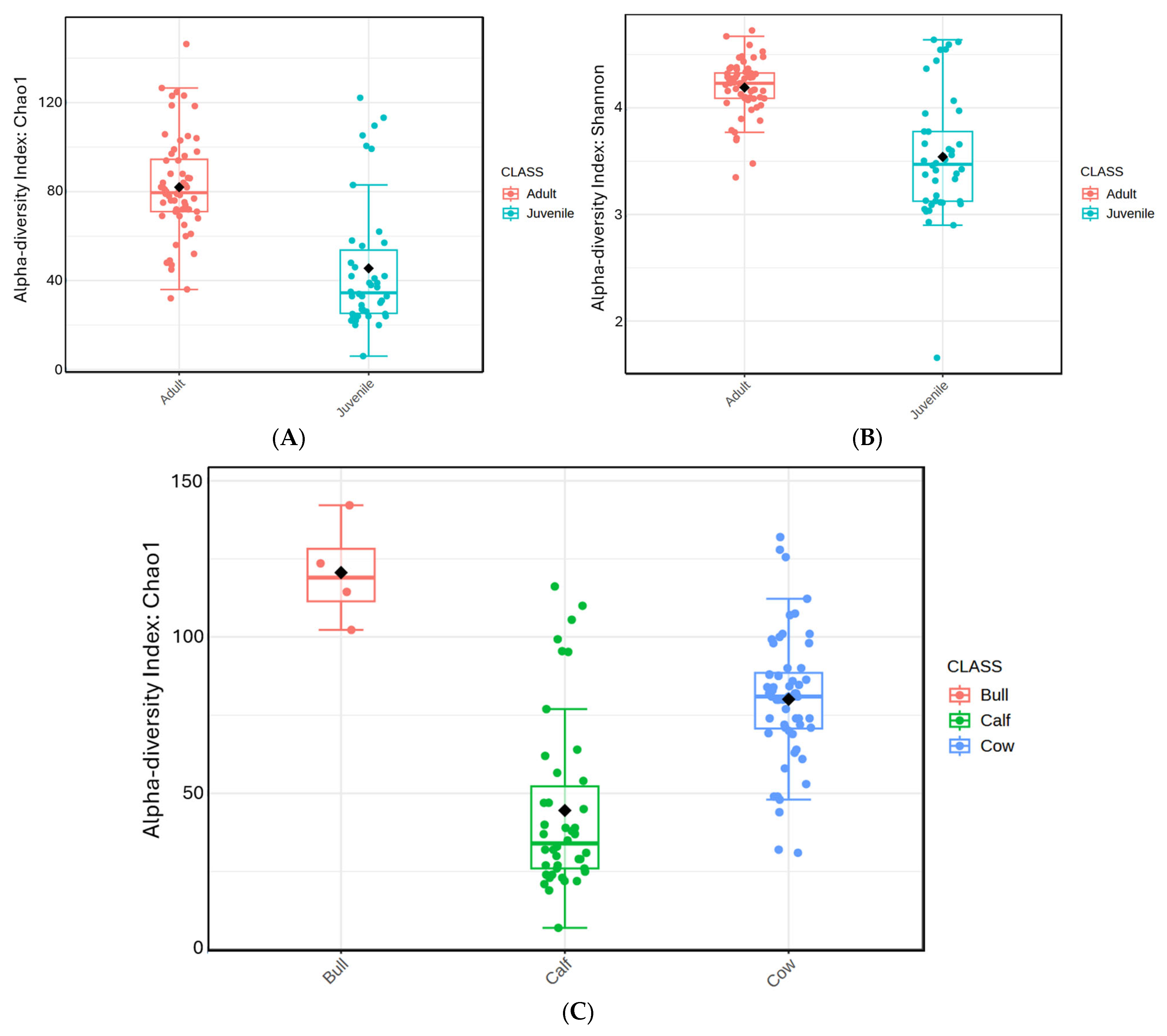
Prokaryotic diversity (
Figure 1A,B) was compared between adults and juveniles, as well as among the groups of cows, calves, and bulls. Adult animals had significantly greater Chao1 species richness when compared to juveniles by Mann–Whitney test (
p < 0.0001). Bulls had the greatest species richness compared to both cows (
p < 0.003) and calves (
p < 0.002) via Kruskal–Wallis ANOVA. Cows also demonstrated greater species richness when compared to calves (
p < 0.0001).
