Helicases at Work: The Importance of Nucleic Acids Unwinding Under Cold Stress
Abstract
1. Introduction
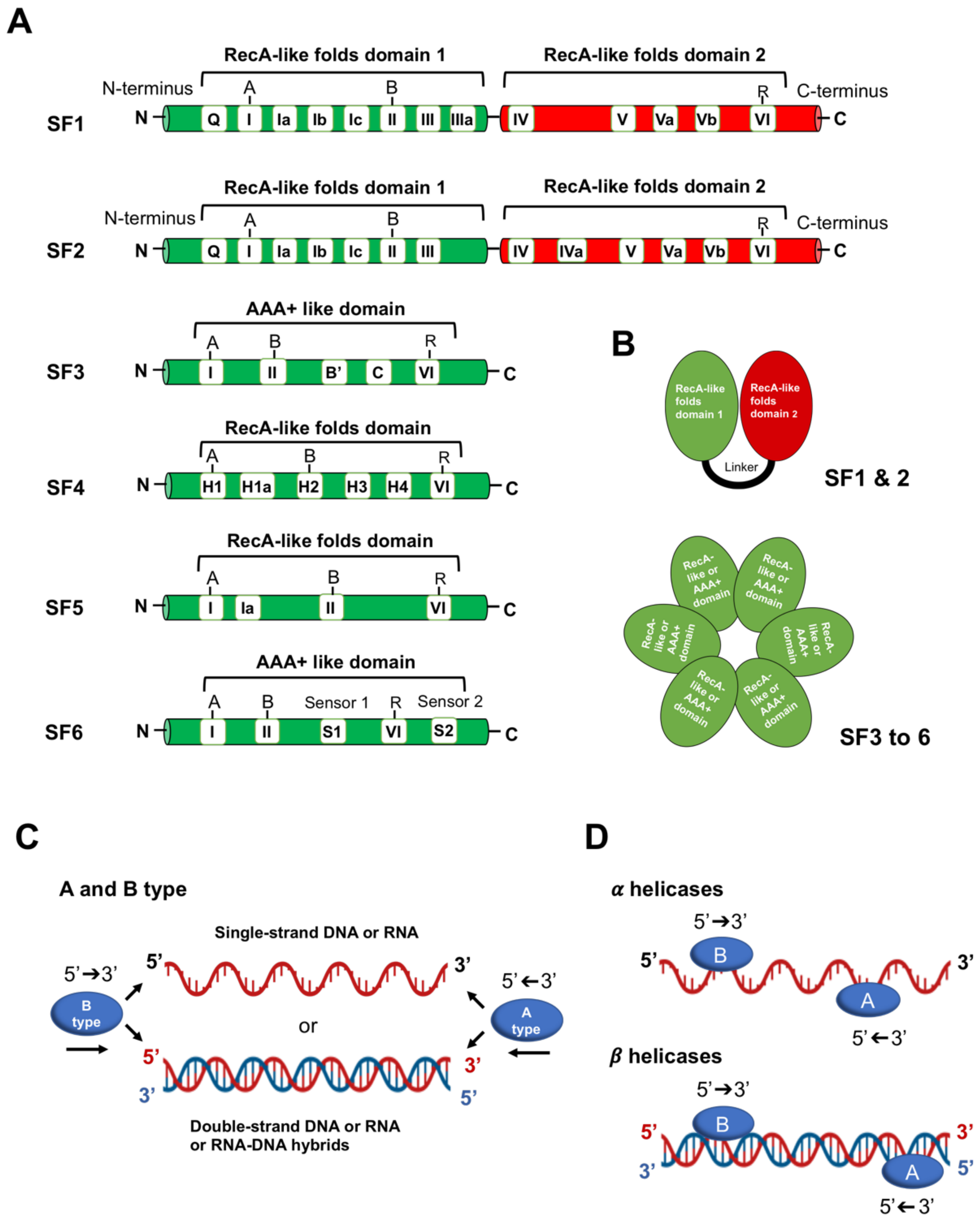
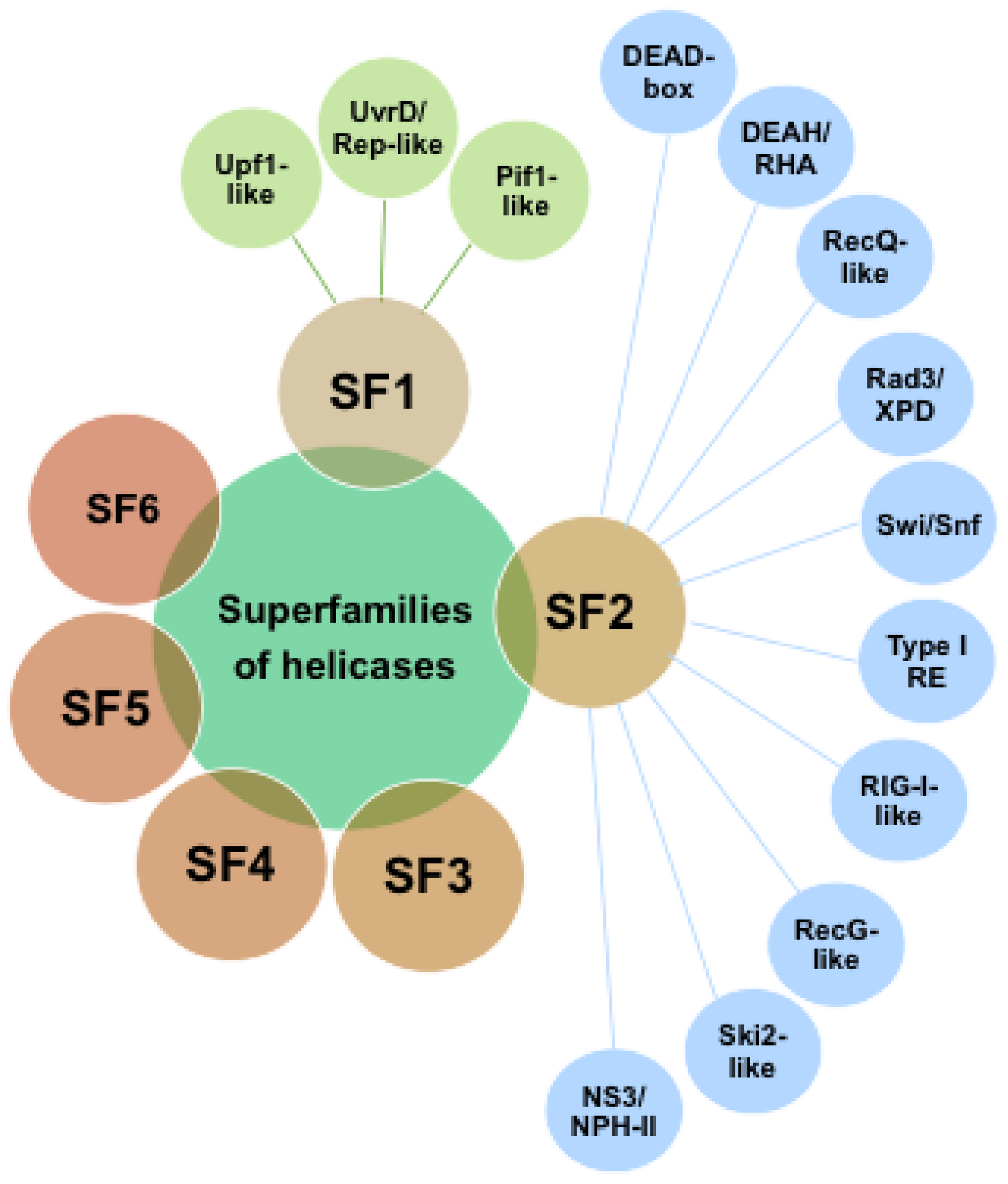
2. Helicases
3. DNA Metabolism at Low Temperatures
3.1. DNA Replication at Subzero Temperature
3.2. DNA Topology
3.3. DNA Replication and Double-Strand Break Repair
4. RNA Metabolism at Low Temperatures
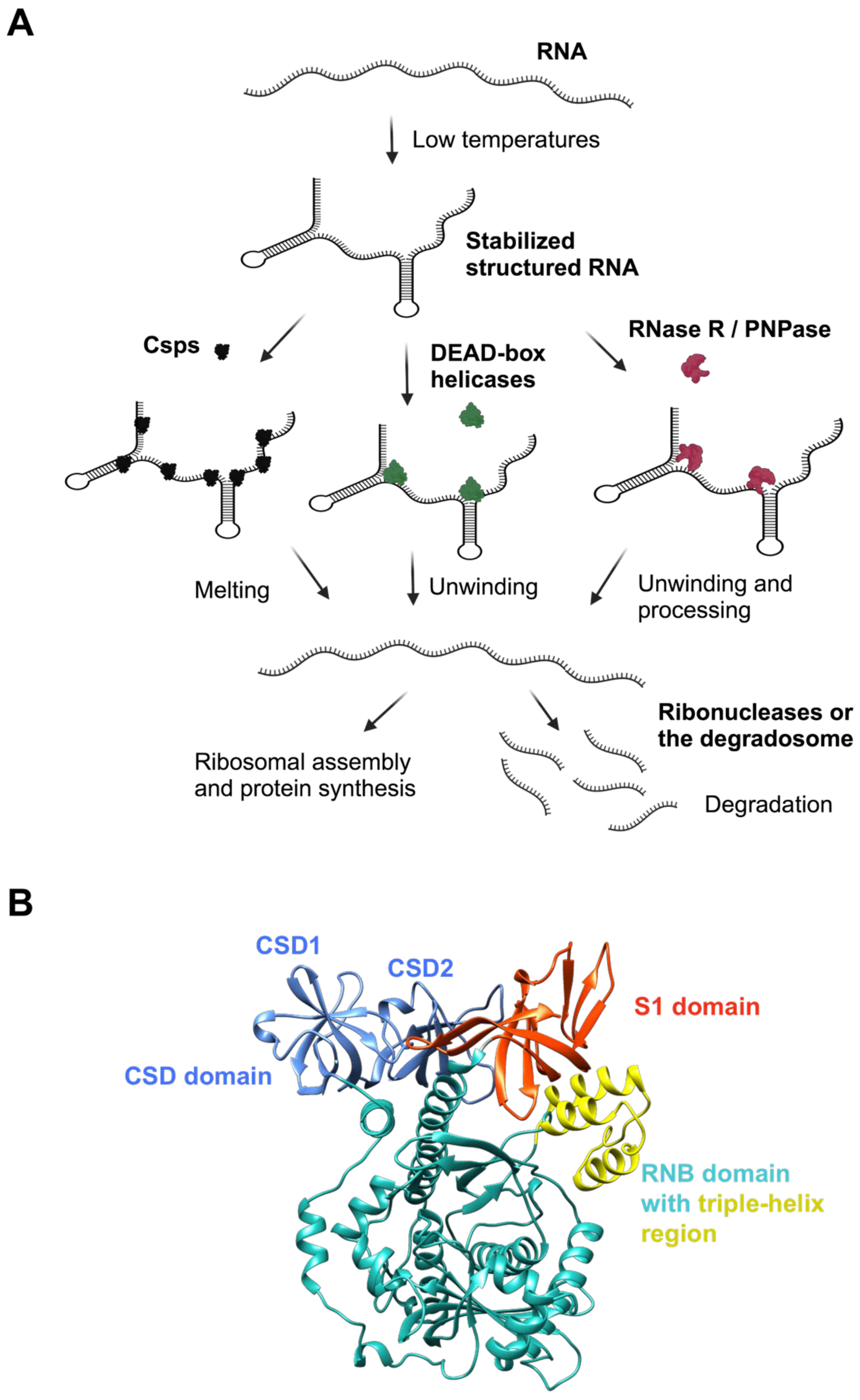
4.1. Cold Shock Response
4.2. DEAD-Box RNA Helicases
4.3. The RNA Degradosome Complex
4.4. Ribonuclease R (RNase R)
5. Cold Stress: Generation of Reactive Oxygen Species
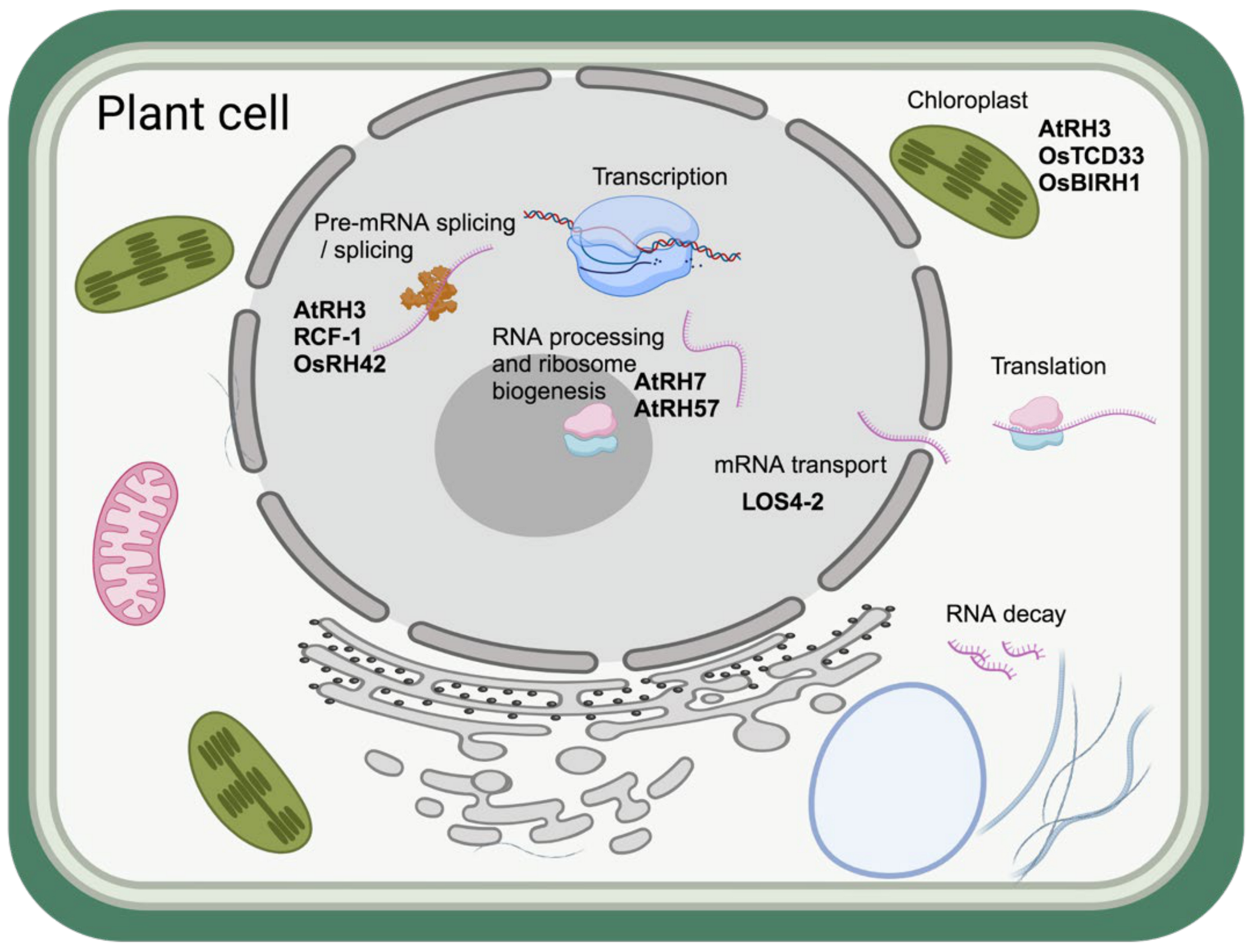
6. Role of Helicases in Cold Adaptation of Plants
7. Significance of Nucleic Acids Unwinding over Degradation in Survival of Microorganisms at Low Temperatures
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Mykytczuk, N.C.S.; Foote, S.J.; Omelon, C.R.; Southam, G.; Greer, C.W.; Whyte, L.G. Bacterial Growth at −15 °C; Molecular Insights from the Permafrost Bacterium Planococcus halocryophilus Or1. ISME J. 2013, 7, 1211–1226. [Google Scholar] [CrossRef] [PubMed]
- Kashefi, K.; Holmes, D.E.; Reysenbach, A.-L.; Lovley, D.R. Use of Fe(III) as an Electron Acceptor to Recover Previously Uncultured Hyperthermophiles: Isolation and Characterization of Geothermobacterium ferrireducens Gen. Nov., Sp. Nov. Appl. Environ. Microbiol. 2002, 68, 1735–1742. [Google Scholar] [CrossRef] [PubMed]
