DNA as a Double-Coding Device for Information Conversion and Organization of a Self-Referential Unity
Abstract
:1. Introduction
2. DNA Is a Source of Two Distinct Types of Information
3. Coupling of Logically Distinct Types of Information
4. Switching Between Alternative Gene Expression Programs
5. Analog/Digital Information Conversion Operates as a Regulatory Device in Living Systems of Diverse Structural Complexity
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kaufman, L.H. Self reference and recursive forms. J. Social. Biol. Struct. 1987, 10, 53–72. [Google Scholar] [CrossRef]
- Razeto-Barry, P. Autopoiesis 40 years later. A review and a reformulation. Orig. Life Evol. Biosph. 2012, 42, 543–567. [Google Scholar] [CrossRef] [PubMed]
- Ruijtenberg, S.; van den Heuvel, S. Coordinating cell proliferation and differentiation: Antagonism between cell cycle regulators and cell type-specific gene expression. Cell Cycle 2016, 15, 196–212. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Papenfort, K.; Bassler, B.L. Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 2016, 14, 576–588. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Antunez-Lamas, M.; Cabrera, E.; Lopez-Solanilla, E.; Solano, R.; González-Melendi, P.; Chico, J.M.; Toth, I.; Birch, P.; Pritchard, L.; Liu, H.; et al. Bacterial chemoattraction towards jasmonate plays a role in the entry of Dickeya dadantii through wounded tissues. Mol. Microbiol. 2009, 74, 662–671. [Google Scholar] [CrossRef] [PubMed]
- Río-Álvarez, I.; Muñoz-Gómez, C.; Navas-Vásquez, M.; Martínez-García, P.M.; Antúnez-Lamas, M.; Rodríguez-Palenzuela, P.; López-Solanilla, E. Role of Dickeya dadantii 3937 chemoreceptors in the entry to Arabidopsis leaves through wounds. Mol Plant Pathol. 2015, 16, 685–698. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jiang, X.; Zghidi-Abouzid, O.; Oger-Desfeux, C.; Hommais, F.; Greliche, N.; Muskhelishvili, G.; Nasser, W.; Reverchon, S. Global transcriptional response of Dickeya dadantii to environmental stimuli relevant to the plant infection. Environ. Microbiol. 2016, 18, 3651–3672. [Google Scholar] [CrossRef] [PubMed]
- Hallgren, J.; Koonce, K.; Felletti, M.; Mortier, J.; Turco, E.; Jonas, K. Phosphate starvation decouples cell differentiation from DNA replication control in the dimorphic bacterium Caulobacter crescentus. PLoS Genet. 2023, 19, e1010882. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Barrows, J.M.; Goley, E.D. Synchronized Swarmers and Sticky Stalks: Caulobacter crescentus as a Model for Bacterial Cell Biology. J. Bacteriol. 2023, 205, e0038422. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dudin, O.; Geiselmann, J.; Ogasawara, H.; Ishihama, A.; Lacour, S. Repression of flagellar genes in exponential phase by CsgD and CpxR, two crucial modulators of Escherichia coli biofilm formation. J. Bacteriol. 2014, 196, 707–715. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Scolari, V.F.; Bassetti, B.; Sclavi, B.; Lagomarsino, M.C. Gene clusters reflecting macrodomain structure respond to nucleoid perturbations. Mol. Biosyst. 2011, 7, 878–888. [Google Scholar] [CrossRef] [PubMed]
- Sobetzko, P.; Glinkowska, M.; Travers, A.; Muskhelishvili, G. DNA thermodynamic stability and supercoil dynamics determine the gene expression program during the bacterial growth cycle. Mol. Biosyst. 2013, 9, 1643–1651. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Sobetzko, P.; Nasser, W.; Reverchon, S.; Muskhelishvili, G. Chromosomal “stress-response” domains govern the spatiotemporal expression of the bacterial virulence program. mBio 2015, 6, e00353-15. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- SantaLucia, J., Jr. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl. Acad. Sci. USA 1998, 95, 1460–1465. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Packer, M.J.; Dauncey, M.P.; Hunter, C.A. Sequence-dependent DNA structure: Dinucleotide conformational maps. J. Mol. Biol. 2000, 295, 71–83. [Google Scholar] [CrossRef] [PubMed]
- Young, R.T.; Czapla, L.; Wefers, Z.O.; Cohen, B.M.; Olson, W.K. Revisiting DNA Sequence-Dependent Deformability in High-Resolution Structures: Effects of Flanking Base Pairs on Dinucleotide Morphology and Global Chain Configuration. Life 2022, 12, 759. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- El Hassan, M.A.; Calladine, C.R. The assessment of the geometry of dinucleotide steps in double-helical DNA; a new local calculation scheme. J. Mol. Biol. 1995, 251, 648–664. [Google Scholar] [CrossRef] [PubMed]
- Calladine, C.R.; Drew, H.R.; Luisi, B.F.; Travers, A.A. Understanding DNA. The Molecule & How It Works, 3rd ed.; Elsevier Academic Press: Amsterdam, The Netherlands, 2004. [Google Scholar]