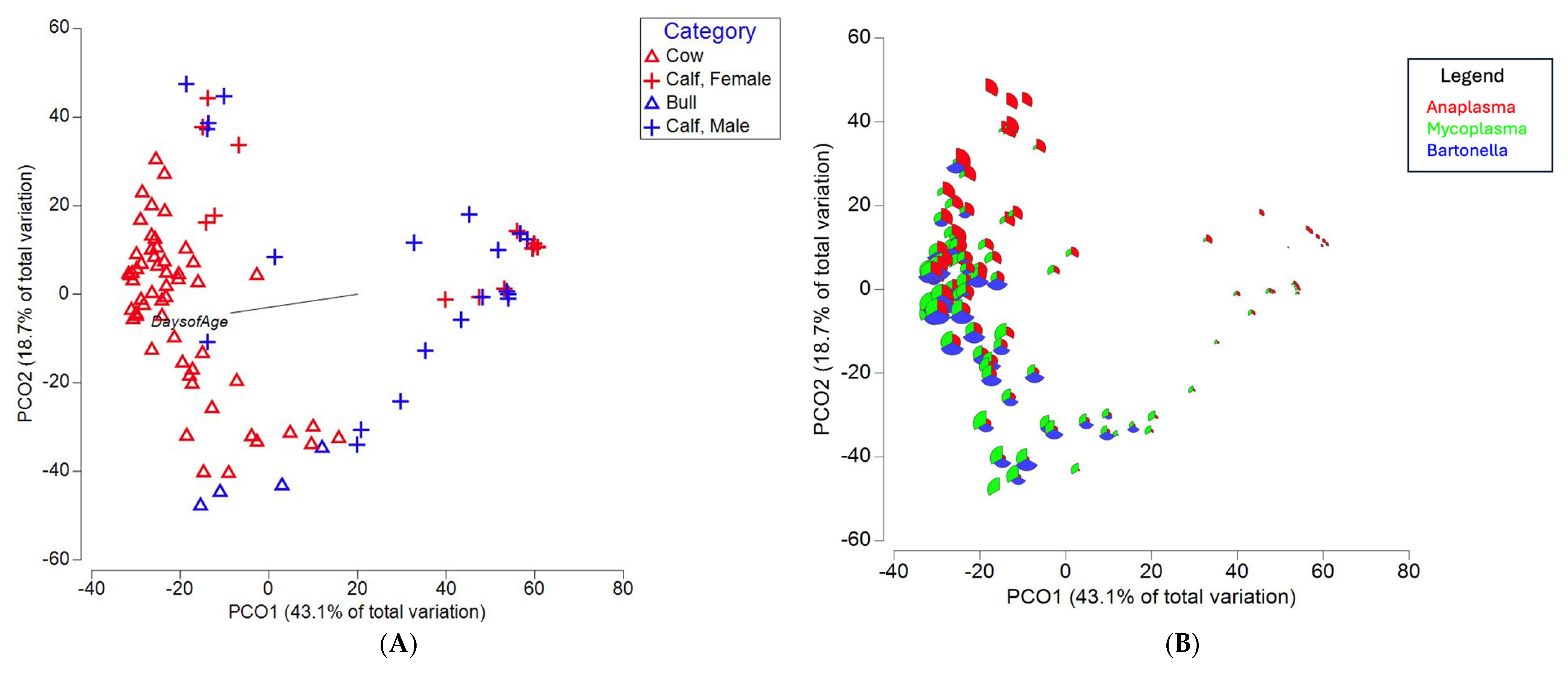
An evaluation of beta diversity among the top three blood-associated genera revealed that cows and bulls clustered separately, with calves overlapping with both groups, eventually moving closer to adult clusters as their age in days increased (
Figure 2A). An additional PCoA revealed (
Figure 2B) how the relative abundance of the top three blood-associated genera changed within individuals across clusters, with
Mycoplasma and
Bartonella sequence counts increasing as animal age increased.