- Pavankumar, T.L.; Mittal, P.; Hallsworth, J.E. Molecular Insights into the Ecology of a Psychrotolerant Pseudomonas syringae. Environ. Microbiol. 2021, 23, 3665–3681. [Google Scholar] [CrossRef] [PubMed]
- Georlette, D.; Blaise, V.; Collins, T.; D’Amico, S.; Gratia, E.; Hoyoux, A.; Marx, J.-C.; Sonan, G.; Feller, G.; Gerday, C. Some like It Cold: Biocatalysis at Low Temperatures. FEMS Microbiol. Rev. 2004, 28, 25–42. [Google Scholar] [CrossRef]
- Berry, E.D.; Foegeding, P.M. Cold Temperature Adaptation and Growth of Microorganisms. J. Food Prot. 1997, 60, 1583–1594. [Google Scholar] [CrossRef]
- Ruelland, E.; Zachowski, A. How Plants Sense Temperature. Environ. Exp. Bot. 2010, 69, 225–232. [Google Scholar] [CrossRef]
- Ding, Y.; Shi, Y.; Yang, S. Advances and Challenges in Uncovering Cold Tolerance Regulatory Mechanisms in Plants. New Phytol. 2019, 222, 1690–1704. [Google Scholar] [CrossRef]
- Akhter, Z.; Bi, Z.; Ali, K.; Sun, C.; Fiaz, S.; Haider, F.U.; Bai, J. In Response to Abiotic Stress, DNA Methylation Confers EpiGenetic Changes in Plants. Plants 2021, 10, 1096. [Google Scholar] [CrossRef]