- Palecek, E. Local supercoil-stabilized DNA structures. Crit. Rev. Biochem. Mol. Biol. 1991, 26, 151–226. [Google Scholar] [CrossRef] [PubMed]
- Irobalieva, R.N.; Fogg, J.M.; Catanese, D.J., Jr.; Sutthibutpong, T.; Chen, M.; Barker, A.K.; Ludtke, S.J.; Harris, S.A.; Schmid, M.F.; Chiu, W.; et al. Structural diversity of supercoiled DNA. Nat. Commun. 2015, 6, 8440. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, Q.; Irobalieva, R.N.; Chiu, W.; Schmid, M.F.; Fogg, J.M.; Zechiedrich, L.; Pettitt, B.M. Influence of DNA sequence on the structure of minicircles under torsional stress. Nucleic Acids Res. 2017, 45, 7633–7642. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pyne, A.L.B.; Noy, A.; Main, K.H.S.; Velasco-Berrelleza, V.; Piperakis, M.M.; Mitchenall, L.A.; Cugliandolo, F.M.; Beton, J.G.; Stevenson, C.E.M.; Hoogenboom, B.W.; et al. Base-pair resolution analysis of the effect of supercoiling on DNA flexibility and major groove recognition by triplex-forming oligonucleotides. Nat. Commun. 2021, 12, 1053. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Muskhelishvili, G.; Travers, A. The regulatory role of DNA supercoiling in nucleoprotein complex assembly and genetic activity. Biophys. Rev. 2016, 8 (Suppl. S1), 5–22. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hsieh, L.S.; Burger, R.M.; Drlica, K. Bacterial DNA supercoiling and [ATP]/[ADP]. Changes associated with a transition to anaerobic growth. J. Mol. Biol. 1991, 219, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Drlica, K. Control of bacterial DNA supercoiling. Mol. Microbiol. 1992, 6, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Van Workum, M.; van Dooren, S.J.; Oldenburg, N.; Molenaar, D.; Jensen, P.R.; Snoep, J.L.; Westerhoff, H.V. DNA supercoiling depends on the phosphorylation potential in Escherichia coli. Mol. Microbiol. 1996, 20, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Snoep, J.L.; van der Weijden, C.C.; Andersen, H.W.; Westerhoff, H.V.; Jensen, P.R. DNA supercoiling in Escherichia coli is under tight and subtle homeostatic control, involving gene-expression and metabolic regulation of both topoisomerase I and DNA gyrase. Eur. J. Biochem. 2002, 269, 1662–1669. [Google Scholar] [CrossRef] [PubMed]
- Schultz, S.G.; Solomon, A.K. Cation transport in Escherichia coli. I. Intracellular Na and K concentrations and net cation movement. J. Gen. Physiol. 1961, 45, 355–369. [Google Scholar] [CrossRef] [PubMed]
- Hempfling, W.P.; Höfer, M.; Harris, E.J.; Pressman, B.C. Correlation between changes in metabolite concentrations and rate of ion transport following glucose addition to Escherichia coli B. Biochim. Biophys. Acta 1967, 141, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Anderson, P.; Bauer, W. Supercoiling in closed circular DNA: Dependence upon ion type and concentration. Biochemistry 1978, 17, 594–601. [Google Scholar] [CrossRef] [PubMed]
- Vologodskii, A.V.; Cozzarelli, N.R. Conformational and thermodynamic properties of supercoiled DNA. Annu. Rev. Biophys. Biomol. Struct. 1994, 23, 609–643. [Google Scholar] [CrossRef] [PubMed]
- Rybenkov, V.V.; Vologodskii, A.V.; Cozzarelli, N.R. The effect of ionic conditions on DNA helical repeat, effective diameter and free energy of supercoiling. Nucleic Acids Res. 1997, 25, 1412–1418. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Xu, Y.C.; Bremer, H. Winding of the DNA helix by divalent metal ions. Nucleic Acids Res. 1997, 25, 4067–4071. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kusano, S.; Ding, Q.; Fujita, N.; Ishihama, A. Promoter selectivity of Escherichia coli RNA polymerase E sigma 70 and E sigma 38 holoenzymes. Effect of DNA supercoiling. J. Biol. Chem. 1996, 271, 1998–2004. [Google Scholar] [CrossRef] [PubMed]
- Schneider, R.; Travers, A.; Muskhelishvili, G. FIS modulates growth phase-dependent topological transitions of DNA in Escherichia coli. Mol. Microbiol. 1997, 26, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Bordes, P.; Conter, A.; Morales, V.; Bouvier, J.; Kolb, A.; Gutierrez, C. DNA supercoiling contributes to disconnect sigmaS accumulation from sigmaS-dependent transcription in Escherichia coli. Mol. Microbiol. 2003, 48, 561–571. [Google Scholar] [CrossRef] [PubMed]
- Fulcrand, G.; Dages, S.; Zhi, X.; Chapagain, P.; Gerstman, B.S.; Dunlap, D.; Leng, F. DNA supercoiling, a critical signal regulating the basal expression of the lac operon in Escherichia coli. Sci. Rep. 2016, 6, 19243. [Google Scholar] [CrossRef]
