Abstract
Inflammatory bowel disease (IBD) is a group of chronic inflammatory disorders that affects many individuals throughout their lives. Ulcerative colitis (UC) and Crohn’s disease (CD) are two major forms of IBD. Until the early 1990s, a murine model of spontaneous chronic colitis was unavailable. As a major breakthrough in the basic research field of IBD, three genetically manipulated murine chronic colitis models, including interleukin (IL)-2 knockout (KO), IL-10 KO, and T cell receptor alpha chain (TCRα) KO models, were established in 1993. Since then, complicated immunobiological mechanisms during the development of UC have been gradually discovered by utilizing a wide variety of murine models of IBD, including the TCRα KO mouse model. In particular, it has been recognized that four major factors, including enteric, environmental, and immunological factors as well as enteric microbiota are highly and mutually involved in the pathogenesis of UC. As a pioneer of the TCRα KO murine model of UC, our group has identified that the interactions between the unique TCRα-β+ T cell population and antigen-presenting cells, including dendritic cells and B cells, play a key role for the development and regulation of UC-like chronic colitis, respectively. Here we have summarized clinically proven pathogenic and regulatory factors which have been identified by this novel TCRα KO murine model of UC in the past nearly three decades.
1. Introduction
It is apparent that animal models are indispensable to analyze the pathogenesis of IBD efficiently and mechanistically. In 1993, the first genetically manipulated murine models of spontaneous chronic colitis including IL-2 KO, IL-10 KO, and T cell receptor alpha chain (TCRα) KO mice were established in the same issue of Cell [1,2,3]. Among the colitis models, TCRα KO mice spontaneously develop Th2-mediated UC-like colitis by 6 months of age [3,4]. The severity of colitis in TCRα KO mice changes depending on the animal facilities; under germ-free conditions, but not specific-pathogen free (SPF) conditions, the onset of chronic colitis was completely suppressed in those mice [5,6]. IL-2 is one of the key cytokines in the regulation of immune responses and this cytokine is crucial for the development of a regulatory T cell population [7].
Currently, at least 66 different kinds of genetically modified spontaneous IBD animal models are established, and these models are well-summarized elsewhere [8,9,10]. Among these IBD animal models, most of them develop CD-like intestinal inflammation, and only limited animal models, including TCRα KO, IL-2 KO, IL-2 receptor (IL-2R) KO, Gαi2 KO mice, Mdr1a (multidrug resistant 1a) KO mice, and WASP (Wiskott Aldrich Syndrome Protein) KO mice, develop UC-like colitis [7]. Wasp encodes a cytoplasmic protein involved in regulating actin cytoskeleton, which is absent/defective in patients with Wiskott Aldrich syndrome, a minor population of whom suffer from gut inflammation [11]. It stands to reason that both IL-2 KO and IL-2R KO mice develop chronic inflammation because IL-2 binds with an IL-2R complex and subsequently activates important immune signaling cascades including JAK/STAT and Ras/MAPK [12]. Nearly 50% of IL-2 KO mice die before 9 weeks of age with severe systemic inflammation, such as severe hemolytic anemia, splenomegaly, and lymphadenopathy preceding colitis. In contrast, TCRα KO mice do not develop systemic inflammation and the inflammation is restricted to the colonic mucosa, suggesting this model is one of the best spontaneous Th2-type colitis models at present [9,10]. Here we will discuss the recent clinical advancement of how the TCRα KO murine colitis model can help identify pathogenic and regulatory factors during the development of UC.
2. Pathogenic Populations, Factors and Pathways in TCRα KO Mice during the Development of UC-like Colitis
2.1. Pathogenic T Cell Population
T cell differentiation involves positive and negative selection in the thymus, followed by MHC class II-restricted helper CD4 T cells and MHC class I-restricted cytotoxic CD8 T cells [13]. TCRs expressed on T cells are randomly selected by rearrangement of the α and β genes during the T cell differentiation process, and the receptors are composed of α chain and β chain heterodimers.
TCRα KO mice have been shown to develop few CD4 or CD8 single positive T cells and, with age, develop a population of CD4 positive T cells expressing TCRβ homodimers [14] or pre-TCR (pTα-β heterodimer) [15]. Therefore, when these mice are raised in an SPF facility, they spontaneously develop UC-like symptoms by 6 months of age, even in the absence of Helicobacter infection.
At the beginning, other researchers were skeptical that this animal model of IBD would be truly effective in elucidating the mechanism of pathogenesis, as T cells with an abnormal TCR phenotype expressing TCRα−β+, which had been identified in TCRα KO mice, had not been confirmed to exist in humans [14]. However, in 2011, Morgan et al. found for the first time that there were patients with homozygous G-to-A mutations in the exon 3 region of TRAC (TCRα subunit constant gene) [16]. This suggests that T cells expressing TCRα−β+, which are found in TCRα KO mice that spontaneously develop colitis with aging, are also present in humans, and that clonal expansion of T cells expressing only the TCRβ chain is one of the causes of the pathogenesis. Interestingly, these patients have been found to develop complex immunodeficiency diseases, such as respiratory infections, otitis media, candidiasis, diarrhea, and stunted growth in infancy [16]. With this research background, TCRα KO mice have been frequently used as animal models of human chronic colitis and have proven to be a useful tool for the identification of many pathogenic and protective factors in UC [3,4,17,18].
T cells expressing TCRα−β+, the diversity of which is restricted to TCR Vb8.2+, are increased in colonic lamina propria (LP) of TCRα KO mice with colitis compared to those mice without colitis. In addition, a restricted TCR repertoire results in a loss of tolerance to enteric bacteria, leading to a decrease in peripherally derived regulatory T cells (Tregs) while leading to hyperactivation of migratory dendritic cells (DCs) [19]. This result suggests that maintaining the diversity of TCRs expressed on Tregs may lead to the loss of control of DCs, which are the headquarters of immunity, and may also target the gut microbiota, causing dysbiosis of the gut microbiota. Generally, colonic epithelial cells and intestinal bacteria are kept spatially separated by mucosal layers, which prevent direct interactions between the two. However, a gap in the repertoire of TCRs expressed on T cells, one of the many immune cells, may not only affect intestinal homeostasis, but may also contribute to an exaggerated response of intestinal epithelial cells [20].
2.2. TCR Repertoire Analysis in IBD
Until about 20 years ago, polymerase chain reaction (PCR) was the most commonly used method to analyze the TCR repertoire expressed in T cells [21]. However, this method can only analyze a very limited number of TCR repertoires, and it is very difficult to comprehensively analyze and clarify all TCR repertoires. Recently, with the advent of next-generation sequencing (NGS), large-scale TCR gene sequencing has become possible, and more detailed sequence information on specificity and diversity can be revealed. As a result, in the gastrointestinal tract of UC patients, their TCR clones are amplified while their TCR repertoire is significantly reduced. However, the repertoire of TCR-γδ has not been altered [22]. It has also been reported that the administration of a monoclonal antibody against α4β7 integrin (vedolizumab) does not affect the TCR repertoire of T cells localized in LPs. Werner et al. examined the TCR repertoire of peripheral blood and colonic tissue samples from treatment-naïve pediatric UC patients and healthy controls and found a marked increase in clones of T cells expressing the TCR β chain in pediatric UC patients and an inverse correlation between disease severity and diversity of the intestinal repertoire [23]. It is also well known that the TCRs expressed on T cells in healthy individuals are highly diverse and vary among individuals. Furthermore, healthy monozygotic (MZ) twins have a more similar TCR repertoire than their nonconsanguineous counterparts [24]. Moreover, Rosati et al. performed a repertoire analysis of TCRα and TCRβ in peripheral blood lymphocytes (PBL) of 28 pairs of MZ twins and found features associated with IBD, disease activity, and smoking habits [25]. They found that active IBD patients have less repertoire sharing than inactive IBD patients and healthy twins, and that the V genes TRBV5-1 and TRBV7-2 are mainly utilized as unique chronotypes in IBD patients [25]. Additionally, smoking has been shown to affect the peripheral TCR repertoire in patients with UC, with fewer shared chronotypes compared to nonsmokers [25]. This suggests that even in MZ twins who originally have similar TCR repertoires, casual daily habits, such as smoking, can affect TCR repertoires, and the fact that siblings with the same genetic background have not developed IBD suggests that a combination of lifestyle habits may increase the risk of developing IBD.
It is believed that TCR clonal diversity is maintained in UC patients compared to CD patients [26]. In addition, it was shown that there is no difference in the concomitant use of TRBV-J between patients with active UC and those in remission (UC-R) [27]. Moreover, Hegazy et al. have shown that intestinal reactive T cells from adult patients with CD and UC have a memory phenotype and a diverse T cell receptor Vβ repertoire in peripheral blood mononuclear cells and intestinal tissues. Furthermore, IL-17A, IFNγ, and TNF produced by these cells stimulate the inflammatory responses of intestinal stromal and epithelial cells [28]. One commonality that has been observed in TCR repertoire analyses by many researchers is that TCR diversity is reduced, and a limited number of clones are expanded in UC patients. However, the TCR repertoires used are diverse and the antigens recognized by T cells are not expected to be identical. The summarized data of the TCR repertoire analysis of pediatric and adult IBD patients using NGS is shown in Table 1.

Table 1.
Summary of TCRβ usage studies in human IBD.
Vedolizumab, an anti-integrin α4β7 Ab, has been used as a treatment for adult patients with moderately to severely active UC and CD. Integrin α4β7, which is highly expressed on activated T cells, is thought to be involved in lymphocyte homing by adhering to MAdCAM-1, which is expressed on vascular endothelial cells in the intestinal tract. Therefore, Zeissig et al. and Gamliel et al. examined the effect of vedolizumab on the phenotype of mucosal intrinsic layer T cells. Vedolizumab has been found not to affect the repertoire of T cells in LP or leukocyte trafficking in vivo [29,30]. However, administration of vedolizumab has been shown to alter macrophage populations, thereby markedly altering the expression of molecules related to microbial sensing, chemoattraction, and modulation of innate effector responses [29]. Therefore, the homing of T cells expressing restricted TCRs present in the intestine of IBD patients may be the cause of the effect of those localized in the intestine, rather than of worsening intestinal inflammation, and the effect may not be immediate because it takes time for vedolizumab to change macrophage populations.
2.3. Galectin-4
Galectin-4 consists of two distinct carbohydrate recognition domains (CRDs) and has unique glycan-binding specificity, including the ability to interact with 3′-O-sulfated immature core 1 O-glycans [31]. It is known to be secreted from the basement membrane and apical side of intestinal epithelial cells [32,33,34]. Although the expression of galectin-4 under inflammatory conditions is not different from that in steady state, only CD4+ T cells in the intestine can specifically stimulate galectin-4, which is thought to be involved in the exacerbation of colitis [32]. Hokama et al. demonstrated that the galectin-4-mediated stimulation of CD4+ T cells is associated with an exacerbation of chronic colitis in TCRα KO mice [32]. Galectin-4 can bind specifically to lipid rafts on CD4+ T cells and activate protein kinase Cθ (PKCθ)-related signaling cascades [32,35].