- Abdel-Monem, M.; Dürwald, H.; Hoffmann-Berling, H. Enzymic Unwinding of DNA. 2. Chain Separation by an ATP-Dependent DNA Unwinding Enzyme. Eur. J. Biochem. 1976, 65, 441–449. [Google Scholar] [CrossRef]
- Abdel-Monem, M.; Hoffmann-Berling, H. Enzymic Unwinding of DNA. 1. Purification and Characterization of a DNA-Dependent ATPase from Escherichia coli. Eur. J. Biochem. 1976, 65, 431–440. [Google Scholar] [CrossRef]
- Mackay, V.; Linn, S. Selective Inhibition of the Dnase Activity of the recBC Enzyme by the DNA Binding Protein from Escherichia coli. J. Biol. Chem. 1976, 251, 3716–3719. [Google Scholar] [CrossRef] [PubMed]
- Geider, K.; Berthold, V.; Abdel-Monem, M.; Hoffman-Berling, H. Control of DNA Structure by Proteins. In The Single-Stranded DNA Phages; Cold Spring Harbor Monogrpah archive; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1978; Volume 8, pp. 379–387. [Google Scholar]
- Singleton, M.R.; Dillingham, M.S.; Wigley, D.B. Structure and Mechanism of Helicases and Nucleic Acid Translocases. Annu. Rev. Biochem. 2007, 76, 23–50. [Google Scholar] [CrossRef] [PubMed]
- Fairman-Williams, M.E.; Guenther, U.-P.; Jankowsky, E. SF1 and SF2 Helicases: Family Matters. Curr. Opin. Struct. Biol. 2010, 20, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Gilhooly, N.S.; Gwynn, E.J.; Dillingham, M.S. Superfamily 1 Helicases. Front. Biosci. (Schol. Ed.) 2013, 5, 206–216. [Google Scholar] [CrossRef] [PubMed]