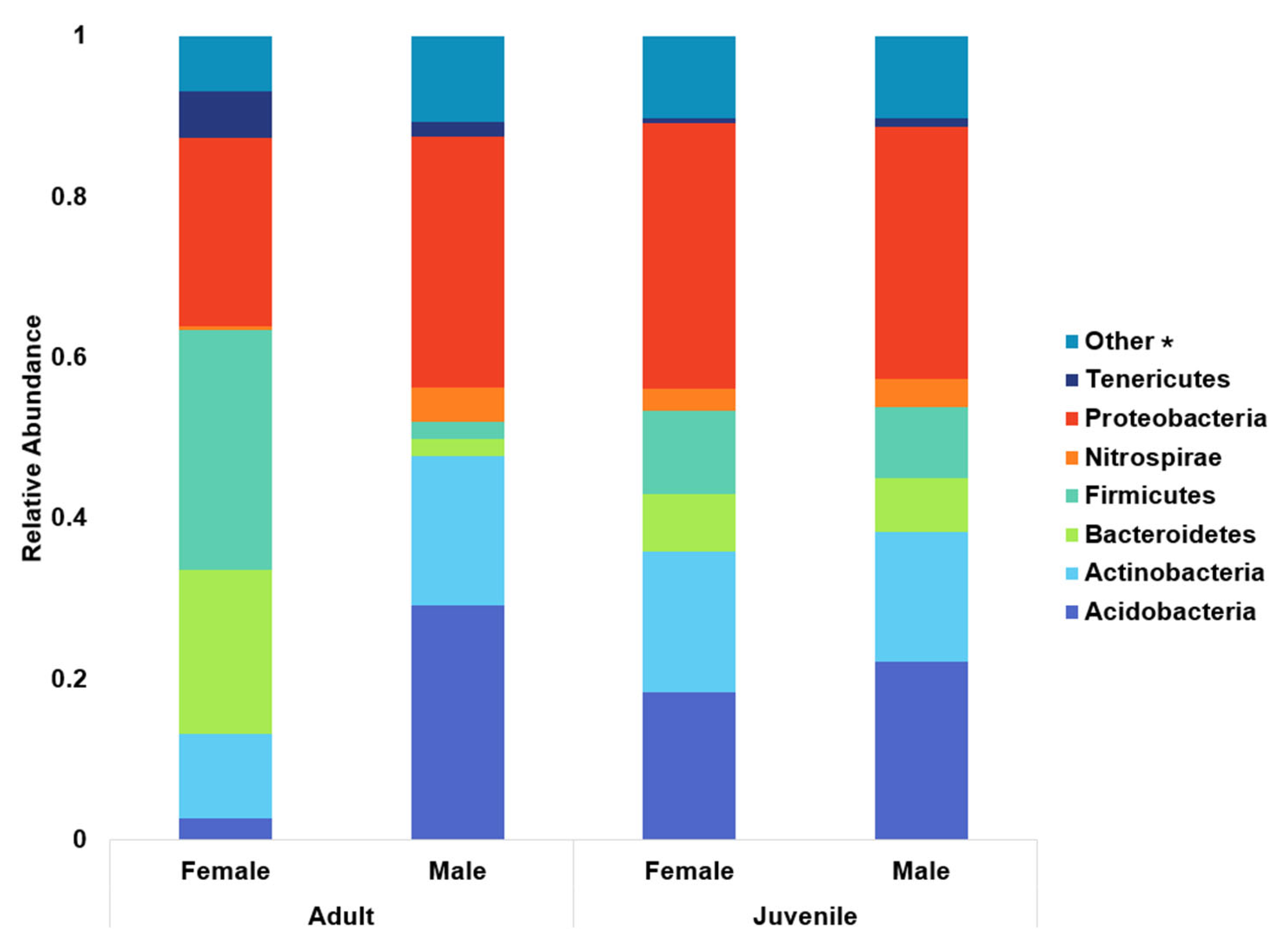
To analyze the relative abundance of the top eight phyla, animals were split into groups of adult females, adult males, female calves, and male calves (
Figure 3). Tenericutes (the phylum containing the genus
Mycoplasma) abundance was lower in calves than in adults of either sex. When compared to adult females, calves had a significant −2.2 log fold change (
p = 0.0003). Compared to adult males, calves tended to have a 4 log fold decrease (
p = 0.055). Males had a greater presence of Acidobacteria, with a 3.8 log fold increase over females (
p < 0.0001). Adult males (bulls) had the greatest number of Acidobacteria overall, with an 8.2 log fold change over adult female cows (
p < 0.0001). Nitrospirae and Firmicutes abundances were significantly greater in bulls, with 6.42 log fold change (
p = 0.008) and a 2 log fold change (
p < 0.0001), respectively.