2.4. Gut Dysbiosis
The adult gastrointestinal tract is known to be composed of about 1000 different species of intestinal bacteria with a total number of 109 to 1012 bacteria [36]. The healthy human intestinal tract has a mucin layer on top of the colonic mucosa, which is further covered by multicellular communities referred to as biofilms. The biofilm that exists in the gastrointestinal tract is composed of not only intestinal bacteria, but also phagocytes, nucleic acid elements, fibrin mesh, and host immunoglobulins [37,38,39,40,41]. The biofilm is not only important for the maintenance of intestinal homeostasis, but also important for the stability and resilience of its community, resistance to the establishment of exogenous pathogens, and the maturation of host defenses [36]. Four research groups have reported that a clear distinction can be made between bacteria in feces and on mucosal surfaces in terms of genetics and habitus [42,43,44,45]. Moreover, there are taxonomic differences and reduced overall diversity of mucosa-associated gut microbiota in IBD patients. Specifically, the presence of the enterotoxins Bacteroides fragilis and Pseudomonas aeruginosa in patients with UC and CD [46,47,48,49], and the reduced presence of Faecalibacterium prausnitzii in CD [50,51] have been observed. The oxygen-sensitive F. prausnitzii is a typical butyrate-producing bacterium that settles in the human intestine and produces an anti-inflammatory effect by promoting the differentiation of IL-10-producing regulatory T (Treg) cells [52]. Thus, it has been suggested that the intestinal microbiota that coexists with us not only contributes to the maintenance of health, but may also be closely associated to the development of disease. Here, we present some selected factors affecting the intestinal microbiota in TCRα KO mice models.
It has been observed that TCRα KO mice bred in SPF facilities develop colitis dramatically less frequently than mice raised in conventional facilities [53]. Under conventional conditions, natural antibodies produced by B-1 cells seem to respond to the microbiota and regulate Th2-mediated colitis in nonhygienic environments [53]. Under SPF conditions, Helicobacter-free TCRα KO mice have been shown to develop colitis. However, it is different in TCRβ KO mice or TCRα × TCRβ double KO (DKO) mice. In addition, H. hepaticus infection is sufficient to cause chronic proliferative enteritis in TCRαβ DKO mice.
In a recent study of factors affecting gut bacteria, Devkota et al. showed that mice fed a diet high in milk-derived fat had an increased proportion of taurine-conjugated bile acids compared to those on a low-fat diet [54]. Moreover, Bilophila wadsworthia, a sulfite-reducing bacterium, proliferates by utilizing the sulfur contained in taurine to produce hydrogen sulfide, a toxic metabolite. These changes were associated with the development of colitis in mice lacking the susceptibility gene. This suggests that a Western diet, altered host metabolites, abnormalities in the bacterial flora, and inflammation are associated in genetically susceptible hosts. In addition, the alteration of the gut microbiota leads to immune dysregulation and autoimmune diseases, and the metabolites of gut bacteria, diet, and antibiotics are associated with the regulation of epigenetic mechanisms [55,56]. In 2019, Song et al. reported that bile acids are crucial for the differentiation of RORγ-positive Tregs in the mouse colon, and it has been suggested that this is linked to their anti-inflammatory effects [57]. In addition, there are numerous reports of altered bile acid profiles in feces from IBD patients [36]. Alterations in the bile acid profile can be predicted from the dysbiosis of the intestinal microbiota found in patients with IBD. The reason for this is that some bile acids are metabolized by intestinal bacteria and converted to secondary bile acids. Moreover, secondary bile acids metabolized by intestinal bacteria are considered to be more toxic than primary bile acids, and excessive secondary bile acid production should be taken into account. In contrast, it has been recently discovered that bile acids also have a significant function in homeostasis, such as stimulating the secretion of blood glucose regulating hormones via receptors expressed in humans.
Bjarnason et al. and Kurahara et al. reported that nonsteroidal anti-inflammatory drugs (NSAIDs) induce colitis in humans [58,59]. NSAIDs then induce apoptosis of colonic epithelial cells, allowing luminal bacteria to invade the colonic mucosa [60]. It has been demonstrated that administration of piroxicam, one of the well-known NSAIDs, to mouse models of UC induces colitis within 14 days, but not in WT mice. It has also been shown that piroxicam-induced induction of proinflammatory cytokines (e.g., IL-1β, IL-17, TNFα, and IFN-γ) can be inhibited by dexamethasone, thus preventing the development of colitis [61]. As summarized in Figure 1, many factors directly or indirectly influence gut microbiota dysbiosis.
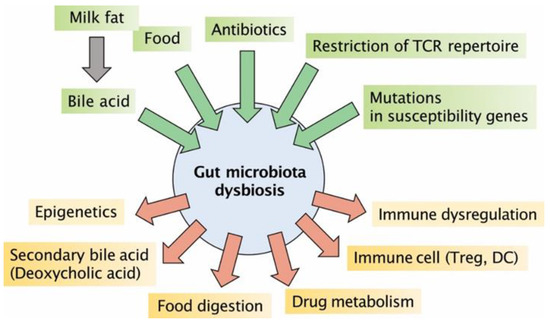
Figure 1.
Factors influencing gut microbiota dysbiosis: Milk fat of animal origin, food, antibiotics, the presence of T cells expressing only a restricted number of TCR repertoires, and mutations in susceptibility genes have been shown to be factors that affect dysbiosis of the gut microbiota. The changes in the intestinal bacterial layer caused by these various factors are closely related to secondary bile acids produced by metabolism by gut microbiota, epigenetics, metabolism of dietary fibers and drugs, and differentiation and dysregulation of immune cells. DC, dendritic cells; TCR, T cell receptor.
2.5. Lympoif Follicles in Cecal Patches (Appendix)
UC causes recurring episodes of chronic inflammation in the mucosal layer of the intestines, beginning in the rectum and extending to various parts of the colon. Colectomy is required in 10% to 15% in moderate to severe disease courses over 5 to 10 years due to many factors, such as stricture, dysplasia, and colorectal cancer. About 10% of UC patients need colectomy within the first year of the diagnosis, and up to 30% of them require colectomy at some point in their life [62,63,64]. Although the etiology of UC is not fully revealed, multiple risk factors are related. Environmental factors, genetic factors, mucosal barrier dysfunction, gut immune responses, and other factors are related to the cause of UC [65]. There are a number of risk factors that give negative effects on clinical courses of patients with UC. However, appendectomy is suggested to be inversely related to the risk of developing UC.
In 1996, our group demonstrated that resection of cecal patches at a young age (3–5 weeks) suppressed the development of IBD, but not at an older age (>6 weeks) in TCRα KO mice [18,66]. In 2017, a national cohort study was conducted in Sweden [67]. It screened more than 63,000 UC patients and demonstrated that appendectomy before the onset of UC, for appendicitis early in life (before 20 years of age) and at any age for diagnoses other than appendicitis, is related to a lower risk of colectomy as well as a milder disease course. In contrast, appendectomy for appendicitis after the onset of UC seemed to be related to a worse disease course. However, some recent reports have shown opposite results. In 2018, Stellingwerf et al. evaluated 13 studies, which collectively included 73,323 UC patients. They demonstrated that there was no significant difference in colectomy rates between patients who underwent appendectomy and those who did not [68].
For a long time, the human appendix had been regarded as a vestigial organ. However, recent studies have revealed some important functions of the human appendix. Firstly, the appendix is a well-organized lymphoid tissue that directly attaches to the cecum where it serves as a reservoir of commensal bacteria [69,70,71]. It might help to replenish and maintain commensal organisms after episodes of colitis or diarrhea. In addition, the generation of IgA-producing B cells seems to be one of the functions of the appendix [72]. However, currently the exact roles of the appendix are still largely enigmatic. UC manifests in a decrease of goblet cells and a destroyed mucosal barrier, which allows pathological intestinal bacteria to invade and boost the inflammation in UC. Therefore, appendectomy for appendicitis after the diagnosis of UC may relate to a more severe disease course and higher rate of colectomy [67]. Furthermore, the appendix is abundant in natural killer (NK)T cells. While the correlation between NKT cells and the pathogenesis of IBD is unrevealed, the aberrant Th2 response in UC harbors IL-13 producing NKT cells. IL-13 damages the intestinal epithelial cell barrier, allowing luminal pathogens to enter the mucosa and cause inflammation [73]. The appendix has a larger number of NKT cells compared to the colon and the small intestine, and the number decreases with age [74]. This phenomenon explains why an appendectomy in early life can prevent the onset of UC (Figure 2).
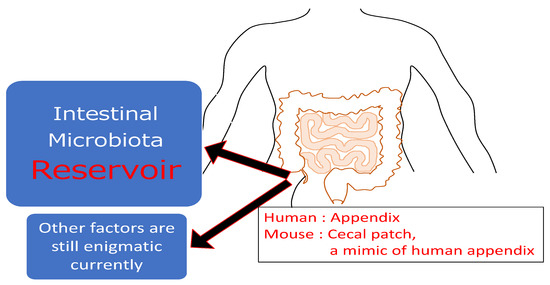
Figure 2.
The roles of the appendix (cecal patch): The appendix is a narrow, worm-shaped sac, which has well-organized lymphoid tissue that directly attach it and is a reservoir of commensal bacteria. Currently, other factors are still highly enigmatic about this organ.
2.6. Chitinase 3-like 1
By utilizing DNA microarray analysis in colonic epithelial cells derived from TCRα KO mice with or without colitis as well as DSS-induced colitis in C57BL/6 mice, our group has identified the unexpected colitis-associated molecule Chitinase 3-like 1 (CHI3L1), which belongs to the glycoside hydrolase 18 family of chitinases [75]. CHI3L1 is not expressed or secreted under healthy conditions, but specifically has been induced on colonic epithelial cells and macrophages under gut inflammatory conditions and plays a pathogenic role in both acute and chronic colitis by enhancing potentially pathogenic bacterial adhesion and invasion on/into those cells [75,76,77]. In particular, N-glycosylated CHI3L1 facilitates CD patient-derived adherent invasive Escherichia coli (AIEC) adhesion to colonic epithelial cells by interacting with bacterial chitinase A (ChiA) via the specific chitin binding domain [77]. The increased expression of CHI3L1 is required for epithelial restitution and survival by promoting the proliferation of these cells under inflammatory conditions as well as in precancerous states [77].