- Medagli, B.; Onesti, S. Structure and Mechanism of Hexameric Helicases. Adv. Exp. Med. Biol. 2013, 767, 75–95. [Google Scholar] [CrossRef]
- Fernandez, A.J.; Berger, J.M. Mechanisms of Hexameric Helicases. Crit. Rev. Biochem. Mol. Biol. 2021, 56, 621–639. [Google Scholar] [CrossRef]
- West, S.C. The RuvABC Proteins and Holliday Junction Processing in Escherichia coli. J. Bacteriol. 1996, 178, 1237–1241. [Google Scholar] [CrossRef]
- Geggier, S.; Kotlyar, A.; Vologodskii, A. Temperature Dependence of DNA Persistence Length. Nucleic Acids Res. 2011, 39, 1419–1426. [Google Scholar] [CrossRef]
- Dohnalová, H.; Matoušková, E.; Lankaš, F. Temperature-Dependent Elasticity of DNA, RNA, and Hybrid Double Helices. Biophys. J. 2024, 123, 572–583. [Google Scholar] [CrossRef]
- Xue, Y.; Braslavsky, I.; Quake, S.R. Temperature Effect on Polymerase Fidelity. J. Biol. Chem. 2021, 297, 101270. [Google Scholar] [CrossRef]
- Tuorto, S.J.; Darias, P.; McGuinness, L.R.; Panikov, N.; Zhang, T.; Häggblom, M.M.; Kerkhof, L.J. Bacterial Genome Replication at Subzero Temperatures in Permafrost. ISME J. 2014, 8, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, T.; Kataoka, K.; Ogata, Y.; Inoue, R.; Sekimizu, K. Increase in Negative Supercoiling of Plasmid DNA in Escherichia coli Exposed to Cold Shock. Mol. Microbiol. 