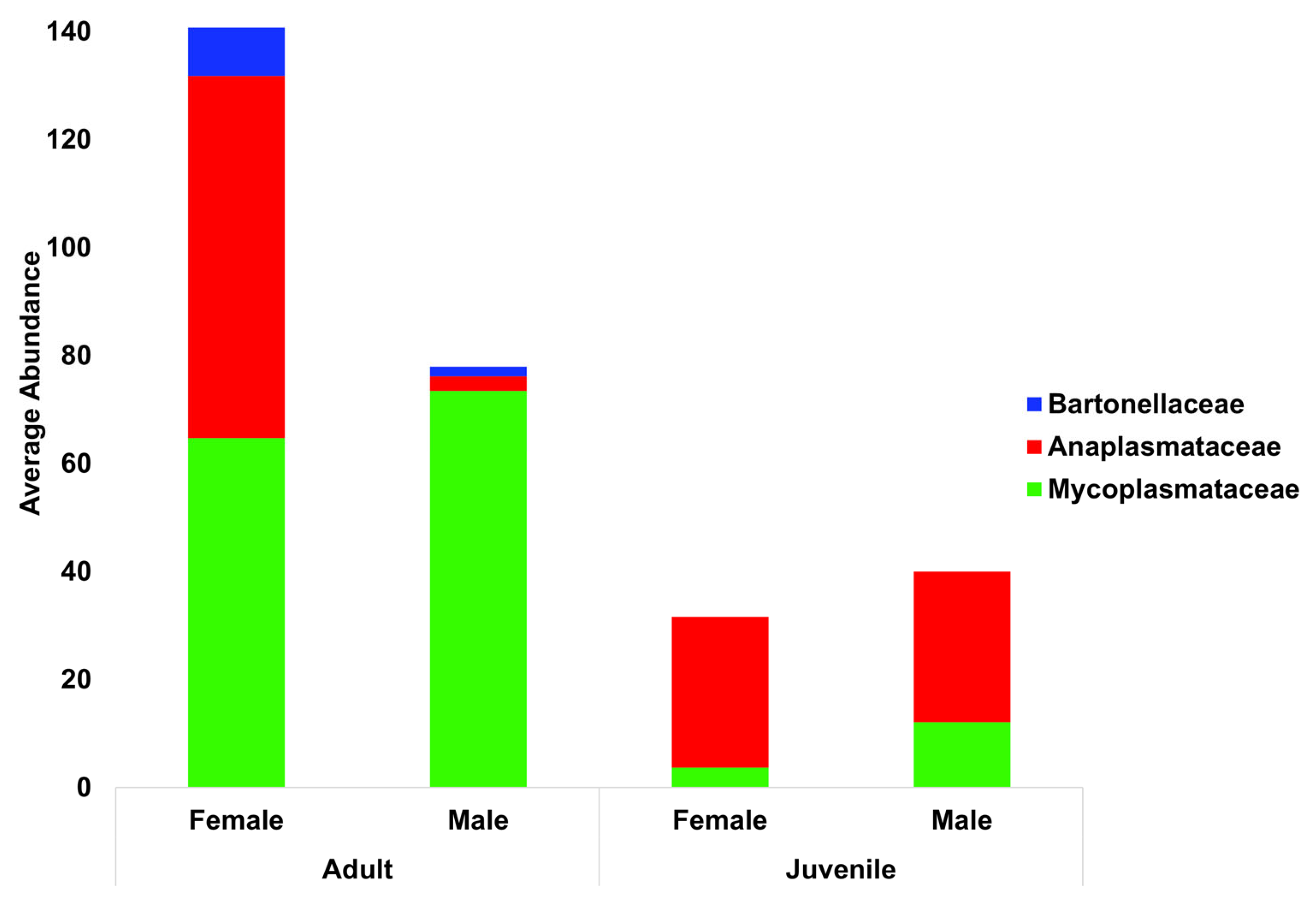
The relative abundance of blood parasite families was also separated into groups of calves, cows, and bulls (
Figure 4). There was a notable scarcity of Anaplasmataceae in adult male bulls, with a 10.5 log fold reduction when compared to females (
p < 0.0001) and a 10.7 log fold reduction compared to calves (
p < 0.0001). Adults of both sexes were more similar when compared to calves, with juveniles of both sexes having less of both Bartonellaceae (
p < 0.0001) and Mycoplasmataceae (
p < 0.0001) compared to their adult counterparts.