Enhanced CHI3L1 expression is likely to be a useful biomarker for predicting malignant transformation in IBD patients [78,79]. Fecal CHI3L1 levels seem to be useful not only for predicting the severity and activity of mucosal inflammation but also for detecting the presence of malignancy in IBD patients [77,80]. High endogenous CHI3L1 expression is also associated with an increased proliferation rate and may promote spontaneous development of polypoid formation in the colon [81]. It has been reported that serum CHI3L1 levels were significantly elevated in patients with severe cases of asthma. Interestingly, a promoter SNP (single-nucleotide polymorphism) of 131C to G in CHI3L1 was associated with an elevated serum CHI3L1 level with significance (P = 1.1 × 10−13) in those patients [82]. By a proteomics assay using Olink proximity extension analysis, CHI3L1 is one of 16 markers in sputum that can distinguish well-controlled asthma patients (n = 23) from poorly controlled (n = 25) ones [83]. This finding supports a link between sputum neutrophil biomarkers and loss of asthma control [83]. It would be worthwhile to perform proteomics analysis of serum and/or feces in IBD patients as well, for the purpose of seeking potentially useful biomarkers.
2.7. TNFR2 Signaling Pathway
In IBD, TNF is one of the key cytokines which is involved in a wide range of pathogenic processes. In fact, anti-TNF strategies, including chimeric monoclonal antibody (infliximab) and fully human monoclonal antibody (adalimumab), are approved in the therapy of both pediatric and adolescent patients with IBD [84]. TNF produces multiple effects including cell proliferation and cell death through distinct signaling pathways resulting from binding to TNF receptor type II (TNFR2) and TNFR type I (TNFR1), respectively [85,86]. Using the RiboQuant multi-probe ribonuclease protection assay, we found increased TNFR2 expression in the colonic epithelial cell compartment in TCRα KO mice with colitis as compared to those mice without colitis or C57Bl/6 WT mice [87]. In human colonic epithelial cell lines, COLO205 and DLD-1, the combination of TNFα with IL-6 was able to upregulate TNFR2 expression, although TNFα alone had no effect on the expression, suggesting an important role of the IL-6/STAT3-mediated pathway in the TNFR2 upregulation [87]. Recently, meta-analysis of gene expression microarray data in WBC and colon biopsies obtained from pediatric UC patients revealed increased expression levels of TNFR2, but not TNFR1 or TNF [88] in those samples. In addition, soluble TNFR2 have been shown to correlate with disease activity in adult IBD [89]. Taken together, specific blockade of the TNF/TNFR2 interaction in acquired immune pathways seems to be important for a safer and more effective therapeutic strategy for patients with UC.
2.8. PKCθ Signaling Pathway
PKCθ, which is expressed mainly in T cells and skeletal muscles, is a family of serine/threonine kinases that plays a key role in immunological synapse-associated signaling pathways, including NF-κB (nuclear factor kappa B), NFAT (nuclear factor of activated T cells) and AP1 [90,91]. Development of chronic colitis in TCRα KO mice was inhibited by the absence of PKCθ. Colonic CD4+ T cells derived from TCRα × PKCθ DKO mice produce less IL-2 as well as Th2-related molecules (IL-4, IL-13, and GATA3) than TCRα KO mice [92]. In addition to the Th2 colitis model, Nagahama et al. demonstrated the importance of the PKCθ signaling pathway in the CD45RB cell transfer colitis model, suggesting that PKCθ plays a common and fundamental role for the induction of both Th1- and Th2-colitis by activating CD4+ T cells under chronic inflammatory conditions in the gut.
2.9. NK Cells
NK cells, which possess cytotoxic functions, play critical roles in both innate and adaptive immune systems. Originally, Mizoguchi et al. reported that CD3− NK1.1+ cells produce IFNγ, but both TCRα-β+ and TCRγδ+ T cells produce IL-4 in the hyperplastic mesenteric lymph nodes isolated from TCRα KO mice [4]. In this study, the authors note that IL-4 production goes in advance to that of IFNγ, which finally induces the presence of both Th1 (IgG2a) and Th2 (IgG1) autoantibody production [4]. Clinically, circulating NK cells from IBD patients produce large amounts of proinflammatory cytokines and IL-17A, but have less killing capacity [93]. In fact, NK cells play a pivotal role in the antagonistic response to intestinal bacterial infections by producing IFNγ, but this function may be reduced or altered in IBD patients [4,94]. Interestingly, NK cells seem to be involved in the pathogenesis of the development of malignancies including colitis-associated cancers (CAC); autophagy in NK cells inhibits chronic colitis but seems to promote CAC [95,96].
2.10. Myeloid Dendritic-like Cells
Within the immune system, macrophages and DCs usually detect and respond to external pathogens. However, because the intestinal mucosa is continuously exposed to antigens, they apply a tolerogenic function to maintain homeostasis. Consequently, the destruction of the tolerogenic function can lead to the onset of IBD.
Kamada et al. identified a unique human intestinal macrophage that expresses both macrophage subsets (CD14, CD33, CD68) and DC markers (CD205, CD209) [97]. The number of these myeloid dendritic-like cells is considerably increased in patients with CD compared to controls. In addition, these cells produce a larger amount of proinflammatory cytokines and IL-23. IFN-γ induces further differentiation of these myeloid dendritic-like cells, which in turn produce an increased amount of IL-23 and activate the Th17 cell response. This positive feedback loop contributes to chronic inflammation in CD patients [97]. Our group also discovered that IL-4 and IFNγ deficient TCRα KO mice tend to generate intestinal granulomas, which is a characteristic feature of CD. The mice also had unique myeloid dendritic-like cells that had both DC subset CD11 and macrophage marker F4/80 [98]. These myeloid dendritic-like cells produce a large amount of IL-23, and we demonstrated that the production of IL-23 directly induced granulomatous formation.
Barman et al. further classified the myeloid dendritic-like cells in human colonic LP. This study showed that an increased number of CD14+ CD163high CD160low cells were confirmed in UC patients. In contrast, UC patients had fewer CD14+ CD163high CD160high cells. Moreover, it demonstrated that CD163high CD160high cells inhibited effector T cell proliferation, and the suppressive activity of CD163high CD160high cells are essential to control the UC disease course [99].
3. Regulatory Populations, Factors, and Pathways in TCRα KO Mice during the Development of UC-like Colitis
3.1. Regulatory B Cells (Bregs)
B cells are a major immune population, which are thought to play a pathogenic role in acquired immune responses by producing autoantibodies under the conditions of autoimmune disorders [100,101]. However, in 1996 Janeway’s group demonstrated for the first time that an immunoregulatory B cell population, which can produce regulatory cytokine IL-10, exists in the recovery phase of a mouse model of acute experimental autoimmune encephalitis (EAE) [102]. In the same EAE model, our group showed the existence of regulatory B cells and named this population Bregs, a population which contributed highly to efficiently suppressing the development of UC-like colitis in TCRα KO mice [103,104]. The transfer of mature B cells led to decreased numbers of the colonic CD4+ TCRα-β+ pathogenic T cell population with a suppression of colitis in B cell (Igμ chain)-deficient TCRα KO mice [105]. Furthermore, the IL-10 producing Bregs in TCRα KO mice are characterized by the upregulation of CD1d, which is involved in the presentation of lipid antigens to T cells [106]. Of note, Bregs have been detected under a wide variety of experimental inflammatory conditions, including EAE, IBD, arthritis, lupus, UV irradiation, and certain infectious diseases [104].
Bregs are specifically induced under inflammatory conditions and are able to effectively suppress the exacerbation of inflammation with regulatory functions through cellular interactions or regulatory cytokine (e.g., IL-10) production independent of immunoglobulins [104]. Some groups of peptides, including IL-1β, IL-6, IFNα, IL-21, IL33, IL-35, BAFF (B cell activating factor), and APRIL (A proliferation-inducing ligand), have been known as Breg-inducing cytokines [107]. Most of above cytokines, except IL-35, are known as proinflammatory cytokines, which are involved in the pathogenesis of autoimmune diseases in mice and humans [107]. Recently, Mauri et al. have reported that the expansion of Bregs occurs under inflammatory conditions with activation of the set of inflammatory signaling cascades in humans [108]. Probably, Bregs are necessary to sustain the progression of inflammatory conditions by regulating the dose of cytokines, microenvironment, co-stimulation, B cell intrinsic factors and so on [107,108,109,110]. Interestingly, Neurath’s group demonstrated that treatment with rituximab, a chimeric monoclonal antibody targeted against CD20, leads to the exacerbation of inflammation in UC patients suggesting a central role of B cells in the maintenance of gut immune tolerance to self [111,112].
It has been reported that enteric microbiota and their metabolic products, such as short chain fatty acid (SCFA), potentially promote B cell differentiation, activation, and maturation at mucosal sites in animal models and in humans [113,114]. Therefore, such gut metabolites may regulate autoinflammatory diseases by acting as the modulator of B cell-intrinsic epigenesis [112].
3.2. IL-22 Signaling Pathway
IL-22 is one of the IL-10 family cytokines also including IL-19, IL-20, IL-24, IL-26 and IFNα [115]. IL-22 binds to the IL-22 receptor (IL-22R) complex, which makes a heterodimer with the IL-22R1 and IL-10RB subunit. The former subunit is also shared with IL-20 and IL-24, and the latter subunit is also used by IL-10, IL-26, and IFNλ [115,116]. In addition to these receptors, IL-22 can bind with soluble IL-22 binding protein (IL-22BP) with extremely high affinity [117]. In the gut, the mucous layer plays an important role as the first line of defense from commensal microflora including potentially pathogenic and non-pathogenic microbes [118]. IL-22 actively supports to maintain the mucous layer by directly inducing the expression of mucin-related genes in intestinal epithelial cells through the activation of the STAT3 signaling pathway [119,120].
IL-22 expression is significantly low in UC and TCRα KO mice as compared to the CD and CD45RB cell transfer model [121]. In 2008, our group performed a breakthrough experiment by utilizing a local gene-delivery system of a full-length mouse IL-22 cDNA expression vector or mock empty vector, either of which are injected into the proximal colon of TCRα KO mice with predetermined severe colitis [119]. Surprisingly, two weeks after the IL-22 gene-delivery, significantly enhanced STAT3 activation of colonic epithelial cells as well as attenuation of colitis in the injected sites were observed as compared to those of mock vector injected sites [119]. As a result, we have proved that IL-22 contributes highly to the improvement of colitis by enhancing the production of membrane-associated mucins including MUC1, MUC3, MUC10 and MUC13.