- Gerganova, V.; Maurer, S.; Stoliar, L.; Japaridze, A.; Dietler, G.; Nasser, W.; Kutateladze, T.; Travers, A.; Muskhelishvili, G. Upstream binding of idling RNA polymerase modulates transcription initiation from a nearby promoter. J. Biol. Chem. 2015, 290, 8095–8109. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Japaridze, A.; Muskhelishvili, G.; Benedetti, F.; Gavriilidou, A.F.; Zenobi, R.; De Los Rios, P.; Longo, G.; Dietler, G. Hyperplectonemes: A Higher Order Compact and Dynamic DNA Self-Organization. Nano Lett. 2017, 17, 1938–1948. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.S.; Haakonsen, D.L.; Zeng, W.; Schumacher, M.A.; Laub, M.T. A Bacterial Chromosome Structuring Protein Binds Overtwisted DNA to Stimulate Type II Topoisomerases and Enable DNA Replication. Cell 2018, 175, 583–597.e23. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tarry, M.J.; Harmel, C.; Taylor, J.A.; Marczynski, G.T.; Schmeing, T.M. Structures of GapR reveal a central channel which could accommodate B-DNA. Sci. Rep. 2019, 9, 16679. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Huang, Q.; Duan, B.; Qu, Z.; Fan, S.; Xia, B. The DNA Recognition Motif of GapR Has an Intrinsic DNA Binding Preference towards AT-rich DNA. Molecules. 2021, 26, 5776. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Xu, W.; Yan, Y.; Artsimovitch, I.; Dunlap, D.; Finzi, L. Positive supercoiling favors transcription elongation through lac repressor-mediated DNA loops. Nucleic Acids Res. 2022, 50, 2826–2835. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Visser, B.J.; Sharma, S.; Chen, P.J.; McMullin, A.B.; Bates, M.L.; Bates, D. Psoralen mapping reveals a bacterial genome supercoiling landscape dominated by transcription. Nucleic Acids Res. 2022, 50, 4436–4449. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Schneider, R.; Lurz, R.; Lüder, G.; Tolksdorf, C.; Travers, A.; Muskhelishvili, G. An architectural role of the Escherichia coli chromatin protein FIS in organising DNA. Nucleic Acids Res. 2001, 29, 5107–5114. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Guo, F.; Adhya, S. Spiral structure of Escherichia coli HUalphabeta provides foundation for DNA supercoiling. Proc. Natl. Acad. Sci. USA 2007, 104, 4309–4314. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Maurer, S.; Fritz, J.; Muskhelishvili, G. A systematic in vitro study of nucleoprotein complexes formed by bacterial nucleoid-associated proteins revealing novel types of DNA organization. J. Mol. Biol. 2009, 387, 1261–1276. [Google Scholar] [CrossRef] [PubMed]
- Dame, R.T.; Kalmykowa, O.J.; Grainger, D.C. Chromosomal macrodomains and associated proteins: Implications for DNA organization and replication in gram negative bacteria. PLoS Genet. 2011, 7, e1002123. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Verma, S.C.; Qian, Z.; Adhya, S.L. Architecture of the Escherichia coli nucleoid. PLoS Genet. 2019, 15, e1008456. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Liu, L.F.; Wang, J.C. Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. USA 1987, 84, 7024–7027. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sobetzko, P. Transcription-coupled DNA supercoiling dictates the chromosomal arrangement of bacterial genes. Nucleic Acids Res. 2016, 44, 1514–1524. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dages, S.; Dages, K.; Zhi, X.; Leng, F. Inhibition of the gyrA promoter by transcription-coupled DNA supercoiling in Escherichia coli. Sci. Rep. 2018, 8, 14759. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- El Houdaigui, B.; Forquet, R.; Hindré, T.; Schneider, D.; Nasser, W.; Reverchon, S.; Meyer, S. Bacterial genome architecture shapes global transcriptional regulation by DNA supercoiling. Nucleic Acids Res. 2019, 47, 5648–5657. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jeong, K.S.; Ahn, J.; Khodursky, A.B. Spatial patterns of transcriptional activity in the chromosome of Escherichia coli. Genome Biol. 2004, 5, R86. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Peter, B.J.; Arsuaga, J.; Breier, A.M.; Khodursky, A.B.; Brown, P.O.; Cozzarelli, N.R. Genomic transcriptional response to loss of chromosomal supercoiling in Escherichia coli. Genome Biol. 2004, 5, R87. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Blot, N.; Mavathur, R.; Geertz, M.; Travers, A.; Muskhelishvili, G. Homeostatic regulation of supercoiling sensitivity coordinates transcription of the bacterial genome. EMBO Rep. 2006, 7, 710–715. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ferrándiz, M.J.; Martín-Galiano, A.J.; Arnanz, C.; Camacho-Soguero, I.; Tirado-Vélez, J.M.; de la Campa, A.G. An increase in negative supercoiling in bacteria reveals topology-reacting gene clusters and a homeostatic response mediated by the DNA topoisomerase I gene. Nucleic Acids Res. 2016, 44, 7292–7303. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Behle, A.; Dietsch, M.; Goldschmidt, L.; Murugathas, W.; Berwanger, L.C.; Burmester, J.; Yao, L.; Brandt, D.; Busche, T.; Kalinowski, J.; et al. Manipulation of topoisomerase expression inhibits cell division but not growth and reveals a distinctive promoter structure in Synechocystis. Nucleic Acids Res. 2022, 50, 12790–12808. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pineau, M.; Martis, B.S.; Forquet, R.; Baude, J.; Villard, C.; Grand, L.; Popowycz, F.; Soulère, L.; Hommais, F.; Nasser, W.; et al. What is a supercoiling-sensitive gene? Insights from topoisomerase I inhibition in the Gram-negative bacterium Dickeya dadantii. Nucleic Acids Res. 2022, 50, 9149–9161. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dorman, C.J.; Ni Bhriain, N.; Higgins, C.F. DNA supercoiling and environmental regulation of virulence gene expression in Shigella flexneri. Nature 1990, 344, 789–792. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, L.S.; Rouviere-Yaniv, J.; Drlica, K. Bacterial DNA supercoiling and [ATP]/[ADP] ratio: Changes associated with salt shock. J. Bacteriol. 1991, 173, 3914–3917. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Crozat, E.; Winkworth, C.; Gaffé, J.; Hallin, P.F.; Riley, M.A.; Lenski, R.E.; Schneider, D. Parallel genetic and phenotypic evolution of DNA superhelicity in experimental populations of Escherichia coli. Mol. Biol. Evol. 2010, 27, 2113–2128. [Google Scholar] [CrossRef] [PubMed]
- Hindré, T.; Knibbe, C.; Beslon, G.; Schneider, D. New insights into bacterial adaptation through in vivo and in silico experimental evolution. Nat. Rev. Microbiol. 2012, 10, 352–365. [Google Scholar] [CrossRef] [PubMed]
- Balke, V.L.; Gralla, J.D. Changes in the linking number of supercoiled DNA accompany growth transitions in Escherichia coli. J. Bacteriol. 1987, 169, 4499–4506. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Berger, M.; Farcas, A.; Geertz, M.; Zhelyazkova, P.; Brix, K.; Travers, A.; Muskhelishvili, G. Coordination of genomic structure and transcription by the main bacterial nucleoid-associated protein HU. EMBO Rep. 2010, 11, 59–64. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sobetzko, P.; Travers, A.; Muskhelishvili, G. Gene order and chromosome dynamics coordinate spatiotemporal gene expression during the bacterial growth cycle. Proc. Natl. Acad. Sci. USA 2012, 109, E42–E50. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hengge-Aronis, R. Stationary phase gene regulation: What makes an Escherichia coli promoter sigmaS-selective? Curr. Opin. Microbiol. 2002, 5, 591–595. [Google Scholar] [CrossRef] [PubMed]
- Klauck, E.; Typas, A.; Hengge, R. The sigmaS subunit of RNA polymerase as a signal integrator and network master regulator in the general stress response in Escherichia coli. Sci. Prog. 2007, 90 Pt 2–3, 103–127. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cameron, A.D.S.; Dillon, S.C.; Kröger, C.; Beran, L.; Dorman, C.J. Broad-scale redistribution of mRNA abundance and transcriptional machinery in response to growth rate in Salmonella enterica serovar Typhimurium. Microb. Genom. 2017, 3, e000127. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Nigatu, D.; Henkel, W.; Sobetzko, P.; Muskhelishvili, G. Relationship between digital information and thermodynamic stability in bacterial genomes. EURASIP J. Bioinform. Syst. Biol. 2016, 2016, 4. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Meyer, S.; Reverchon, S.; Nasser, W.; Muskhelishvili, G. Chromosomal organization of transcription: In a nutshell. Curr. Genet. 2018, 64, 555–565. [Google Scholar] [CrossRef] [PubMed]
- Muskhelishvili, G.; Sobetzko, P.; Geertz, M.; Berger, M. General organisational principles of the transcriptional regulation system: A tree or a circle? Mol. Biosyst. 2010, 6, 662–676. [Google Scholar] [CrossRef] [PubMed]
- Sousa, C.; de Lorenzo, V.; Cebolla, A. Modulation of gene expression through chromosomal positioning in Escherichia coli. Microbiology 1997, 143 Pt 6, 2071–2078. [Google Scholar] [CrossRef] [PubMed]
- Teufel, M.; Henkel, W.; Sobetzko, P. The role of replication-induced chromosomal copy numbers in spatio-temporal gene regulation and evolutionary chromosome plasticity. Front. Microbiol. 2023, 14, 1119878. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pountain, A.W.; Jiang, P.; Yao, T.; Homaee, E.; Guan, Y.; McDonald, K.J.C.; Podkowik, M.; Shopsin, B.; Torres, V.J.; Golding, I.; et al. Transcription-replication interactions reveal bacterial genome regulation. Nature 2024, 626, 661–669. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lang, B.; Blot, N.; Bouffartigues, E.; Buckle, M.; Geertz, M.; Gualerzi, C.O.; Mavathur, R.; Muskhelishvili, G.; Pon, C.L.; Rimsky, S.; et al. High-affinity DNA binding sites for H-NS provide a molecular basis for selective silencing within proteobacterial genomes. Nucleic Acids Res. 2007, 35, 6330–6337. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Montero Llopis, P.; Jackson, A.F.; Sliusarenko, O.; Surovtsev, I.; Heinritz, J.; Emonet, T.; Jacobs-Wagner, C. Spatial organization of the flow of genetic information in bacteria. Nature 2010, 466, 77–81. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kuhlman, T.E.; Cox, E.C. Gene location and DNA density determine transcription factor distributions in Escherichia coli. Mol. Syst. Biol. 2012, 8, 610. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Antipov, S.S.; Tutukina, M.N.; Preobrazhenskaya, E.V.; Kondrashov, F.A.; Patrushev, M.V.; Toshchakov, S.V.; Dominova, I.; Shvyreva, U.S.; Vrublevskaya, V.V.; Morenkov, O.S.; et al. The nucleoid protein Dps binds genomic DNA of Escherichia coli in a non-random manner. PLoS ONE 2017, 12, e0182800. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Reverchon, S.; Meyer, S.; Forquet, R.; Hommais, F.; Muskhelishvili, G.; Nasser, W. The nucleoid-associated protein IHF acts as a ‘transcriptional domainin’ protein coordinating the bacterial virulence traits with global transcription. Nucleic Acids Res. 2021, 49, 776–790. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hardy, C.D.; Cozzarelli, N.R. A genetic selection for supercoiling mutants of Escherichia coli reveals proteins implicated in chromosome structure. Mol. Microbiol. 2005, 57, 1636–1652. [Google Scholar] [CrossRef] [PubMed]
- Martín-Galiano, A.J.; Ferrándiz, M.J.; de la Campa, A.G. Bridging Chromosomal Architecture and Pathophysiology of Streptococcus pneumoniae. Genome Biol. Evol. 2017, 9, 350–361. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yan, Y.; Xu, W.; Kumar, S.; Zhang, A.; Leng, F.; Dunlap, D.; Finzi, L. Negative DNA supercoiling makes protein-mediated looping deterministic and ergodic within the bacterial doubling time. Nucleic Acids Res. 2021, 49, 11550–11559. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cortassa, S.; Aon, M.A. Altered topoisomerase activities may be involved in the regulation of DNA supercoiling in aerobic-anaerobic transitions in Escherichia coli. Mol. Cell Biochem. 1993, 126, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Jaén, K.E.; Sigala, J.C.; Olivares-Hernández, R.; Niehaus, K.; Lara, A.R. Heterogeneous oxygen availability affects the titer and topology but not the fidelity of plasmid DNA produced by Escherichia coli. BMC Biotechnol. 2017, 17, 60. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Alexeeva, S.; Hellingwerf, K.J.; Teixeira de Mattos, M.J. Requirement of ArcA for redox regulation in Escherichia coli under microaerobic but not anaerobic or aerobic conditions. J. Bacteriol. 2003, 185, 204–209. [Google Scholar] [CrossRef]
- Levanon, S.S.; San, K.Y.; Bennett, G.N. Effect of oxygen on the Escherichia coli ArcA and FNR regulation systems and metabolic responses. Biotechnol. Bioeng. 2005, 89, 556–564. [Google Scholar] [CrossRef]
- Mika, F.; Hengge, R. A two-component phosphotransfer network involving ArcB, ArcA, and RssB coordinates synthesis and proteolysis of sigmaS (RpoS) in E. coli. Genes Dev. 2005, 19, 2770–2781. [Google Scholar] [CrossRef]
- Muskhelishvili, G.; Sobetzko, P.; Mehandziska, S.; Travers, A. Composition of Transcription Machinery and Its Crosstalk with Nucleoid-Associated Proteins and Global Transcription Factors. Biomolecules 2021, 11, 924. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Borowiec, J.A.; Gralla, J.D. All three elements of the lac ps promoter mediate its transcriptional response to DNA supercoiling. J. Mol. Biol. 1987, 195, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Auner, H.; Buckle, M.; Deufel, A.; Kutateladze, T.; Lazarus, L.; Mavathur, R.; Muskhelishvili, G.; Pemberton, I.; Schneider, R.; Travers, A. Mechanism of transcriptional activation by FIS: Role of core promoter structure and DNA topology. J. Mol. Biol. 2003, 331, 331–344. [Google Scholar] [CrossRef] [PubMed]
- Travers, A.; Muskhelishvili, G. DNA supercoiling—A global transcriptional regulator for enterobacterial growth? Nat. Rev. Microbiol. 2005, 3, 157–169. [Google Scholar] [CrossRef] [PubMed]