Phylogenic comparison of the ASVs identified in this project revealed observable differences between the ASVs found here and published strains of both
M. wenyonii and
M. c. haemobos (
Figure 5).
A total of 19 unique ASV clusters were identified after rarefaction that likely belong to genus
Mycoplasma. Sequence fragments were not long enough to identify with greater specificity than genus.
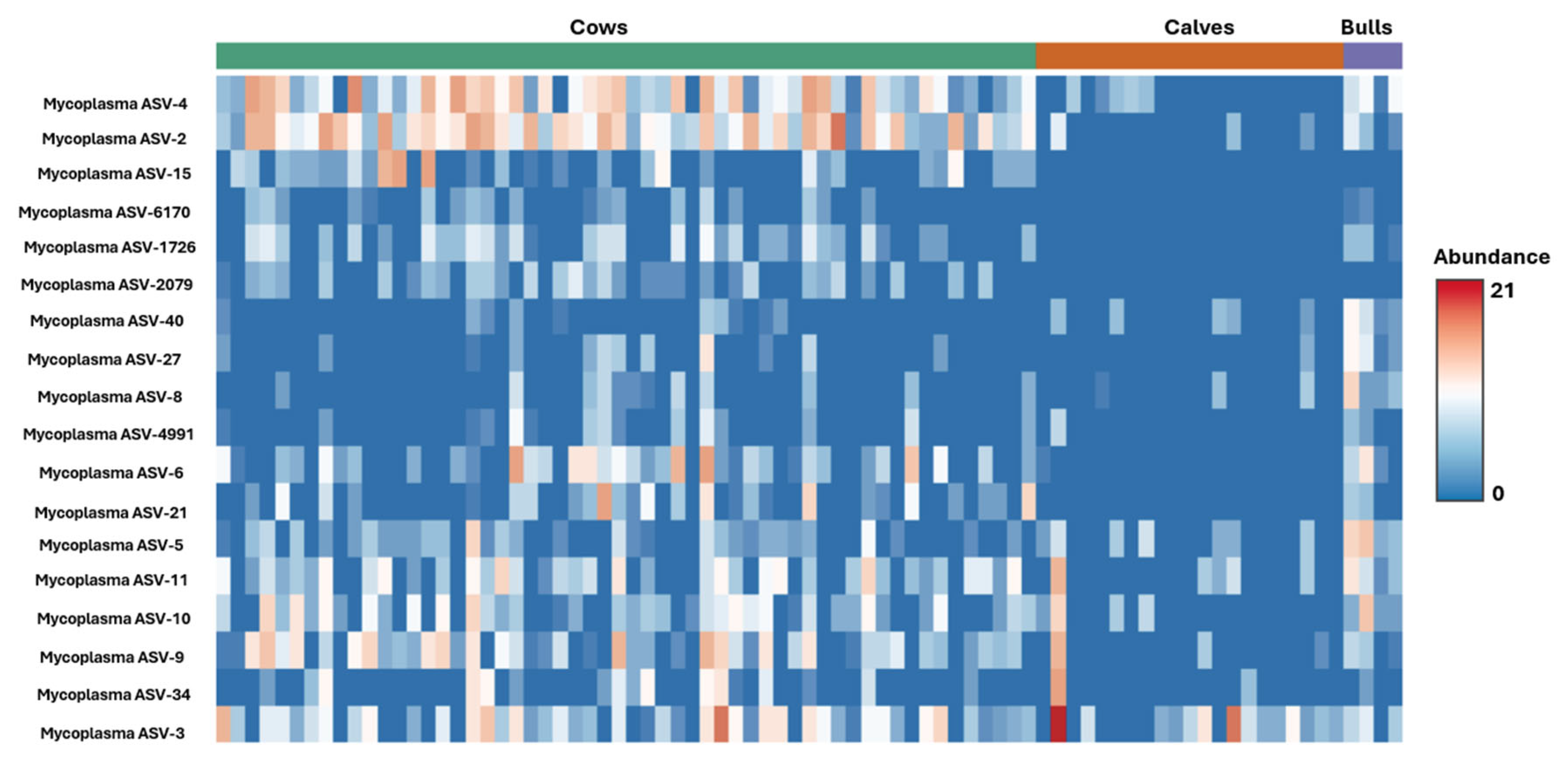
Mycoplasma ASV differences were illustrated with a heatmap image based on differences generated from Kruskal–Wallis ANOVA (
Figure 6). For bulls, ASV 2, 5, 8, 27, and 40 had the most significant differences from cows and calves, while ASV 15 and 2079 stood out distinctly as only appearing in adult females. Calves had fewer
Mycoplasma and
Bartonella ASVs and sequence counts than bulls or cows.