An increased level of IL-22BP expression is determined in the normal colon, whereas the expression is significantly reduced in an acute DSS-induced colitis model [119,122]. The reduction of IL-22BP seems to be associated with the formation of inflammasomes, which are responsible for the activation of inflammatory responses mediated by IL-1 and IL-18 [122]. The supplementation of IL-22BP with a local gene delivery system delayed the recovery from DSS-induced acute colitis by inhibiting IL-22 activity [119]. In contrast, IL-22BP expression is relatively increased in IBD patients, but the expression (mainly on CD4+ T cells) is significantly reduced after anti-TNFα antibody treatment [123]. This result suggests one possibility that anti-TNFα antibody therapy mediated mucosal healing, a current therapeutic goal of IBD, may be caused by increased IL-22 production after the reduction of IL-22BP in the colon of IBD patients [124]. Although the beneficial effects of IL-22 have been closed up clinically, this cytokine has also been called “a sheep in wolf’s clothing” due to its potential ability of excreting inflammatory responses and developing colitis-associated cancer [124,125,126]. Therefore, the beneficial versus deleterious effects of IL-22 should be kept in mind for its clinical application in the near future.
3.3. Muc 1
Muc1 is membrane-bound mucin that is produced in the lungs, intestines, and several other organs. It plays many different roles, such as cell adhesion, cell proliferation, and protection against intestinal bacteria. It is thought that intestinal barrier abnormalities cause higher absorption of luminal antigens through the intestinal epithelium, triggering the immune system and causing mucosal inflammation in people with IBD.
Vancamelbeke et al. analyzed 128 intestinal barrier genes using microarray and quantitative RT-PCR. The results showed that MUC1 and MUC4 play an essential role in the pathogenesis of IBD [127]. Th17 cells serve as an intestinal epithelial barrier by producing IL-17, which upregulates the production of Muc1 as well. Nishida et al. demonstrated that Muc1 works in a negative feedback pathway to prevent an excessive Th17 cell response in TCRα KO mice with chronic colitis [128]. The absence of Muc1 perpetuates the expansion of Lin- cKit- Scal1+ Thy1+ innate lymphoid cells that produce IL-17 and enhance the Th17 cell response [129]. This result supports the concept that commensal microbiota can trigger the formation of an intestinal Th17 cell response.
3.4. Carbon Monoxide
Cigarette smoking has been shown to have a protective effect against UC, but it is a risk factor for CD [130]. Carbon monoxide (CO), one of the major components in cigarette smoke, is produced in the body with free iron during the reaction process of heme iron to form biliverdin by inducible heme oxygenase-1 (HO-1). The expression of HO-1 enzyme, which is important for the generation of endogenous CO, is known to be induced by various factors, such as oxidative stress, ischemia, hypoxia, inflammatory cytokines, endotoxin, and heat shock. This is the only biological reaction in which endogenous CO is produced, which is known to have anti-inflammatory and antiapoptotic effects [36,131]. Administration of cobalt protoporphyrin, an inducer of CO and HO-1, to TCRα KO mice has been shown to reduce apoptosis of colonic epithelial cells and alleviate symptoms of colitis [132]. In contrast, inhibition of heme oxygenase activity increases colonic severity in a 2,4,6-trinitrobenzene sulfonic acid (TNBS)-induced colitis model in rats and increases levels of proinflammatory cytokines in a DSS-induced colitis model in mice [131]. In addition, Mesalazine (5-aminosalicylic acid, 5-ASA), currently used as a basic medication for IBD treatment, is thought to have antibacterial and anti-inflammatory effects. Moreover, one of the mechanisms of action of 5-SAS is thought to be due to its induction of HO-1 expression [39]. Hence, it has been suggested that HO-1, the rate-limiting enzyme of CO, may be a promising therapeutic target for IBD.
3.5. Chitin-Microparticles
Chitin is a polymer form of N-acetylglucosamine (GlcNAc) that is a primary structural component of cell walls of many organisms including fungi, crustaceans, insects, and cephalopod beaks, but mammals and bacteria do not possess chitin [133]. Chitin is the second most abundant polysaccharide in nature next to cellulose [133]. Interestingly, chitin shows size-dependent and pathway-specific immunological effects. Intermediate chitin fragments (40–70 μm in size) trigger inflammation by activating the production of TNFα, IL-17 and IL-23 via the TLR-2 and the MyD88 signaling pathway as an alarm signal [134]. In contrast, chitin microparticles (<10 μm in size) enhance the production of IL-10, a well-known anti-inflammatory cytokine [135]. Nagatani et al. orally administered chitin microparticles or PBS (as a vehicle control) to TCRα KO mice every 3 days for six consecutive weeks starting from their weaning age to determine the prophylactic effects of chitin microparticles in chronic colitis [136]. As a result, chitin microparticle-treated mice showed a significantly milder form of colitis with an increased production of IFNγ by CD4+ T cells as compared to PBS-treated control mice [136]. Furthermore, Louis et al. reported that an intermediate size of chitin particles showed an anti-inflammatory effect after being digested with acidic mammalian chitinases depending on the expression of host TLR2 and CD14 [137]. Clinically, chitin microparticles are well tolerated in healthy volunteers and show a more enhanced anti-inflammatory effect after nasal lipopolysaccharide challenges as compared to the placebo group [138]. Based on the positive effect of chitin microparticles, they may also have a potentially useful therapeutic effect on IBD as well.
3.6. Regeneration/Detoxification-Associated Molecules
To identify the function of colonic epithelial cells with excess elongation during the recovery phase of chronic colitis, our group performed a DNA microarray analysis of freshly isolated colonic epithelial cells from TCRα KO mice with colitis [139]. As a result, genes associated with detoxification and biotransformation, such as multiple drug resistance (MDR) 1a and carbonic anhydrase (CAR)-IV, were significantly downregulated in TCRα KO mice as compared to age-matched C57Bl/6 mice [139]. In contrast, genes-associated with regeneration and cell growth, such as regenerating gene (REG) IIIγ and REG IIIβ, were present in the colonic epithelial cells of TCRα KO mice with chronic colitis, but were not quantified during the recovery phase (day 8) of DSS-induced colitis [139]. Xu et al. proved that the expression of SATA3-associated cytokines, including IL-6, IL-17, and IL-22, was significantly increased with 2% DSS-induced colitis, being positively correlated with the expression of REG IIIβ and REGIIIγ in the colonic tissues of mice [140]. Clinically, in vancomycin-resistant Enterococcus infection patients, Reg III was down-regulated in both fecal microbiota transplantation (FMT) and in groups treated with two Lactobacillus strains (Y74 and HT121), suggesting the possibility of Reg III as a biomarker of colonic inflammation [141,142].
3.7. Elemental Diet
In 2000, Kiyono’s group demonstrated that by suppressing the production of Th2-type of cytokine, elementary diet (ED)-fed TCRα KO mice showed no pathogenic feature of UC-like colitis as compared to regular diet (RD)-fed TCRα KO mice [143]. Interestingly, almost 80% of RD-fed mice were infected with Bacteroides vulgatus, which seemed to be associated with the production of Th2 cytokine production by the colonic pathogenic CD4+ T cell population [143]. Rectal administration of B. vulgatus to ED-fed TCRα KO mice led to development of the Th2-type of colitis, suggesting the ED may suppress the development of colitis by modulating the microenvironment of microbiota in TCRα KO mice.
4. Conclusions
By utilizing the TCRα KO spontaneous murine chronic colitis model, several pathogenic and regulatory factors have been identified. As shown in this review, many findings based on basic science have been connected with the development of clinically useful information for UC treatment. Based on this knowledge, along with continuing works utilizing many other animal models of UC, future researchers should be able to successfully develop prophylactic, diagnostic, and therapeutic strategies against UC and its associated diseases including colitis-associated cancer.