1997, 23, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Fogg, J.M.; Judge, A.K.; Stricker, E.; Chan, H.L.; Zechiedrich, L. Supercoiling and Looping Promote DNA Base Accessibility and Coordination among Distant Sites. Nat. Commun. 2021, 12, 5683. [Google Scholar] [CrossRef] [PubMed]
- Drew, H.R.; Weeks, J.R.; Travers, A.A. Negative Supercoiling Induces Spontaneous Unwinding of a Bacterial Promoter. EMBO J. 1985, 4, 1025–1032. [Google Scholar] [CrossRef] [PubMed]
- Magnan, D.; Bates, D. Regulation of DNA Replication Initiation by Chromosome Structure. J. Bacteriol. 2015, 197, 3370–3377. [Google Scholar] [CrossRef] [PubMed]
- Sinha, A.K.; Pavankumar, T.L.; Kamisetty, S.; Mittal, P.; Ray, M.K. Replication Arrest Is a Major Threat to Growth at Low Temperature in Antarctic Pseudomonas syringae Lz4W. Mol. Microbiol. 2013, 89, 792–810. [Google Scholar] [CrossRef]
- Regha, K.; Satapathy, A.K.; Ray, M.K. RecD Plays an Essential Function during Growth at Low Temperature in the Antarctic Bacterium Pseudomonas syringae Lz4W. Genetics 2005, 170, 1473–1484. [Google Scholar] [CrossRef]