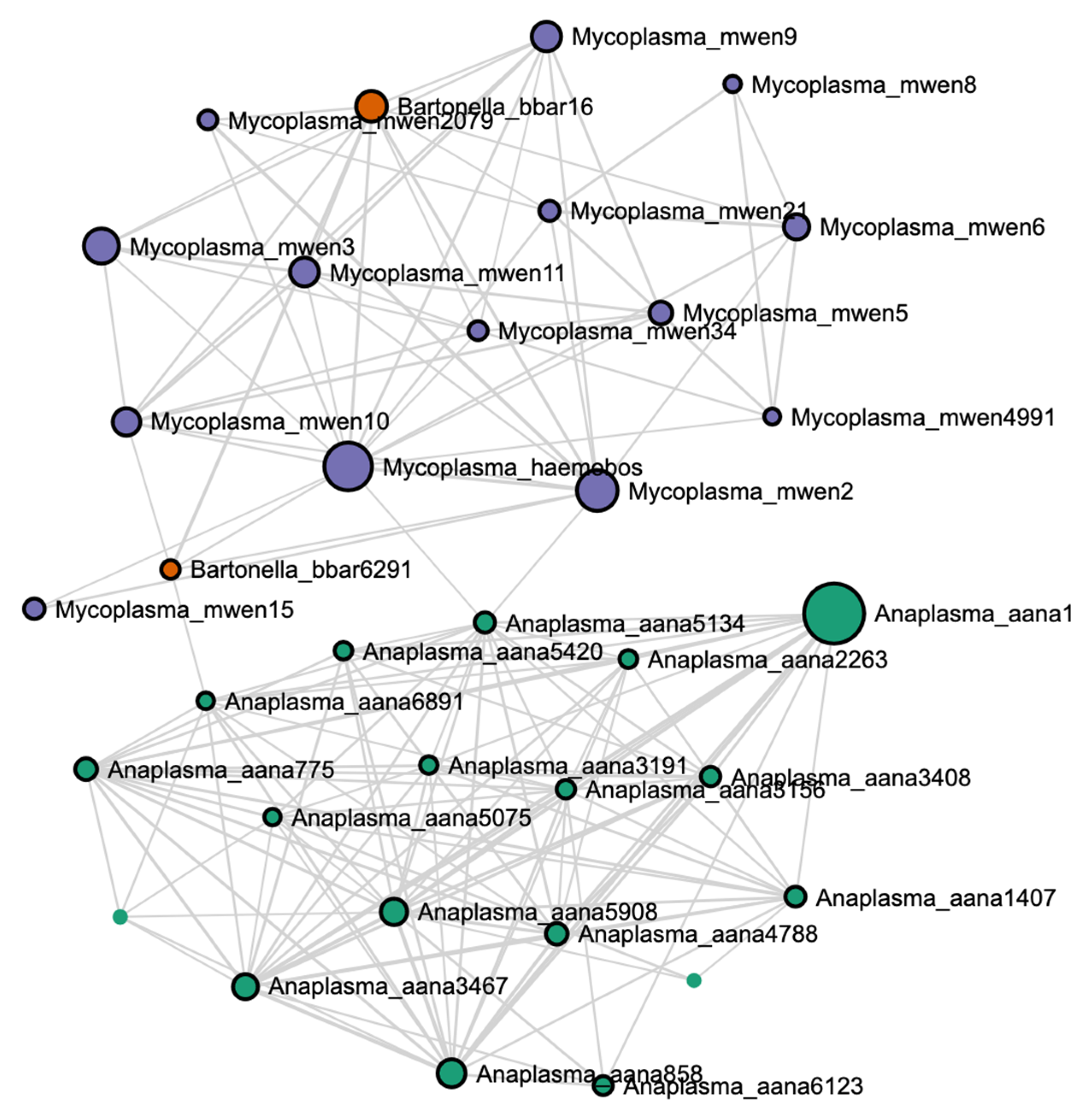
A blood parasite correlation network at the ASV level showed that most
Anaplasma ASVs clustered with each other, apart from both
Mycoplasma and
Bartonella ASVs, which tended to cluster together (
Figure 7).