Author Contributions
Conception, E.M., T.S. and T.O.; writing—original draft preparation, E.M., T.S. and T.O.; writing—review and editing, E.M., T.S. and T.O.; funding acquisition, E.M. All authors have read and agreed to the published version of this manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Acknowledgments
The authors are grateful to Kori Aiken for her professional English edits in preparing this manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sadlack, B.; Merz, H.; Schorle, H.; Schimpl, A.; Feller, A.C.; Horak, I. Ulcerative colitis-like disease in mice with a disrupted interleukin-2 gene. Cell 1993, 75, 253–261. [Google Scholar] [CrossRef]
- Kuhn, R.; Lohler, J.; Rennick, D.; Rajewsky, K.; Muller, W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell 1993, 75, 263–274. [Google Scholar] [CrossRef]
- Mombaerts, P.; Mizoguchi, E.; Grusby, M.J.; Glimcher, L.H.; Bhan, A.K.; Tonegawa, S. Spontaneous development of inflammatory bowel disease in T cell receptor mutant mice. Cell 1993, 75, 274–283. [Google Scholar] [CrossRef]
- Mizoguchi, A.; Mizoguchi, E.; Chiba, C.; Spiekermann, G.M.; Tonegawa, S.; Nagler-Anderson, C.; Bhan, A.K. Cytokine imbalance and autoantibody production in T cell receptor-alpha mutant mice with inflammatory bowel disease. J. Exp. Med. 1996, 183, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Dianda, I.; Hanby, A.M.; Wright, N.A.; Sebesteny, A.; Hayday, A.C.; Owen, M.J. T cell receptor-alpha beta-deficient mice fail to develop colitis in the absence of a microbial environment. Am. J. Pathol. 1997, 150, 91–97. [Google Scholar] [PubMed]
- Gaskins, H.R.; Vondrak-Juergens, G.L.; McCracken, B.A.; Woolsey, J.H. Specific-pathogen-free conditions enhance inflammatory bowel disease in T-cell receptor knockout, but not C3H/HeJBir mice. Lab. Anim. Sci. 1997, 47, 650–655. [Google Scholar] [PubMed]
- Sakaguchi, S.; Sakaguchi, N.; Asano, M.; Itoh, M.; Toda, M. Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a single mechanisms of self-tolerance causes various autoimmune diseases. J. Immunol. 1995, 155, 1151–1164. [Google Scholar]
- Mizoguchi, A.; Takeuchi, T.; Himuro, H.; Okada, T.; Mizoguchi, E. Genetically engineered mouse models for studying inflammatory bowel disease. J. Pathol. 2016, 238, 205–219. [Google Scholar] [CrossRef]
- Mizoguchi, A. Animal models of inflammatory bowel disease. Prog. Mol. Biol. Trans. Sci. 2012, 105, 263–320. [Google Scholar]
- Mizoguchi, E.; Low, D.; Ezaki, Y.; Okada, T. Recent updates on the basic mechanisms and pathogenesis of inflammatory bowel diseases in experimental animal models. Intes. Res. 2020, 18, 151–167. [Google Scholar] [CrossRef] [PubMed]
- Snapper, S.B.; Rosen, F.S.; Mizoguchi, E.; Cohen, P.; Khan, W.; Liu, C.H.; Hagemann, T.L.; Kwan, S.P.; Ferrini, R.; Davidson, L.; et al. Wiskott-Aldrich syndrome protein-deficient mice reveal a role for WASP in T but not B cell activation. Immunity 1998, 9, 81–91. [Google Scholar] [CrossRef]
- Beadling, C.; Johnson, K.W.; Smith, K.A. Isolation of interleukin 2-induced immediate-early genes. Proc. Natl. Acad. Sci. USA 1993, 90, 2719–2723. [Google Scholar] [CrossRef]
- Rothenberg, E.V.; Taghon, T. Molecular genetics of T cell development. Annu. Rev. Immunol. 2005, 23, 601–649. [Google Scholar] [CrossRef]
- Takahashi, I.; Iijima, H.; Katashima, R.; Itakura, M.; Kiyono, H. Clonal expansion of CD4+ TCRββ+ T cells in TCR α-chain-deficient mice by gut-derived antigens. J. Immunol. 1999, 162, 1843–1850. [Google Scholar]
- Barber, D.F.; Passoni, L.; Wen, L.; Geng, L.; Hayday, A.C. The expression in vivo of a second isoform of pT alpha: Implications for the mechanism of pT alpha action. J. Immunol. 1998, 161, 11–16. [Google Scholar] [PubMed]
- Morgan, N.V.; Goddard, S.; Cardno, T.S.; McDonald, D.; Rahman, F.; Barge, D.; Ciupek, A.; Straatman-Iwanowska, A.; Pasha, S.; Guckian, M.; et al. Mutation in the TCRα subunit constant gene (TRAC) leads to a human immunodeficiency disorder characterized by a lack of TCRαβ+ T cells. J. Clin. Investig. 2011, 121, 695–702. [Google Scholar] [CrossRef]
- Matsumura, K.; Nakase, H.; Kosugi, I.; Honzawa, Y.; Yoshino, T.; Matsuura, M.; Kawasaki, H.; Arai, Y.; Iwashita, T.; Nagasawa, T.; et al. Establishment of a novel mouse model of ulcerative colitis with concominanty cytomegalovirus infection: In vivo identification of cytomegalovirus persistent infected cells. Inflamm. Bowel Dis. 2013, 19, 1951–1963. [Google Scholar]
- Mizoguchi, A.; Mizoguchi, E.; Chiba, C.; Bhan, A.K. Role of appendix in the development of inflammatory bowel disease in TCR-α mutant mice. J. Exp. Med. 1996, 184, 707–715. [Google Scholar] [CrossRef]
- Nishio, J.; Baba, M.; Atarashi, K.; Tanoue, T.; Negishi, H.; Yanai, H.; Habu, S.; Hori, S.; Honda, K.; Taniguchi, T. Requirement of full TCR repertoire for regulatory T cells to maintain intestinal homeostasis. Proc. Natl. Acad. Sci. USA 2015, 112, 12770–12775. [Google Scholar] [CrossRef]
- Curciarello, R.; Canziani, K.E.; Docena, G.H.; Muglia, C.I. Contribution of non-immune cells to activation and modulation of the intestinal inflammation. Front. Immunol. 2019, 10, 647. [Google Scholar] [CrossRef] [PubMed]
- Probert, C.S.J.; Saubermann, L.J.; Balk, S.; Blumberg, R.S. Repertoire of the αβ T-cell receptor in the intestine. Immunol. Rev. 2007, 215, 215–225. [Google Scholar] [CrossRef]
- Holtmeier, W.; Hennemann, A.; May, E.; Duchmann, R.; Caspary, W.F. T cell receptor δ repertoire in inflamed and noninflamed colon of patients with IBD analyzed by CDR3 spectratyping. Am. J. Physiol. Gastrointest. Liver Physiol. 2002, 282, 1024–1034. [Google Scholar] [CrossRef] [PubMed]
- Werner, L.; Nunberg, M.Y.; Rechavi, E.; Lev, A.; Braun, T.; Haberman, Y.; Lahad, A.; Shteyer, E.; Schvimer, M.; Somech, R.; et al. Altered T cell receptor beta repertoire patterns in pediatric ulcerative colitis. Clin. Exp. Immunol. 2019, 196, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zvyagin, I.V.; Pogorelyy, M.V.; Ivanova, M.E.; Komech, E.A.; Shugay, M.; Bolotin, D.A.; Shelenkov, A.A.; Kurnosov, A.A.; Staroverov, D.B.; Chudakov, D.M.; et al. Distinctive properties of identical twins’ TCR repertoires revealed by high-throughput sequencing. Proc. Natl. Acad. Sci. USA 2014, 111, 5980–5985. [Google Scholar] [CrossRef]
- Rosati, E.; Pogorelyy, M.V.; Marie Dowds, C.; Moller, F.T.; Sorensen, S.B.; Lebedev, Y.B.; Frey, N.; Schreiber, S.; Spehlmann, M.E.; Andersen, V.; et al. Identification of disease-associated traits and clonotypes in the T Cell receptor repertoire of monozygotic twins affected by inflammatory bowel diseases. J. Crohn Colitis 2020, 14, 778–790. [Google Scholar] [CrossRef]
- Kakuta, Y.; Nakano, T.; Naito, T.; Watanabe, K.; Izumiyama, Y.; Okamoto, D.; Ichikawa, R.; Moroi, R.; Kuroha, M.; Kanazawa, Y.; et al. Repertoire analysis of memory T-cell receptors in Japanese patients with inflammatory bowel disease. JGH Open 2020, 4, 624–631. [Google Scholar] [CrossRef]
- Günaltay, S.; Repsilber, D.; Helenius, G.; Nyhlin, N.; Bohr, J.; Hultgren, O.; Hultgren Hörnquist, E. Oligoclonal T-cell receptor 646 repertoire in colonic biopsies of patients with microscopic colitis and ulcerative colitis. Inflamm. Bowel Dis. 2017, 23, 932–945. [Google Scholar] [CrossRef]
- Hegazy, A.N.; West, N.R.; Stubbington, M.J.T.; Wendt, E.; Suijker, K.I.M.; Datsi, A.; This, S.; Danne, C.; Campion, S.; Duncan, S.H.; et al. Circulating and tissue-resident CD4+ T cells with reactivity to intestinal microbiota are abundant in healthy individuals and function is altered during inflammation. Gastroenterology 2017, 153, 1320–1337. [Google Scholar] [CrossRef]
- Zeissig, S.; Rosati, E.; Dowds, C.M.; Aden, K.; Bethge, J.; Schulte, B.; Pan, W.H.; Mishra, N.; Zuhayra, M.; Marx, M.; et al. Vedolizumab is associated with changes in innate rather than adaptive immunity in patients with inflammatory bowel disease. Gut 2019, 68, 25–39. [Google Scholar] [CrossRef] [PubMed]
- Gamliel, A.; Werner, L.; Pinsker, M.; Salamon, N.; Weiss, B.; Shouval, D.S. Circulating α4β7+ memory T cells in pediatric ibd patients express a polyclonal T cell receptor repertoire. Clin. Exp. Gastroenterol. 2020, 13, 439–447. [Google Scholar] [CrossRef]
- Ideo, H.; Seko, A.; Ohkura, T.; Matta, K.L.; Yamashita, K. High-affinity binding of recombinant human galectin-4 to SO 3 → 3Gal β 1 → 3GalNAc pyranoside. Glycobiology 2002, 12, 199–208. [Google Scholar] [CrossRef]
- Hokama, A.; Mizoguchi, E.; Sugimoto, K.; Shimomura, Y.; Tanaka, Y.; Yoshida, M.; Rietdijk, S.T.; De Jong, Y.P.; Snapper, S.B.; Terhorst, C.; et al. Induced reactivity of intestinal CD4+ T cells with an epithelial cell lectin, galectin-4, contributes to exacerbation of intestinal inflammation. Immunity 2004, 20, 681–693. [Google Scholar] [CrossRef]
- Rechreche, H.; Mallo, G.V.; Montalto, G.; Dagorn, J.C.; Iovanna, J.L. Cloning and expression of the mRNA of human galectin-4, an S-type lectin down-regulated in colorectal cancer. Eur. J. Biochem. 1997, 248, 225–230. [Google Scholar] [CrossRef] [PubMed]