- Satapathy, A.K.; Pavankumar, T.L.; Bhattacharjya, S.; Sankaranarayanan, R.; Ray, M.K. ATPase Activity of RecD Is Essential for Growth of the Antarctic Pseudomonas syringae Lz4W at Low Temperature. FEBS J. 2008, 275, 1835–1851. [Google Scholar] [CrossRef]
- Amundsen, S.K.; Taylor, A.F.; Chaudhury, A.M.; Smith, G.R. recD: The Gene for an Essential Third Subunit of Exonuclease V. Proc. Natl. Acad. Sci. USA 1986, 83, 5558–5562. [Google Scholar] [CrossRef]
- Pavankumar, T.L.; Sinha, A.K.; Ray, M.K. All Three Subunits of RecBCD Enzyme Are Essential for DNA Repair and Low-Temperature Growth in the Antarctic Pseudomonas syringae Lz4W. PLoS ONE 2010, 5, e9412. [Google Scholar] [CrossRef]
- Dillingham, M.S.; Kowalczykowski, S.C. RecBCD Enzyme and the Repair of Double-Stranded DNA Breaks. Microbiol. Mol. Biol. Rev. 2008, 72, 642–671. [Google Scholar] [CrossRef] [PubMed]
- Wiktor, J.; van der Does, M.; Büller, L.; Sherratt, D.J.; Dekker, C. Direct Observation of End Resection by RecBCD during Double-Stranded DNA Break Repair in Vivo. Nucleic Acids Res. 2018, 46, 1821–1833. [Google Scholar] [CrossRef] [PubMed]
- Sivaramakrishnan, P.; Sepúlveda, L.A.; Halliday, J.A.; Liu, J.; Núñez, M.A.B.; Golding, I.; Rosenberg, S.M.; Herman, C. The Transcription Fidelity Factor GreA Impedes DNA Break Repair. Nature 2017, 550, 214–218. [Google Scholar] [CrossRef] [PubMed]