When comparing the average CQ values among groups for the 16S qPCR assay (
Table 1), there were significant differences across groups (Kruskal–Wallis χ2 = 10.319, df = 2,
p = 0.006), with cows being significantly different from calves (
p = 0.027). There was a tendency toward differences in copy number between cows and calves for the 16S dPCR assay (Kruskal–Wallis chi-squared = 5.6091, df = 2,
p = 0.06054; adjusted Dunn’s
p-value = 0.05874288). Statistical differences in bacterial load for the PolC assay were not possible between qPCR and dPCR when comparing the bull, cow, and calf groups due to low numbers of positive qPCR samples. Overall, the group had a higher infection rate when tested broadly for
M. wenyonii infection, with only a few animals testing positive for the INFAP02 strain.
4. Discussion
The blood microbiome is an emerging area of study, from humans to cattle. Traditionally, blood is considered a sterile environment, and yet, recent studies have shown that bacteria are present throughout most tissues, including blood [
26]. When sampling blood, there are many possible routes of contamination during the process. Even with best sampling practices, there is still the possibility of having a plug of dermis from the needle puncture included in the blood sample. Beyond the issue of sampling contamination, there is also room for debate regarding the permanence of such DNA-detected microbial populations. Currently, there is no agreement as to whether these bacteria represent active populations or simply dormant visitors that are being dealt with by the host immune system. Recent studies have shown that resuscitation and subsequent culture of bacteria extracted from the blood of healthy humans is possible [
12]. However, a great deal more evidence is necessary to challenge the prevailing understanding of the sterility of blood. Since the organisms being detected in this study cannot be grown in culture, this could account for their presence being detected only through DNA-based diagnostics.
A recent study conducted in South Africa found a large presence of Firmicutes, Bacteriodetes, Proteobacteria, and Actinobacteria in bovine blood from randomly selected, apparently healthy cattle through 16S rRNA sequencing [
27]. Additionally, a 2021 study by Scarsella and others examined the microbiota of feces, milk, and blood in dairy cattle to determine if differences existed between healthy cattle and those with mastitis [
28]. That study reported similar relative proportions of bacterial phyla from blood samples, with the authors considering the bacteria present as likely to be dormant. Both studies suggest similar findings to the current study, where a large fraction of the microbial DNA detected in blood belongs to known blood pathogens, but not all. With the methods used in this study, it is impossible to determine if the DNA detected through 16S surveying belonged to living organisms.
As ruminants, cattle have robust microbial populations within their rumen that are used to generate fermentation end-products such as microbial cell proteins and VFAs that are necessary for the animal’s metabolism [
29]. According to previous studies, Bacteroidetes and Firmicutes are the most abundant phyla found in the bovine rumen [
30]. This is notable in the context of the current study, as, outside of the phyla responsible for blood pathogens, Bacteroidetes and Firmicutes were some of the most abundant phyla found here. Other standout phyla included Acidobacteria and Actinobacteria, which were differentially abundant in males and females and of which many species thrive in lower pH environments [
31,
32]. Notably, the current study observed a greater abundance of Acidobacteria in adult males than in females or juveniles of either sex. This shift in the microbiome indicated a potential difference in blood pH between bulls and cows. Since blood pH was not directly measured in the study, we have no direct confirmation regarding blood pH differences that may explain the differential abundance of Acidobacteria. There is currently no data supporting pH differences between healthy cattle of different sexes, with pH being highly regulated through homeostatic mechanisms [
33]. It is possible that differences in diet and subsequent volatile fatty acid (VFA) concentration could play a role in explaining the differences in phyla abundance across groups. Dietary factors will influence ruminal microbial communities and have subsequent impacts on blood parameters such as pH downstream, particularly in animals experiencing physiological imbalances [
34]. Additionally, the rumen environment is one that develops over the course of an animal’s life. After birth, young ruminants very quickly acquire necessary cellulolytic bacteria, but the relative proportions of anaerobes and aerobic bacteria change as the animal develops [
29]. The information that was available with the samples included only identification tag number, date of birth, and sex of the animals, but dietary information was not within the scope of the current study.