- Gitt, M.A.; Colnot, C.; Poirier, F.; Nani, K.J.; Barondes, S.H.; Leffler, H. Galectin-4 and galectin-6 are two closely related lectins expressed in mouse gastrointestinal tract. J. Biol. Chem. 1998, 273, 2954–2960. [Google Scholar] [CrossRef]
- Hokama, A.; Mizoguchi, E.; Mizoguchi, A. Roles of galectins in inflammatory bowel disease. World J. Gastroenterol. 2008, 14, 5133–5137. [Google Scholar] [CrossRef]
- Lavelle, A.; Sokol, H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 223–237. [Google Scholar] [CrossRef] [PubMed]
- Motta, J.P.; Flannigan, K.L.; Agbor, T.A.; Beatty, J.K.; Blackler, R.W.; Workentine, M.L.; Da Silva, G.J.; Wang, R.; Buret, A.G.; Wallace, J.L. Hydrogen sulfide protects from colitis and restores intestinal microbiota biofilm and mucus production. Inflamm. Bowel Dis. 2015, 21, 1006–1017. [Google Scholar] [CrossRef]
- Buret, A.; Ward, K.H.; Olson, M.E.; Costerton, J.W. An in vivo model to study the pathobiology of infectious biofilms on biomaterial surfaces. J. Biomed. Mater. Res. 1991, 25, 865–874. [Google Scholar] [CrossRef] [PubMed]
- Palestrant, D.; Holzknecht, Z.E.; Collins, B.H.; Parker, W.; Miller, S.E.; Bollinger, R.R. Microbial biofilms in the gut: Visualization by electron microscopy and by acridine orange staining. Ultrastruct. Pathol. 2004, 28, 23–27. [Google Scholar] [CrossRef]
- Randal Bollinger, R.; Everett, M.L.; Palestrant, D.; Love, S.D.; Lin, S.S.; Parker, W. Human secretory immunoglobulin A may contribute to biofilm formation in the gut. Immunology 2003, 109, 580–587. [Google Scholar] [CrossRef]
- Banwell, J.G.; Howard, R.; Cooper, D.; Costerton, J.W. Intestinal microbial flora after feeding phytohemagglutinin lectins (Phaseolus vulgaris) to rats. Appl. Environ. Microbiol. 1985, 50, 68–80. [Google Scholar] [CrossRef]
- Yasuda, K.; Oh, K.; Ren, B.; Tickle, T.L.; Franzosa, E.A.; Wachtman, L.M.; Miller, A.D.; Westmoreland, S.V.; Mansfield, K.G.; Vallender, E.J.; et al. Biogeography of the intestinal mucosal and lumenal microbiome in the rhesus macaque. Cell Host Microbe 2015, 17, 385–391. [Google Scholar] [CrossRef]
- Zoetendal, E.G.; Von Wright, A.; Vilpponen-Salmela, T.; Ben-Amor, K.; Akkermans, A.D.L.; De Vos, W.M. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl. Environ. Microbiol. 2002, 68, 3401–3407. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Antonopoulos, D.A.; Zhu, X.; Harrell, L.; Hanan, I.; Alverdy, J.C.; Meyer, F.; Musch, M.W.; Young, V.B.; Chang, E.B. Laser capture microdissection and metagenomic analysis of intact mucosa-associated microbial communities of human colon. Appl. Microbiol. Biotechnol. 2010, 88, 1333–1342. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nava, G.M.; Friedrichsen, H.J.; Stappenbeck, T.S. Spatial organization of intestinal microbiota in the mouse ascending colon. ISME J. 2011, 5, 627–638. [Google Scholar] [CrossRef] [PubMed]
- Swidsinski, A.; Loening-Baucke, V.; Lochs, H.; Hale, L.P. Spatial organization of bacterial flora in normal and inflamed intestine: A fluorescence in situ hybridization study in mice. World J. Gastroenterol. 2005, 11, 1131–1140. [Google Scholar] [CrossRef]
- Glasser, A.L.; Boudeau, J.; Barnich, N.; Perruchot, M.H.; Colombel, J.F.; Darfeuille-Michaud, A. Adherent invasive Escherichia coli strains from patients with Crohn′s disease survive and replicate within macrophages without inducing host cell death. Infect. Immun. 2001, 69, 5529–5537. [Google Scholar] [CrossRef]
- Wang, M.; Molin, G.; Ahrné, S.; Adawi, D.; Jeppsson, B. High proportions of proinflammatory bacteria on the colonic mucosa in a young patient with ulcerative colitis as revealed by cloning and sequencing of 16S rRNA genes. Dig. Dis. Sci. 2007, 52, 620–627. [Google Scholar] [CrossRef]
- Palmela, C.; Chevarin, C.; Xu, Z.; Torres, J.; Sevrin, G.; Hirten, R.; Barnich, N.; Ng, S.C.; Colombel, J.F. Adherent-invasive Escherichia coli in inflammatory bowel disease. Gut 2018, 67, 574–587. [Google Scholar] [CrossRef]
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermúdez-Humarán, L.G.; Gratadoux, J.-J.; Blugeon, S.; Bridonneau, C.; Furet, J.-P.; Corthier, G.; et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA 2008, 105, 16731–16736. [Google Scholar] [CrossRef]
- Swidsinski, A.; Loening-Baucke, V.; Vaneechoutte, M.; Doerffel, Y. Active Crohn’s disease and ulcerative colitis can be specifically diagnosed and monitored based on the biostructure of the fecal flora. Inflamm. Bowel Dis. 2008, 14, 147–161. [Google Scholar] [CrossRef] [PubMed]
- Quévrain, E.; Maubert, M.A.; Michon, C.; Chain, F.; Marquant, R.; Tailhades, J.; Miquel, S.; Carlier, L.; Bermúdez-Humarán, L.G.; Pigneur, B.; et al. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn’s disease. Gut 2016, 65, 415–425. [Google Scholar] [CrossRef] [PubMed]
- Shimomura, Y.; Mizoguchi, E.; Sugimoto, K.; Kibe, R.; Benno, Y.; Mizoguchi, A.; Bhan, A.K. Regulatory role of B-1 B cells in chronic colitis. Int. Immunol. 2008, 20, 729–737. [Google Scholar] [CrossRef]
- Devkota, S.; Wang, Y.; Musch, M.W.; Leone, V.; Fehlner-Peach, H.; Nadimpalli, A.; Antonopoulos, D.A.; Jabri, B.; Chang, E.B. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature 2012, 487, 104–108. [Google Scholar] [CrossRef]
- Wu, H.J.; Wu, E. The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes 2012, 3, 4–14. [Google Scholar] [CrossRef]
- Romano, K.A.; Rey, F.E. Is maternal microbial metabolism an early-life determinant of health? Lab. Anim. 2018, 47, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Sun, X.; Oh, S.F.; Wu, M.; Zhang, Y.; Zheng, W.; Geva-Zatorsky, N.; Jupp, R.; Mathis, D.; Benoist, C.; et al. Microbial bile acid metabolites modulate gut RORγ+ regulatory T cell homeostasis. Nature 2020, 577, 410–415. [Google Scholar] [CrossRef] [PubMed]
- Bjarnason, I.; Hayllar, J.; MacPherson, A.J.; Russell, A.S. Side effects of nonsteroidal anti-inflammatory drugs on the small and large intestine in humans. Gastroenterology 1993, 104, 1832–1847. [Google Scholar] [CrossRef]
- Kurahara, K.; Matsumoto, T.; Iida, M.; Honda, K.; Yao, T.; Fujishima, M. Clinical and endoscopic features of nonsteroidal anti-inflammatory drug-induced colonic ulcerations. Am. J. Gastroenterol. 2001, 96, 473–480. [Google Scholar] [CrossRef]
- Hale, L.P.; Gottfried, M.R.; Swidsinski, A. Piroxicam treatment of IL-10-deficient mice enhances colonic epithelial apoptosis and mucosal exposure to intestinal bacteria. Inflamm. Bowel Dis. 2005, 11, 1060–1069. [Google Scholar] [CrossRef]
- Nishiyori, A.; Nagakura, Y.; Ichikawa, K. Piroxicam accelerates development of colitis in T-cell receptor α chain-deficient mice. Eur. J. Pharmacol. 2009, 615, 241–245. [Google Scholar] [CrossRef]
- Feuerstein, J.D.; Isaacs, K.L.; Schneider, Y.; Siddique, S.M.; Falck-Ytter, Y.; Singh, S.; AGA Institute Clinical Guidelines Committee. AGA clinical practice guidelines on the management of moderate to severe ulcerative colitis. Gastroenterology. 2020, 158, 1450–1461. [Google Scholar] [CrossRef]
- Gajendran, M.; Loganathan, P.; Jimenez, G.; Catinella, A.P.; Ng, N.; Umapathy, C.; Ziade, N.; Hashah, J.G. A comprehensive review and update on ulcerative colitis. Dis. Mon. 2019, 65, 100851. [Google Scholar] [CrossRef] [PubMed]
- Andersson, P.; Söderholm, J.D. Surgery in ulcerative colitis: Indication and timing. Dig Dis. 2009, 27, 335–340. [Google Scholar] [CrossRef]
- Kobayashi, T.; Siegmund, B.; Le Berre, C.; Wei, S.C.; Ferrante, M.; Shen, B.; Bernstein, C.N.; Danese, S.; Peyrin-Biroulet, L.; Hibi, T. Ulcerative colitis. Nat. Rev. Dis. Primers 2020, 10, 74. [Google Scholar] [CrossRef]
- Mizoguchi, E.; Mizoguchi, A.; Bhan, A.K. Insights from recent advances in animal models of inflammatory bowel disease. In Molecular Genetics of Inflammatory Bowel Disease; D’Amato, M., Rioux, J.D., Eds.; Springer: New York, NY, USA, 2013; pp. 45–83. [Google Scholar]
- Myrelid, P.; Landerholm, K.; Nordenvall, C.; Pinkney, T.D.; Andersson, R.E. Appendectomy and the risk of coloctomy in ulcerative colitis: A national cohort study. Am. J. Gastroenterol. 2017, 112, 1311–1319. [Google Scholar] [CrossRef] [PubMed]
- Stellingwerf, M.E.; Sahami, S.; Winter, D.C.; Martin, S.T.; D′Haens, G.R.; Cullen, G.; Doherty, G.A.; Mulcahy, H.; Bemelman, W.A.; Buskens, C.J. Prospective cohort study of appendicectomy for treatment of therapy-refractory ulcerative colitis. Br. J. Surg. 2019, 106, 1697–1704. [Google Scholar] [CrossRef]
- Girard-Madoux, M.J.H.; Gomez de Agüero, M.; Ganal-Vonarburg, S.C.; Mooser, C.; Belz, G.T.; Macpherson, A.J.; Vivier, E. The immunological functions of the appendix: An example of redundancy? Semin. Immunol. 2018, 36, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Sahami, S.; Kooji, I.A.; Meijer, S.L.; Van den Brink, G.R.; Buskens, C.J.; Te Velde, A.A. The link between the appendix and ulcerative colitis: Clinical relevance and potential immunological mechanisms. Am. J. Gastroenterol. 2016, 111, 163–169. [Google Scholar] [CrossRef]