- Forquet, R.; Pineau, M.; Nasser, W.; Reverchon, S.; Meyer, S. Role of the Discriminator Sequence in the Supercoiling Sensitivity of Bacterial Promoters. mSystems 2021, 6, e0097821. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Forquet, R.; Nasser, W.; Reverchon, S.; Meyer, S. Quantitative contribution of the spacer length in the supercoiling-sensitivity of bacterial promoters. Nucleic Acids Res. 2022, 50, 7287–7297. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Klein, C.A.; Teufel, M.; Weile, C.J.; Sobetzko, P. The bacterial promoter spacer modulates promoter strength and timing by length, TG-motifs and DNA supercoiling sensitivity. Sci. Rep. 2021, 11, 24399. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rochman, M.; Aviv, M.; Glaser, G.; Muskhelishvili, G. Promoter protection by a transcription factor acting as a local topological homeostat. EMBO Rep. 2002, 3, 355–360. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Olivares-Zavaleta, N.; Jáuregui, R.; Merino, E. Genome analysis of Escherichia coli promoter sequences evidences that DNA static curvature plays a more important role in gene transcription than has previously been anticipated. Genomics 2006, 87, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Balas, D.; Fernández-Moreira, E.; De La Campa, A.G. Molecular characterization of the gene encoding the DNA gyrase A subunit of Streptococcus pneumoniae. J. Bacteriol. 1998, 180, 2854–2861. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hirvonen, C.A.; Ross, W.; Wozniak, C.E.; Marasco, E.; Anthony, J.R.; Aiyar, S.E.; Newburn, V.H.; Gourse, R.L. Contributions of UP elements and the transcription factor FIS to expression from the seven rrn P1 promoters in Escherichia coli. J. Bacteriol. 2001, 183, 6305–6314. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Vijayan, V.; Zuzow, R.; O’Shea, E.K. Oscillations in supercoiling drive circadian gene expression in cyanobacteria. Proc. Natl. Acad. Sci. USA 2009, 106, 22564–22568. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Muskhelishvili, G.; Forquet, R.; Reverchon, S.; Meyer, S.; Nasser, W. Coherent Domains of Transcription Coordinate Gene Expression During Bacterial Growth and Adaptation. Microorganisms 2019, 7, 694. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Le, T.B.; Imakaev, M.V.; Mirny, L.A.; Laub, M.T. High-resolution mapping of the spatial organization of a bacterial chromosome. Science 2013, 342, 731–734. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Le, T.B.; Laub, M.T. Transcription rate and transcript length drive formation of chromosomal interaction domain boundaries. EMBO J. 2016, 35, 1582–1595. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Booker, B.M.; Deng, S.; Higgins, N.P. DNA topology of highly transcribed operons in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 2010, 78, 1348–1364. [Google Scholar] [CrossRef] [PubMed]
- Ito, D.; Kawamura, H.; Oikawa, A.; Ihara, Y.; Shibata, T.; Nakamura, N.; Asano, T.; Kawabata, S.I.; Suzuki, T.; Masuda, S. ppGpp functions as an alarmone in metazoa. Commun. Biol. 2020, 3, 671. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Potrykus, K.; Cashel, M. (p)ppGpp: Still magical? Annu. Rev. Microbiol. 2008, 62, 35–51. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, D.; Collier, J. Effects of (p)ppGpp on the progression of the cell cycle of Caulobacter crescentus. J. Bacteriol. 2014, 196, 2514–2525. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kraemer, J.A.; Sanderlin, A.G.; Laub, M.T. The Stringent Response Inhibits DNA Replication Initiation in E. coli by Modulating Supercoiling of oriC. mBio 2019, 10, e01330-19. [Google Scholar] [CrossRef] [PubMed]
- Cashel, M. Inhibition of RNA polymerase by ppGpp, a nucleotide accumulated during the stringent response to aminoacid starvation in E. coli. Cold Spring Harb. Symp. Quant. Biol. 1970, 35, 407–413. [Google Scholar] [CrossRef]
- Ptashne, M. A Genetic Switch, 2nd ed.; Blackwell Scientific Publications & Cell Press: Hoboken, NY, USA, 1992. [Google Scholar]
- Laganenka, L.; Sander, T.; Lagonenko, A.; Chen, Y.; Link, H.; Sourjik, V. 2019. Quorum Sensing and Metabolic State of the Host Control Lysogeny-Lysis Switch of Bacteriophage T1. MBio 2019, 10, e01884-19. [Google Scholar] [CrossRef]
- Norregaard, K.; Andersson, M.; Sneppen, K.; Nielsen, P.E.; Brown, S.; Oddershede, L.B. Effect of supercoiling on the λ switch. Bacteriophage 2014, 4, e27517. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ding, Y.; Manzo, C.; Fulcrand, G.; Leng, F.; Dunlap, D.; Finzi, L. DNA supercoiling: A regulatory signal for the λ repressor. Proc. Natl. Acad. Sci. USA 2014, 111, 15402–15407. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kahramanoglou, C.; Prieto, A.I.; Khedkar, S.; Haase, B.; Gupta, A.; Benes, V.; Fraser, G.M.; Luscombe, N.M.; Seshasayee, A.S. Genomics of DNA cytosine methylation in Escherichia coli reveals its role in stationary phase transcription. Nat. Commun. 2012, 3, 886. [Google Scholar] [CrossRef] [PubMed]