Many genera are known to be responsible for bacteremia, including the genera found in this study (
Anaplasma,
Mycoplasma, and
Bartonella). These intra-erythrocytic parasites have frequently been found in seemingly healthy cattle [
27,
35]. However, they have also been implicated in severe disease [
36]. The existence of multiple concurrent infections may contribute a valuable piece of the puzzle to understanding the relationship between latent infection and acute disease. An outbreak in Switzerland among dairy cattle of fatal hemolytic anemia led to an investigation that uncovered as many as five blood parasites present within the acutely ill animals [
36]. While
Anaplasma marginale was considered to be the key player in the outbreak,
Mycoplasma were implicated in the expansion of the disease [
36]. A 2021 study investigating the connection between
Mycoplasma and mastitis found similar results—
Mycoplasma populations were greater in cows with mastitis, yet
Mycoplasma was not detected in milk samples, suggesting that
Mycoplasma may impact host immune response but is not directly responsible for mastitis [
28]. A 2022 study in sheep noticed a correlation between
Mycoplasma fermentans and the foot-rot-causing pathogen
Dichelobacter nodosus. The study was able to determine that
M. fermentans-infected skin cells were unable to produce a satisfactory inflammatory response when exposed to
D. nodosus, leading to the conclusion that
M. fermentans infection leads to susceptibility to other pathogens [
37]. These studies highlight the necessity of understanding how pathogenic species interact with one another. When viewing the correlation network in the current study, it can be seen that
Anaplasma and
Mycoplasma appeared largely uncorrelated to one another. This may reflect a lack of coordinated interaction and could explain why the herd appeared visually healthy despite having a large presence of erythrocytic infectious agents. It was also noted that relative abundance of
Anaplasma was greatest in calves, while
Mycoplasma numbers were greatest in bulls. This could be related to cumulative exposure events, or could possibly be related to pH differences, as discussed previously. Further investigation is warranted to investigate mechanisms that might explain differences in blood parasite abundance within a single cattle herd.
In addition to the coinfection model contributing to the development of acute disease from latent infection, bacterial load also appears to play a role. The
Mycoplasma wenyonii bacterial load was greater in herds that experienced fatal anemia outbreaks than in those that had not [
19]. A study of 22 German cattle herds by Ade and others similarly revealed a robust bacterial load of hemotrophic
Mycoplasma. Of the 19 samples that identified positively for
M. wenyonii in the German study, they had an average bacterial load of 4.29 × 10
5 bacteria/mL blood [
9]. These estimates are similar to the findings of the current study, which place the burden of infection at a similar order of magnitude.
Mode of transmission is yet another grey area when it comes to hemotrophic
Mycoplasma. Blood-sucking arthropods have long been considered the most likely suspect, but currently non-vector-specific blood and bodily fluid contact is considered the primary post-natal source of infectious transmission [
1]. It is possible that higher contact with both blood and bodily fluid from other infected animals could explain the greater relative quantity of
M. wenyonii in the adult animals in this study, as breeding animals do come into greater contact with such sources of infection during sexual reproduction.
The possibility of placental transfer of hemotrophic
Mycoplasma has not been fully investigated. It appears that placental transfer may be possible, but not overwhelming. A 2011 study found that 10.5% of calves born to hemoplasma-infected dams were born infected, as tested through precolostral blood samples taken immediately after birth [
38]. This may explain the lower relative abundance of
Mycoplasma in calves compared to adult female cows found in this study. Conversely, it has been noted that
Anaplasma presence is higher in cattle under three years of age [
39]. This supports the findings of this study that
Anaplasma was more abundant in calves than in adults. This differential abundance across age groups may also help to explain the disparate clustering seen in this study.
Both dPCR and qPCR were used for this study to facilitate comparison between more traditional qPCR techniques and the newer dPCR technology. From this study, it was evident that dPCR was much more effective at detecting low copy number targets and is likely to be used in future studies of blood pathogens.
Of the
M. wenyonii DNA detected in this study, there appears to be considerable diversity within the population. Upon phylogenetic comparison, it was seen that many of the
M. wenyonii ASVs detected were not identical to published strains. Despite hypothesizing that the INIFAP02 strain, which was originally described from samples associated with Chihuahua, Mexico [
7], would be more abundant in Texas due to geographic proximity, this was not found to be the case. The PCR results did show that the INIFAP02 strain was present but was not the primary agent responsible for
M. wenyonii infection in this herd. Additionally, the phylogenetic analyses also support this, showing that the ASVs generated from this study were more closely related to the Massachusetts strain than the INIFAP02 strain.