- Vitetta, L.; Chen, J.; Clarke, S. The vermiform appendix: An immunological organ sustaining a microbiome inoculum. Clin. Sci. 2019, 133, 1–8. [Google Scholar] [CrossRef]
- Masahata, K.; Umemoto, E.; Kayama, H.; Kotani, M.; Nakamura, S.; Kurakawa, T.; Kikuta, J.; Gotoh, K.; Motooka, D.; Sato, S.; et al. Generation of colonic IgA-secreting cells in the caecal patch. Nat. Commun. 2014, 5, 3704. [Google Scholar] [CrossRef]
- Palm, N.W.; De Zoete, M.R.; Cullen, T.W.; Barry, N.A.; Stefanowski, J.; Hao, L.; Degnan, P.H.; Hu, J.; Peter, I.; Zhang, W.; et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 2014, 158, 1000–1010. [Google Scholar] [CrossRef] [PubMed]
- Heller, F.; Florian, P.; Bojarski, C.; Richter, J.; Christ, M.; Hillenbrand, B.; Mankertz, J.; Gitter, A.H.; Bürgel, N.; Fromm, M.; et al. Interleukin-13 is the key effector Th2 cytokine in ulcerative colitis that affects epithelial tight junctions, apoptosis, and cell restitution. Gastroenterology 2005, 129, 550–564. [Google Scholar] [CrossRef] [PubMed]
- Olszak, T.; An, D.; Zeissig, S.; Vera, M.P.; Richter, J.; Franke, A.; Glickman, J.N.; Siebert, R.; Baron, R.M.; Kasper, D.L.; et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science 2012, 336, 489–493. [Google Scholar] [CrossRef]
- Mizoguchi, E. Chitinase 3-like 1 exacerbates intestinal inflammation by enhancing bacterial adhesion and invasion in colonic epithelial cells. Gastroenterology 2006, 130, 398–411. [Google Scholar] [CrossRef] [PubMed]
- Low, D.; Tran, H.T.; Lee, I.A.; Dreux, N.; Reinecker, H.C.; Darfeuille-Michaud, A.; Barnich, N.; Mizoguchi, E. Chitin-bindingdomains of Escherichia coli chiA mediate interactions with intestinal epithelial cells in mice with colitis. Gastroenterology 2013, 145, 602–612. [Google Scholar] [CrossRef] [PubMed]
- Low, D.; Subramaniam, R.; Lin, L.; Aomatsu, T.; Mizoguchi, A.; Ng, A.; DeGruttola, A.K.; Lee, C.G.; Elias, J.A.; Andoh, A.; et al. Chitinase 3-like 1 induces survival and proliferation of intestinal epithelial cells during chronic inflammation and colitis-associated cancer by regulating S100A9. Oncotarget 2015, 6, 36535–36550. [Google Scholar] [CrossRef]
- Chen, C.C.; Pekow, J.; Llado, V.; Kanneganti, M.; Lau, C.W.; Mizoguchi, A.; Mino-Kenudson, M.; Bissonnette, M.; Mizoguchi, E. Chitinase 3-like 1 (CHI3L1/YKL-40) expression in colonic epithelial cells as a potentially novel marker for colitis-associated neoplasia. Am. J. Pathol. 2011, 179, 1494–1503. [Google Scholar] [CrossRef]
- Mizoguchi, E.; Subramaniam, R.; Okada, T.; Mizoguchi, A. A review of selected IBD biomarkers: From animal models to bedside. Diagnostics 2021, 11, 207. [Google Scholar] [CrossRef]
- Aomatsu, T.; Imaeda, H.; Matsumoto, K.; Kimura, E.; Yoden, A.; Tamai, H.; Fijiyama, Y.; Mizoguchi, E.; Andoh, A. Fecal chitinase 3-like 1 is a novel biological marker for disease activity in pediatric inflammatory bowel disease patients. Aliment. Pharmacol. Ther. 2011, 34, 941–948. [Google Scholar] [CrossRef]
- Low, D.; Poltrak, A.; DeGruttola, A.K.; Mizoguchi, A.; Mino-Kenudson, M.; Mizoguchi, E. High endogenous expression of Chitinase 3-like 1 and excessive epithelial proliferation with colonic tumor formation in MOLF/EiJ mice. PLoS ONE 2015, 10, e0139149. [Google Scholar] [CrossRef] [PubMed]
- Ober, C.; Tan, Z.; Sun, Y.; Possick, J.D.; Pan, L.; Nicolae, R.N.; Radford, S.; Parry, R.R.; Heinzmann, A.; Deichmann, K.A.; et al. Effect of variation in CHI3L1 on serum YKL-40 level, risk of asthma, and lung function. N. Engl. J. Med. 2008, 358, 1682–1691. [Google Scholar] [CrossRef]
- Kasaian, M.T.; Lee, J.; Brennan, A.; Danto, S.I.; Black, K.E.; Fitz, L.; Dixon, A.E. Proteomics analysis of serum and sputum analytes distinguishes controlled and poorly controlled asthmatics. Clin. Exp. Allergy 2018, 48, 814–824. [Google Scholar] [CrossRef] [PubMed]
- Aardoom, M.A.; Veereman, G.; de Ridder, L. A review on the use of anti-TNF in children and adolescents with inflammatory bowel disease. Int. J. Mol. Sci. 2019, 20, 2529. [Google Scholar] [CrossRef] [PubMed]
- Wallach, D.; Varfotomeev, E.E.; Malinin, N.L.; Goltsev, Y.V.; Kovalenko, A.V.; Boldin, M.P. Tumor necrosis factor receptor and Fas signaling mechanisms. Ann. Rev. Immunol. 1999, 17, 331–367. [Google Scholar] [CrossRef] [PubMed]
- Schall, T.J.; Lewis, M.; Koller, K.J.; Lee, A.; Rice, G.C.; Wong, G.H.W.; Gatanaga, T.; Granger, G.A.; Lentz, R.; Raab, H.; et al. Molecular cloning and expression of a receptor for human tumor necrosis factor. Cell 1990, 61, 361–370. [Google Scholar] [CrossRef]
- Mizoguchi, E.; Mizoguchi, A.; Takedatsu, H.; Caro, E.; de Jong, Y.P.; Ooi, C.J.; Xavier, R.J.; Terhorst, C.; Podolsky, D.K.; Bhan, A.K. Role of tumor necrosis factor receptor 2 (TNFR2) in colonic epithelial hyperplasia and chronic intestinal inflammation in mice. Gastroenterology 2002, 122, 134–144. [Google Scholar] [CrossRef]
- Li, X.; Lee, E.J.; Gawel, D.R.; Lilja, S.; Schafer, S.; Zhang, H.; Benson, M. Meta-analysis of expression profiling data indicates need for combinational biomarkers in pediatric ulcerative colitis. J. Immunol. Res. 2020, 2020, 8279619. [Google Scholar] [CrossRef]
- Spoettl, T.; Hausmann, M.; Klebl, F.; Dirmeier, A.; Klump, B.; Hoffmann, J.; Herfarth, H.; Timmer, A.; Rogler, G. Serum soluble TNF receptor I and II levels correlate with diseaseactivity in IBD patients. Inflamm. Bowel Dis. 2007, 13, 727–732. [Google Scholar] [CrossRef]
- Altman, A.; Villalba, M. Protein kinase C-θ (PKCθ): It′s all about location, location, location. Immunol. Rev. 2003, 192, 53–63. [Google Scholar] [CrossRef]
- Pfeifhofer, C.; Kofler, K.; Gruber, T.; Ghaffari Tabrizi, N.; Lutz, C.; Maly, K.; Leitges, M.; Baier, G. Protein kinase Cθ affects Ca2+ mobilization and NFAT cell activation in primary mouse T cells. J. Exp. Med. 2003, 197, 1525–1535. [Google Scholar] [CrossRef] [PubMed]
- Nagahama, K.; Ogawa, A.; Shirane, K.; Shimomura, Y.; Sugimoto, K.; Mizoguchi, A. Protein kinase θ plays a fundamental role in different types of chronic colitis. Gastroenterology 2008, 134, 459–469. [Google Scholar] [CrossRef] [PubMed]
- Zaiats Bittencourt, V.; Jones, F.; Tosetto, M.; Doherty, G.A.; Ryan, E.J. Dysregulation of metabolic pathways in circulating natural killer cells isolated from inflammatory bowel disease patients. J. Cron Colitis 2021, in press. [Google Scholar] [CrossRef]
- Poggi, A.; Benelli, R.; Vene, R.; Costa, D.; Ferrari, N.; Tosetti, F.; Zocchi, M.R. Human gut-associated natural killer cells in health and disease. Front. Immunol. 2019, 10, 961. [Google Scholar] [CrossRef]
- Egawa, S.; Hiwatashi, N. Natural killer cell activity in patients with inflammatory bowel disease. J. Clin. Lab. Immunol. 1986, 20, 187–192. [Google Scholar] [PubMed]
- Wu, Y.; Yao, J.; Xie, J.; Liu, Z.; Zhou, Y.; Pan, H.; Han, W. The role of autophagy in colitis-associated colorectal cancer. Signal Transduct. Target. Ther. 2018, 3, 31. [Google Scholar] [CrossRef]
- Kamada, N.; Hisamatsu, T.; Okamoto, S.; Chinen, H.; Kobayashi, T.; Sato, T.; Sakuraba, A.; Kitazume, M.T.; Sugita, A.; Koganei, K.; et al. Unique CD14 intestinal macrophages contribute to the pathogenesis of crohn disease via IL-23/IFN-gamma axis. J. Clin. Investig. 2008, 118, 2269–2280. [Google Scholar]
- Mizoguchi, A.; Ogawa, A.; Takedatsu, H.; Sugimoto, K.; Shimomura, Y.; Shirane, K.; Nagahama, K.; Nagaishi, T.; Mizoguchi, E.; Blumberg, R.S.; et al. Dependence of intestinal granuloma formation on unique myeloid DC-like cells. J. Clin. Investig. 2007, 117, 605–615. [Google Scholar] [CrossRef]
- Barman, S.; Kayama, H.; Okuzaki, D.; Ogino, T.; Osawa, H.; Matsuno, H.; Mizushima, T.; Mori, M.; Nishimura, J.; Takeda, K. Identification of a human intestinal myeloid cell subset that regulates gut homeostasis. Int. Immunol. 2016, 28, 533–545. [Google Scholar] [CrossRef] [PubMed]
- Murakami, M.; Honjo, T. Transgenic mouse models for B cell dominant autoimmune disease. Curr. Opin. Immunol. 1997, 9, 846–850. [Google Scholar] [CrossRef]
- Martin, F.; Chan, A.C. Pathogenic roles of B cells in human autoimmunity: Insights from the clinic. Immunity 2004, 20, 517–527. [Google Scholar] [CrossRef]
- Wolf, S.D.; Dittel, B.N.; Hardardottir, F.; Janeway, C.A., Jr. Experimental autoimmune encephalomyelitis induction in genetically B cell-deficient mice. J. Exp. Med. 1996, 184, 2271–2278. [Google Scholar] [CrossRef]
- Mizoguchi, A.; Mizoguchi, E.; Smith, R.N.; Preffer, F.I.; Bhan, A.K. Suppressive role of B cells in chronic colitis of T cell receptor α mutant mice. J. Exp. Med. 1997, 186, 1749–1756. [Google Scholar] [CrossRef]
- Mizoguchi, A.; Bhan, A.K. A case for regulatory B cells. J. Immunol. 2006, 176, 705–710. [Google Scholar] [CrossRef]
- Mizoguchi, E.; Mizoguchi, A.; Preffer, F.I.; Bhan, A.K. Regulatory role of mature B cells in a murine model of inflammatory bowel disease. Int. Immunol. 2000, 12, 597–605. [Google Scholar] [CrossRef]