- Kahramanoglou, C.; Seshasayee, A.S.; Prieto, A.I.; Ibberson, D.; Schmidt, S.; Zimmermann, J.; Benes, V.; Fraser, G.M.; Luscombe, N.M. Direct and indirect effects of H-NS and Fison global gene expression control in Escherichia coli. Nucleic Acids Res. 2011, 39, 2073–2091. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Prieto, A.I.; Kahramanoglou, C.; Ali, R.M.; Fraser, G.M.; Seshasayee, A.S.; Luscombe, N.M. Genomic analysis of DNA binding and gene regulation by homologous nucleoid-associated proteins IHF and HU in Escherichia coli K12. Nucleic Acids Res. 2012, 40, 3524–3537. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Japaridze, A.; Yang, W.; Dekker, C.; Nasser, W.; Muskhelishvili, G. DNA sequence-directed cooperation between nucleoid-associated proteins. iScience 2021, 24, 102408. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hommais, F.; Krin, E.; Laurent-Winter, C.; Soutourina, O.; Malpertuy, A.; Le Caer, J.P.; Danchin, A.; Bertin, P. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol. Microbiol. 2001, 40, 20–36. [Google Scholar] [CrossRef] [PubMed]
- Berger, M.; Gerganova, V.; Berger, P.; Rapiteanu, R.; Lisicovas, V.; Dobrindt, U. Genes on a Wire: The Nucleoid-Associated Protein HU Insulates Transcription Units in Escherichia coli. Sci. Rep. 2016, 6, 31512. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rashid, F.M.; Dame, R.T. 2024: A “nucleoid space” odyssey featuring H-NS. Bioessays 2024, 26, e2400098. [Google Scholar] [CrossRef] [PubMed]
- Ali Azam, T.; Iwata, A.; Nishimura, A.; Ueda, S.; Ishihama, A. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 1999, 181, 6361–6370. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Conway, C.; Beckett, M.C.; Dorman, C.J. The DNA relaxation-dependent OFF-to-ON biasing of the type 1 fimbrial genetic switch requires the Fis nucleoid-associated protein. Microbiology 2023, 169, 001283. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Fujioka, M.; Jaynes, J.B.; Goto, T. Early even-skipped stripes act as morphogenetic gradients at the single cell level to establish engrailed expression. Development 1995, 121, 4371–4382. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Nakamura, Y.; Tomonari, S.; Kawamoto, K.; Yamashita, T.; Watanabe, T.; Ishimaru, Y.; Noji, S.; Mito, T. Evolutionarily conserved function of the even-skipped ortholog in insects revealed by gene knock-out analyses in Gryllus bimaculatus. Dev. Biol. 2022, 485, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Sonnenschein, N.; Geertz, M.; Muskhelishvili, G.; Hütt, M.T. Analog regulation of metabolic demand. BMC Syst. Biol. 2011, 5, 40. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pérez-Mojica, J.E.; Enders, L.; Walsh, J.; Lau, K.H.; Lempradl, A. Continuous transcriptome analysis reveals novel patterns of early gene expression in Drosophila embryos. Cell Genom. 2023, 3, 100265. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gemkow, M.J.; Dichter, J.; Arndt-Jovin, D.J. Developmental regulation of DNA-topoisomerases during Drosophila embryogenesis. Exp. Cell Res. 2001, 262, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Marr, A.G. Growth rate of Escherichia coli. Microbiol. Rev. 1991, 55, 316–333. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Koyama, T.; Mirth, C.K. Unravelling the diversity of mechanisms through which nutrition regulates body size in insects. Curr. Opin. Insect Sci. 2018, 25, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Moshkin, Y.M.; Chalkley, G.E.; Kan, T.W.; Reddy, B.A.; Ozgur, Z.; van Ijcken, W.F.; Dekkers, D.H.; Demmers, J.A.; Travers, A.A.; Verrijzer, C.P. Remodelers organize cellular chromatin by counteracting intrinsic histone-DNA sequence preferences in a class-specific manner. Mol. Cell Biol. 2012, 32, 675–688. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kosmidis, K.; Hütt, M.T. The E. coli transcriptional regulatory network and its spatial embedding. Eur. Phys. J. E Soft Matter. 2019, 42, 30. [Google Scholar] [CrossRef] [PubMed]
- Kosmidis, K.; Jablonski, K.P.; Muskhelishvili, G.; Hütt, M.T. Chromosomal origin of replication coordinates logically distinct types of bacterial genetic regulation. NPJ Syst. Biol. Appl. 2020, 6, 5. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ricci, D.P.; Melfi, M.D.; Lasker, K.; Dill, D.L.; McAdams, H.H.; Shapiro, L. Cell cycle progression in Caulobacter requires a nucleoid-associated protein with high AT sequence recognition. Proc. Natl. Acad. Sci. USA 2016, 113, E5952–E5961. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Biondi, E.G.; Reisinger, S.J.; Skerker, J.M.; Arif, M.; Perchuk, B.S.; Ryan, K.R.; Laub, M.T. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature 2006, 444, 899–904. [Google Scholar] [CrossRef] [PubMed]
- Laub, M.T.; Shapiro, L.; McAdams, H.H. Systems biology of Caulobacter. Annu. Rev. Genet. 2007, 41, 429–441. [Google Scholar] [CrossRef] [PubMed]