- Mizoguchi, A.; Mizoguchi, E.; Takedatsu, H.; Blumberg, R.S.; Bhan, A.K. Chronic intestinal inflammatory condition generates IL-10-producing regulatory B cell subset characterized by CD1d-upregulation. Immunity 2002, 16, 219–230. [Google Scholar] [CrossRef]
- Mohd Jaya, F.N.; Garcia, S.G.; Borras, F.E.; Chan, G.C.F.; Franquesa, M. Paradoxical role of Breg-inducing cytokines in autoimmune diseases. J. Trans. Autoimm. 2019, 2, 100011. [Google Scholar] [CrossRef] [PubMed]
- Mauri, C.; Menon, M. Human regulatory B cells in health and disease; therapeutic potential. J. Clin. Investig. 2017, 127, 772–779. [Google Scholar] [CrossRef] [PubMed]
- Menon, M.; Blair, P.A.; Isenberg, D.A.; Mauri, C. A regulatory feedback between plasmacytoid dendritic cells and regulatory B cells is aberrant in systemic lupus erythematosus. Immunity 2016, 44, 683–697. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.; Brand, D.; Zheng, S.G. Targeting IL-2: An unexpected effect in treating immunological diseases. Signal Transduct. Target. Ther. 2018, 3, 2. [Google Scholar] [CrossRef]
- Goetz, M.; Atreya, R.; Ghalibafian, M.; Galle, P.R.; Neurath, M.F. Exacerbation of ulcerative colitis after rituximab salvage therapy. Inflamm. Bowel Dis. 2007, 13, 1365–1368. [Google Scholar] [CrossRef]
- Zouali, M. B lymphocytes, the gastrointestinal tract and autoimmunity. Autoimm. Rev. 2021, 20, 102777. [Google Scholar] [CrossRef] [PubMed]
- Fagarasan, S.; Muramatsu, M.; Suzuki, R.; Nagaoka, H.; Hirai, H.; Honjo, T. Critical roles of activation induced cytidine deaminase in the homeostasis of gut flora. Science 2002, 298, 1424–1427. [Google Scholar] [CrossRef]
- Kim, M.; Qie, Y.; Park, J.; Kim, C.H. Gut microbial metabolites fuel host antibody responses. Cell Host Microbe 2016, 20, 202–214. [Google Scholar] [CrossRef]
- Wang, X.; Wong, K.; Ouyang, W.; Rutz, S. Targeting IL-10 family cytokines for the treatment of human diseases. Cold Spring Harb. Perspect. Biol. 2019, 11, a028548. [Google Scholar] [CrossRef]
- Ouyang, W.; O′Garra, A. IL-10 family cytokines IL-10 and IL-22: From basic science to clinical translation. Immunity 2019, 50, 871–879. [Google Scholar] [CrossRef]
- Dumoutier, L.; Lejeune, D.; Colau, D.; Renauld, J.C. Cloning and characterization of IL-22 binding protein, a natural antagonist of IL-10-related T cell-derived inducible factor/IL-22. J. Immunol. 2001, 166, 7090–7095. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.E.; Hansson, G.C. Immunological aspects of intestinal mucus and mucins. Nat. Rev. Immunol. 2016, 16, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, K.; Ogawa, A.; Mizoguchi, E.; Shimomura, Y.; Andoh, A.; Bhan, A.K.; Blumberg, R.S.; Xavier, R.J.; Mizoguchi, A. IL-22 ameliorates intestinal inflammation in a mousemedel of ulcerative colitis. J. Clin. Investig. 2008, 118, 534–544. [Google Scholar]
- Lindemans, C.A.; Calafiore, M.; Mertelsmann, A.M.; O′Connor, M.H.; Dudakov, J.A.; Jenq, R.R.; Velardi, E.; Young, L.F.; Smith, O.M.; Lawrence, G.; et al. Interleukin-22 proteins intestinal-stem-cell-mediated epithelial regeneration. Nature 2015, 528, 560–564. [Google Scholar] [CrossRef]
- Andoh, A.; Zhang, Z.; Inatomi, O.; Fujino, S.; Deguchi, Y.; Araki, Y.; Tsujikawa, T.; Kitoh, K.; Kim–Mitsuyama, S.; Takayanagi, A.; et al. Interleukin-22, a member of the Il-10 subfamily, induces inflammatory responses in colonic subepithelial myofibroblasts. Gastroenterology 2005, 129, 969–984. [Google Scholar] [CrossRef]
- Huber, S.; Gagliani, N.; Zenewicz, L.A.; Huber, F.J.; Bosurgi, L.; Hu, B.; Hedl, M.; Zhang, W.; O′Connor, W.; Murphy, A.J.; et al. IL-22BP is regulated by the inflammasome and modulates tumorigenesis in the intestine. Nature 2012, 491, 259–263. [Google Scholar] [CrossRef]
- Pelczar, P.; Witkowski, M.; Perez, L.G.; Kempski, J.; Hammel, A.G.; Brockmann, L.; Kleinschmidt, D.; Wende, S.; Haueis, C.; Bedke, T.; et al. A pathogenic role for T cell-derived IL-22BP in inflammatory bowel disease. Science 2016, 354, 358–362. [Google Scholar] [CrossRef]
- Mizoguchi, A.; Yano, A.; Himuro, H.; Ezaki, Y.; Sadanaga, T.; Mizoguchi, E. Clinical importance of IL-22 cascade in IBD. J. Gastroenterol. 2018, 53, 465–474. [Google Scholar] [CrossRef] [PubMed]
- Sonnenberg, G.F.; Fouser, L.A.; Artis, D. Border patrol: Regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat. Immunol. 2011, 12, 383–390. [Google Scholar] [CrossRef] [PubMed]
- Keir, M.E.; Yi, Y.; Lu, T.; Ghilardi, N. The role of IL-22 in intestinal health and disease. J. Exp. Med. 2020, 217, e20192195. [Google Scholar] [CrossRef]
- Vancamelbeke, M.; Vanuytsel, T.; Farré, R.; Verstockt, S.; Ferrante, M.; Assche, G.V.; Rutgeerts, P.; Schuit, F.; Vermeire, S.; Arijs, I.; et al. Genetic and transcriptomic bases of intestinal epithelial barrier dysfunction in inflammatory bowel disease. Inflamm. Bowel Dis. 2017, 23, 1718–1729. [Google Scholar] [CrossRef] [PubMed]
- Nishida, A.; Lau, C.W.; Zhang, M.; Andoh, A.; Shi, H.N.; Mizoguchi, E.; Mizoguchi, A. The membrane-bound mucin Muc1 regulates T helper 17-cell responses and colitis in mice. Gastroenterology 2012, 142, 865–874. [Google Scholar] [CrossRef]
- Buonocore, S.; Ahern, P.P.; Uhlig, H.H.; Ivanov, I.I.; Littman, D.R.; Maloy, K.J.; Powrie, F. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature. 2010, 464, 1371–1375. [Google Scholar] [CrossRef]
- Atarashi, K.; Tanoue, T.; Oshima, K.; Suda, W.; Nagano, Y.; Nishikawa, H.; Fukuda, S.; Saito, T.; Narushima, S.; Hase, K.; et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013, 500, 232–236. [Google Scholar] [CrossRef]
- Sheikh, S.Z.; Hegazi, R.A.; Kobayashi, T.; Onyiah, J.C.; Russo, S.M.; Matsuoka, K.; Sepulveda, A.R.; Li, F.; Otterbein, L.E.; Plevy, S.E. An anti-inflammatory role for carbon monoxide and heme oxygenase-1 in chronic Th2-mediated murine colitis. J. Immunol. 2011, 186, 5506–5513. [Google Scholar] [CrossRef]
- Atarashi, K.; Tanoue, T.; Shima, T.; Imaoka, A.; Kuwahara, T.; Momose, Y.; Cheng, G.; Yamasaki, S.; Saito, T.; Ohba, Y.; et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011, 331, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Debono, M.; Gordee, R.S. Antibiotics that inhibit fungal cell wall development. Ann. Rev. Microbiol. 1994, 48, 471–497. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, C.A.; Hartl, D.; Lu, W.; Lee, C.G.; Elias, J.A. TLR-2 and IL-17A in chitin-induced macrophage activation and acute inflammation. J. Immunol. 2008, 181, 4279–4286. [Google Scholar] [CrossRef]
- Lee, C.G.; Da Silva, C.A.; Lee, J.Y.; Hartl, D.; Elias, J.A. Chitin regulation of immune responses: An old molecule with new roles. Curr. Opin. Immunol. 2008, 20, 684–689. [Google Scholar] [CrossRef]
- Nagatani, K.; Wang, S.; Llado, V.; Lau, C.W.; Li, Z.; Mizoguchi, A.; Nagler, C.R.; Shibata, Y.; Reinecker, H.C.; Mora, R.J.; et al. Chitin microparticles for the control of intestinal inflammation. Inflamm. Bowel Dis. 2012, 18, 1698–1710. [Google Scholar] [CrossRef] [PubMed]
- Louis, P.; Mercer, B.; Cirone, A.M.; Brust, C.; Lee, Z.J.; Esiobu, N.; Li, Z.; Wei, J.; Dorey, C.K.; Shibata, Y.; et al. Dietary chitin particles called mimetic fungi ameliorate colitis in toll-like receptor 2/CD14-and sex-dependent manner. Infect. Immun. 2019, 87, e00006-19. [Google Scholar] [CrossRef] [PubMed]
- Sigsgaard, T.; Thorne, P.S.; Schlunssen, V.; Bonlokke, J.; Riddervold, I.S.; Hoppe, K.A.; Andersen, N.T.; Mackenzie, N.M. The change in nasal inflammatory markers after intranasal challenges with particulate chitin and lipopolysaccharide: A randomized, double-blind, placebo-controlled, crossover study with a positive control. Int. Forum Allergy Rhinol. 2015, 5, 16–23. [Google Scholar] [CrossRef]
- Mizoguchi, E.; Xavier, R.J.; Reinecker, H.C.; Uchino, H.; Bhan, A.K.; Podolsky, D.K.; Mizoguchi, A. Colonic epithelial function phenotype varies with type and phase of experimental colitis. Gastroenterology 2003, 125, 148–161. [Google Scholar] [CrossRef]
- Xu, X.; Fukui, H.; Ran, Y.; Wang, X.; Inoue, Y.; Ebisudani, N.; Nishimura, H.; Tomita, T.; Oshima, T.; Watari, J.; et al. The link between type III Reg and STAT3-associated cytokines in inflamed colonic tissues. Mediat. Inflamm. 2019, 2019, 7859460. [Google Scholar] [CrossRef]
- Li, X.; Song, L.; Zhu, S.; Xiao, Y.; Huang, Y.; Hua, Y.; Chu, Q.; Ren, Z. Two strains of Lactobacilli effectively decrease the colonization of VRE in a mouse model. Front. Cell Infect. Microbiol. 2019, 9, 6. [Google Scholar] [CrossRef] [PubMed]
- Kishi, D.; Takahashi, I.; Kai, Y.; Tamagawa, H.; Iijima, H.; Obunai, S.; Nezu, R.; Ito, T.; Matsuda, H.; Kiyono, H. Alteration of V beta usage and cytokine production of CD4+ TCR beta beta homodimer T cells by elimination of Bacteroides vulgatus prevents colitis in TCR alpha-chain-deficient mice. J. Immunol. 2000, 165, 5891–5899. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).