- Bhowmick, T.; Ghosh, S.; Dixit, K.; Ganesan, V.; Ramagopal, U.A.; Dey, D.; Sarma, S.P.; Ramakumar, S.; Nagaraja, V. Targeting Mycobacterium tuberculosis nucleoid-associated protein HU with structure-based inhibitors. Nat. Commun. 2014, 5, 4124. [Google Scholar] [CrossRef] [PubMed]
- Suarez, M.A.; Valencia, J.; Cadena, C.C.; Maiti, R.; Datta, C.; Puerto, G.; Isaza, J.H.; San Juan, H.; Nagaraja, V.; Guzman, J.D. Diarylethenes Display In Vitro Anti-TB Activity and Are Efficient Hits Targeting the Mycobacterium tuberculosis HU Protein. Molecules 2017, 22, 1245. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sitarek, P.; Merecz-Sadowska, A.; Sikora, J.; Dudzic, M.; Wiertek-Płoszaj, N.; Picot, L.; Śliwiński, T.; Kowalczyk, T. Flavonoids and their derivatives as DNA topoisomerase inhibitors with anti-cancer activity in various cell models: Exploring a novel mode of action. Pharmacol. Res. 2024, 209, 107457. [Google Scholar] [CrossRef] [PubMed]
- Paul, A.; Nanjunda, R.; Wilson, W.D. Binding to the DNA Minor Groove by Heterocyclic Dications: From AT Specific to GC Recognition Compounds. Curr. Protoc. 2023, 3, e729. [Google Scholar] [CrossRef] [PubMed]
- Guo, P.; Farahat, A.A.; Paul, A.; Kumar, A.; Boykin, D.W.; Wilson, W.D. Extending the σ-Hole Motif for Sequence-Specific Recognition of the DNA Minor Groove. Biochemistry 2020, 59, 1756–1768. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Paul, A.; Guo, P.; Boykin, D.W.; Wilson, W.D. A New Generation of Minor-Groove-Binding-Heterocyclic Diamidines That Recognize G·C Base Pairs in an AT Sequence Context. Molecules 2019, 24, 946. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Metcalfe, N.B. How important is hidden phenotypic plasticity arising from alternative but converging developmental trajectories, and what limits it? J. Exp. Biol. 2024, 227 (Suppl. S1), jeb246010. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Moreno-Gámez, S.; Kiviet, D.J.; Vulin, C.; Schlegel, S.; Schlegel, K.; van Doorn, G.S.; Ackermann, M. Wide lag time distributions break a trade-off between reproduction and survival in bacteria. Proc. Natl. Acad. Sci. USA 2020, 117, 18729–18736. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hales, C.N.; Barker, D.J. The thrifty phenotype hypothesis. Br. Med. Bull. 2001, 60, 5–20. [Google Scholar] [CrossRef] [PubMed]
- Neel, J.V. Diabetes mellitus: A “thrifty” genotype rendered detrimental by “progress”? 1962. Bull World Health Organ. 1999, 77, 694–703; discussion 692–693. [Google Scholar] [PubMed] [PubMed Central]
- Westman, C.A.; Goldbach, L.; Wagner, A. The adaptive Landscapes of Three Global Escherichia coli Transcriptional Regulators. bioRxiv 2024. bioRxiv:11.623025. [Google Scholar] [CrossRef]
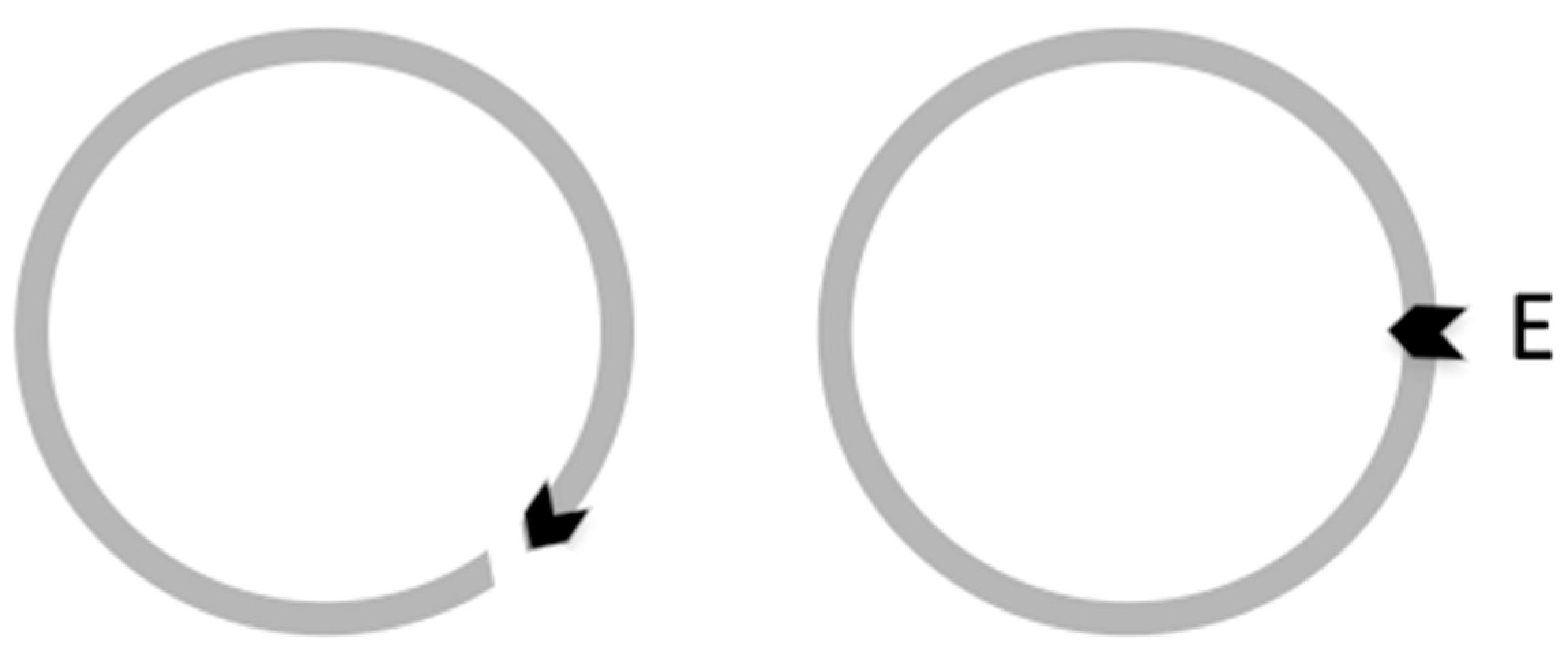

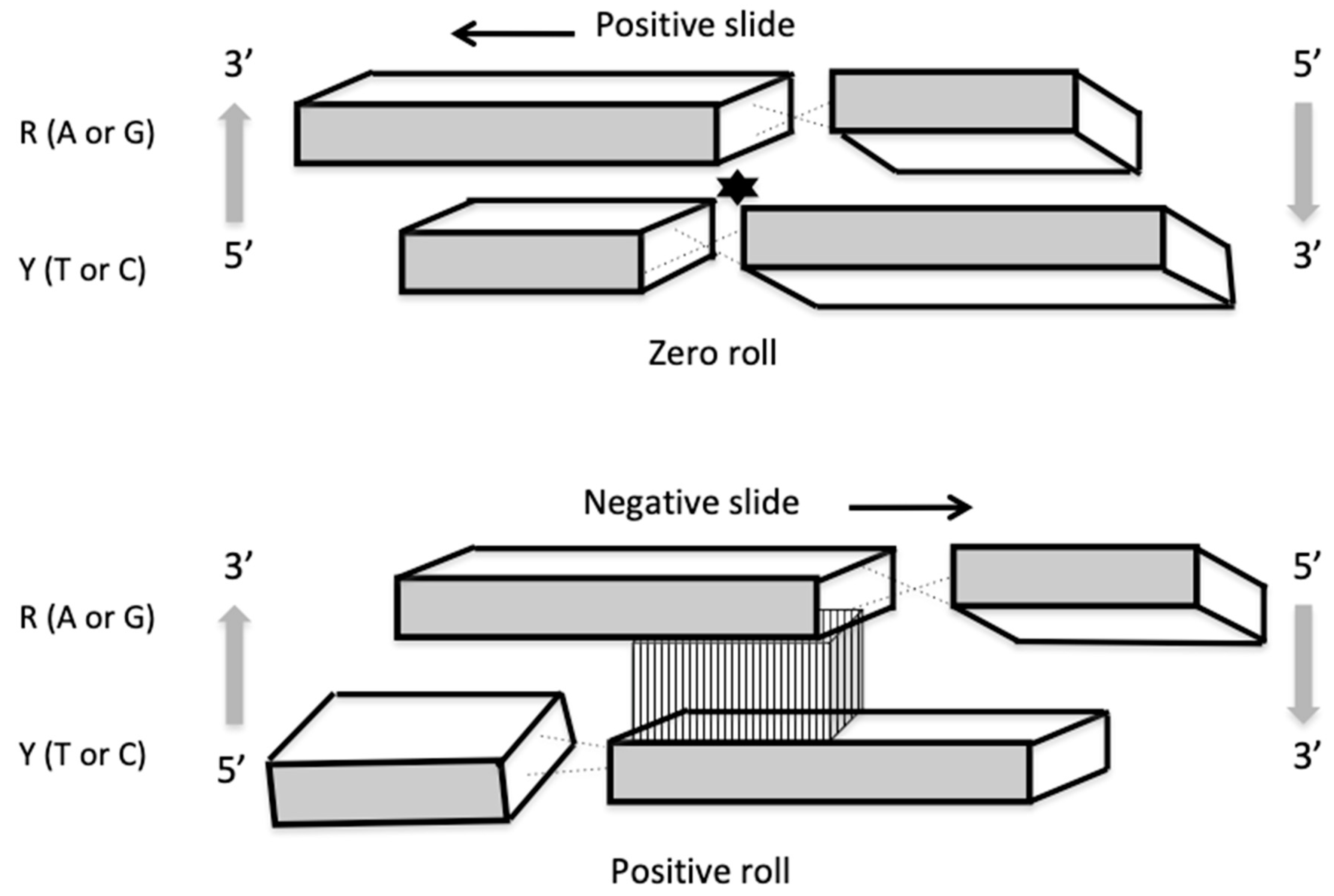
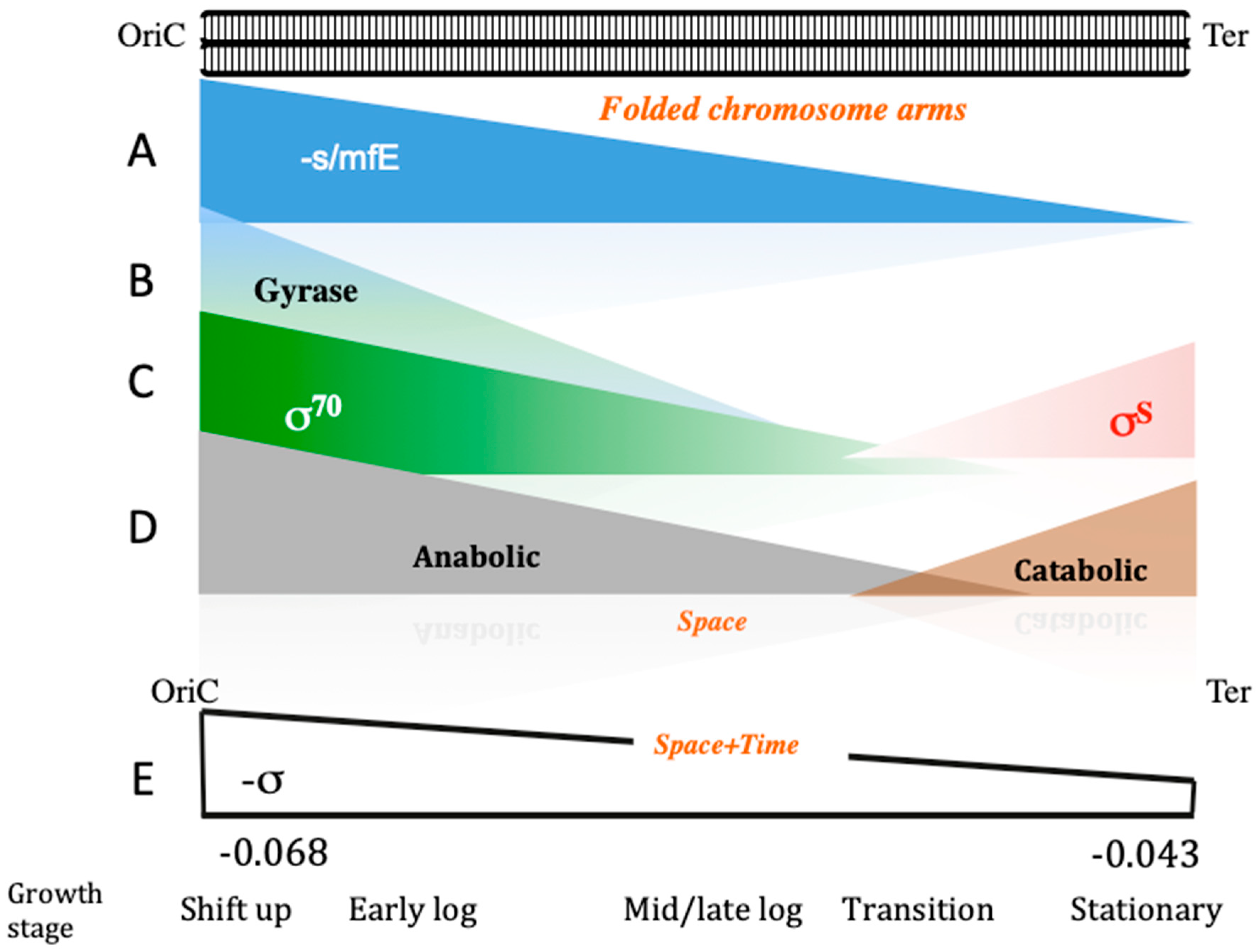
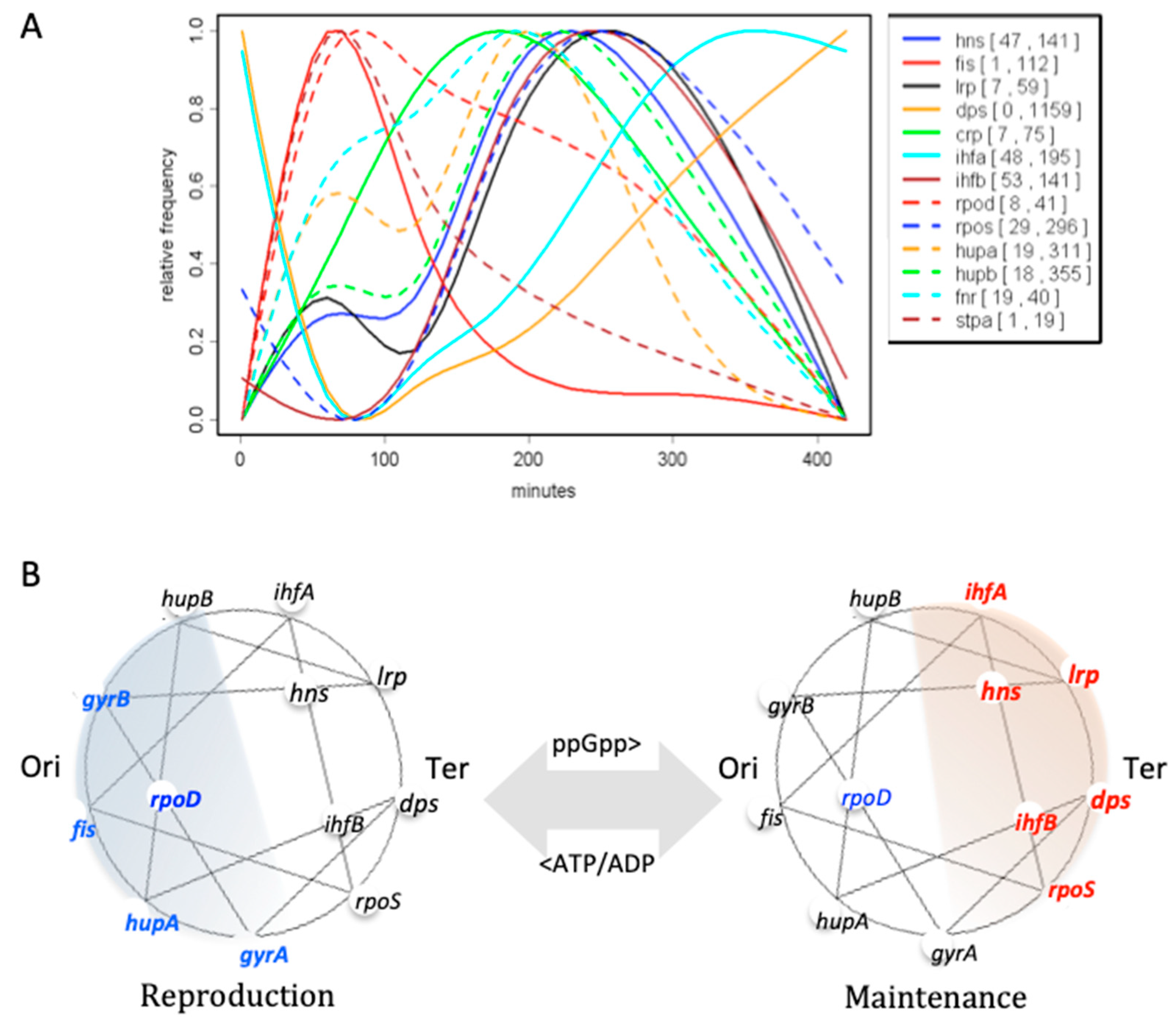
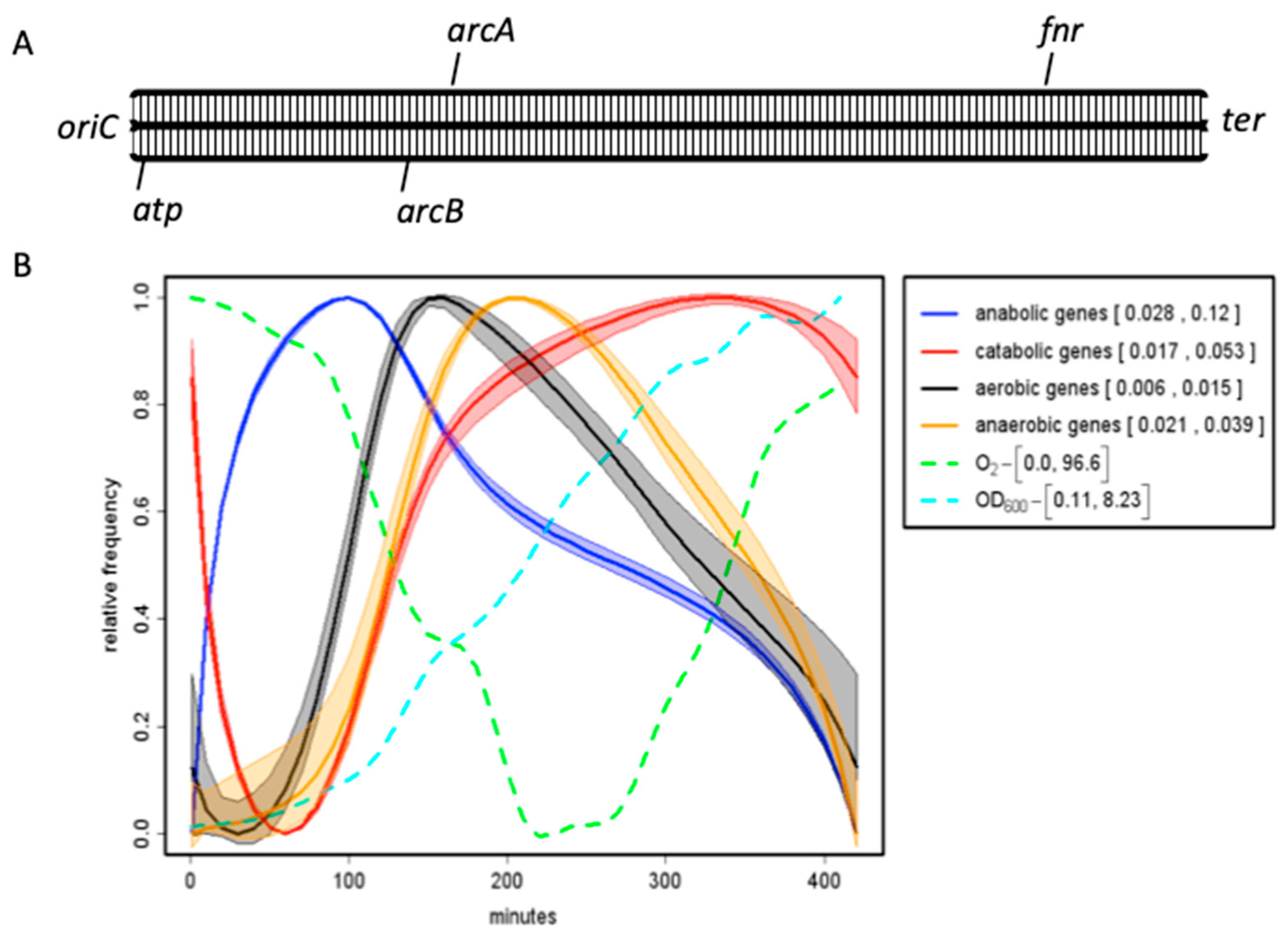

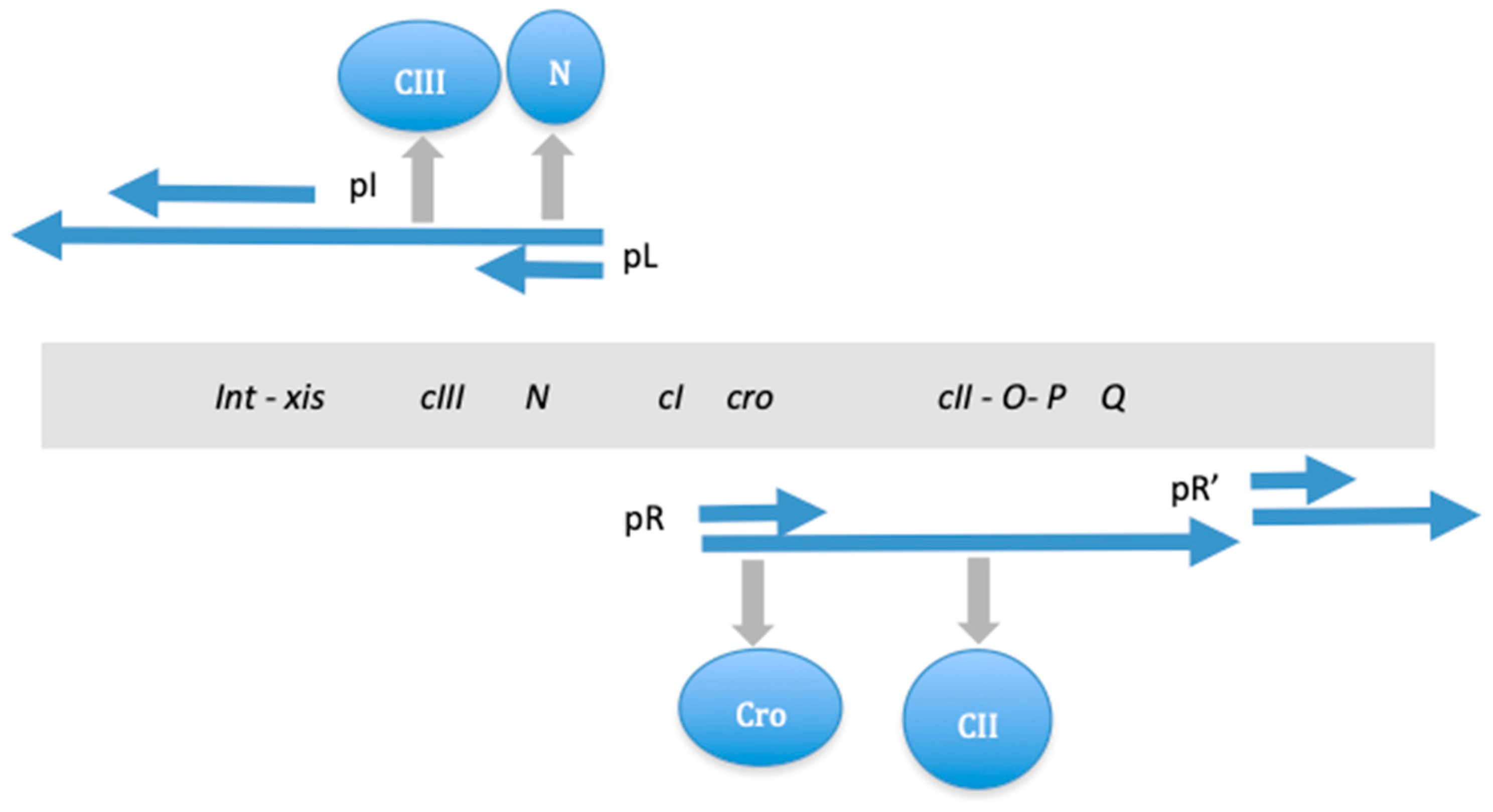
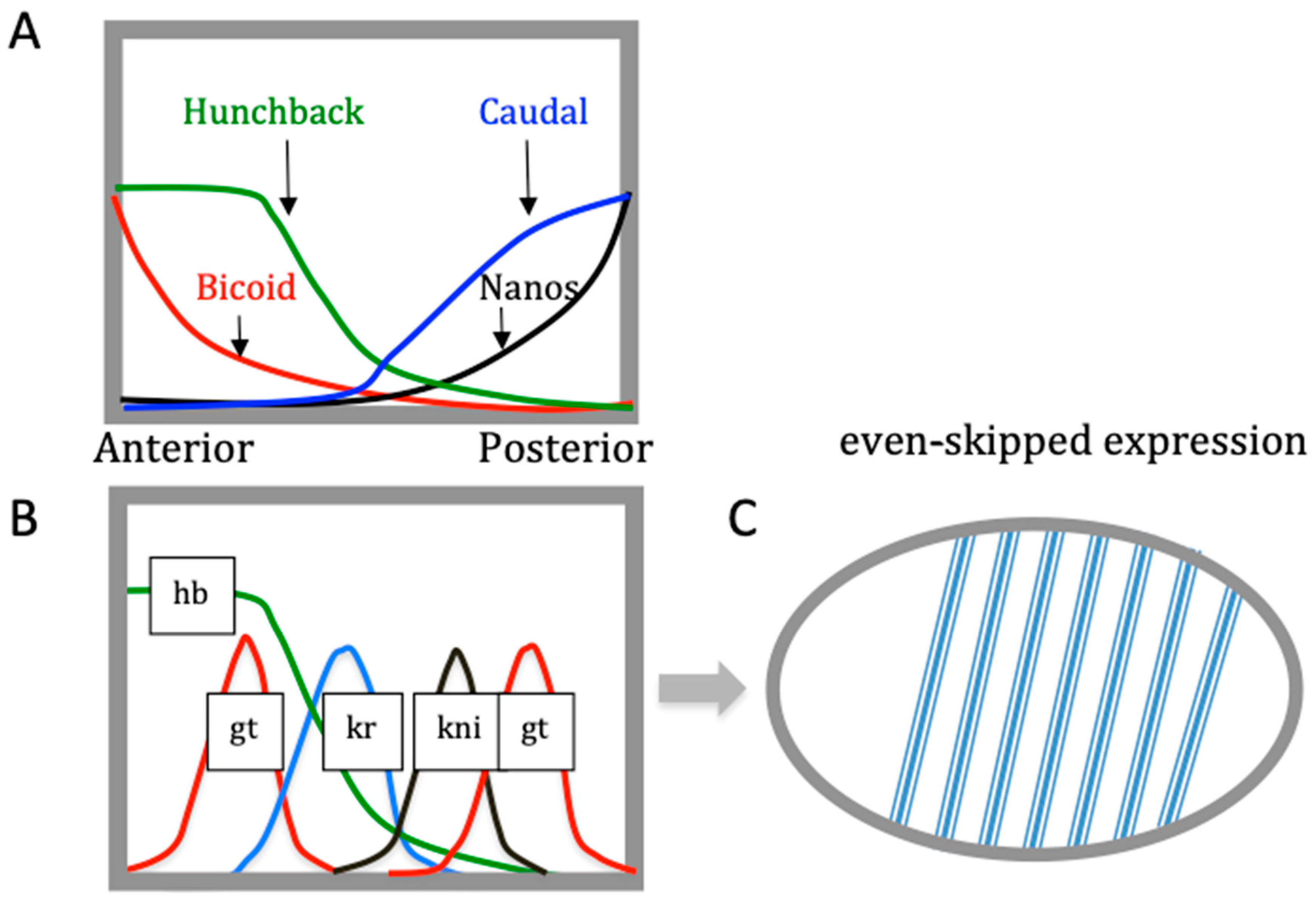
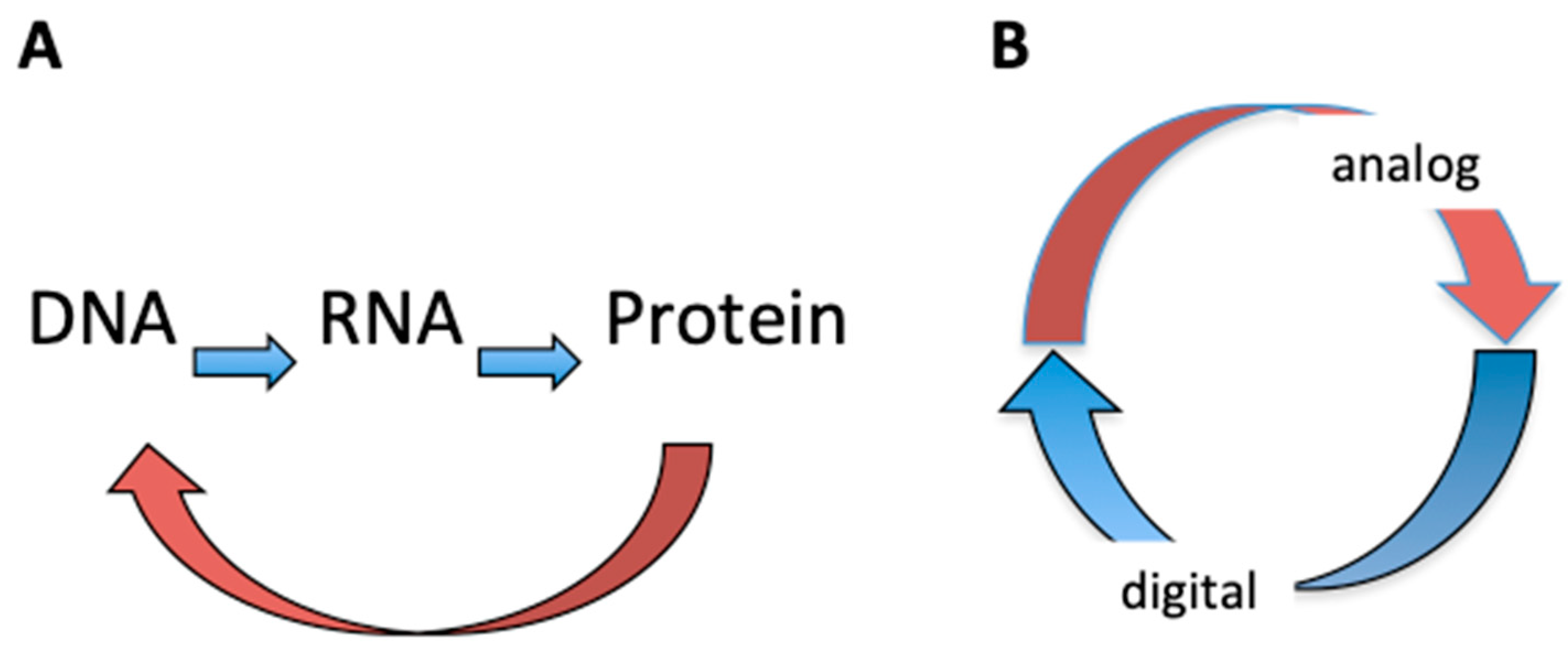
| Base Steps | kcal/mol |
|---|---|
| AA/TT | −1.02 |
| AT/AT | −0.73 |
| TA/TA | −0.60 |
| CA/TG | −1.38 |
| GT/AC | −1.43 |
| CT/AG | −1.16 |
| GA/CT | −1.46 |
| CG/CG | −2.09 |
| GC/GC | −2.28 |
| GG/CC | −1.77 |
| Protein | Exponential Phase | Early Stationary Phase | ||
|---|---|---|---|---|
| No./Cell | Concn (μM) | No./Cell | Concn (μM) | |
| Dps | 8000 | 7 | 120,000 | 100 |
| FIS | 60,000 | 50 | Not detectable | |
| H-NS | 20,000 | 17 | 15,000 | 13 |
| HU | 55,000 | 45 | 25,000 | 20 |
| IHF | 10,000 | 8 | 50,000 | 41 |
| StpA | 25,000 | 28 | 15,000 | 17 |
| Totals | 155 | 191 | ||
| LacI (LacR) | 10 | |||
| RNAP | 4000–6000 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muskhelishvili, G.; Nasser, W.; Reverchon, S.; Travers, A. DNA as a Double-Coding Device for Information Conversion and Organization of a Self-Referential Unity. DNA 2024, 4, 473-493. https://doi.org/10.3390/dna4040032
Muskhelishvili G, Nasser W, Reverchon S, Travers A. DNA as a Double-Coding Device for Information Conversion and Organization of a Self-Referential Unity. DNA. 2024; 4(4):473-493. https://doi.org/10.3390/dna4040032
Chicago/Turabian StyleMuskhelishvili, Georgi, William Nasser, Sylvie Reverchon, and Andrew Travers. 2024. "DNA as a Double-Coding Device for Information Conversion and Organization of a Self-Referential Unity" DNA 4, no. 4: 473-493. https://doi.org/10.3390/dna4040032
APA StyleMuskhelishvili, G., Nasser, W., Reverchon, S., & Travers, A. (2024). DNA as a Double-Coding Device for Information Conversion and Organization of a Self-Referential Unity. DNA, 4(4), 473-493. https://doi.org/10.3390/dna4040032