- Dillingham, M.S.; Spies, M.; Kowalczykowski, S.C. RecBCD Enzyme Is a Bipolar DNA Helicase. Nature 2003, 423, 893–897. [Google Scholar] [CrossRef]
- Phillips, R.J.; Hickleton, D.C.; Boehmer, P.E.; Emmerson, P.T. The RecB Protein of Escherichia coli Translocates along Single-Stranded DNA in the 3′ to 5′ Direction: A Proposed Ratchet Mechanism. Mol. Gen. Genet. 1997, 254, 319–329. [Google Scholar] [CrossRef]
- Yeeles, J.T.P.; Dillingham, M.S. The Processing of Double-Stranded DNA Breaks for Recombinational Repair by Helicase-Nuclease Complexes. DNA Repair 2010, 9, 276–285. [Google Scholar] [CrossRef]
- Yu, M.; Souaya, J.; Julin, D.A. The 30-kDa C-Terminal Domain of the RecB Protein Is Critical for the Nuclease Activity, but Not the Helicase Activity, of the RecBCD Enzyme from Escherichia coli. Proc. Natl. Acad. Sci. USA 1998, 95, 981–986. [Google Scholar] [CrossRef]
- Korangy, F.; Julin, D.A. Kinetics and Processivity of ATP Hydrolysis and DNA Unwinding by the RecBC Enzyme from Escherichia coli. Biochemistry 1993, 32, 4873–4880. [Google Scholar] [CrossRef]
- Wu, C.G.; Bradford, C.; Lohman, T.M. Escherichia coli RecBC Helicase Has Two Translocase Activities Controlled by a Single ATPase Motor. Nat. Struct. Mol. Biol. 2010, 17, 1210–1217. [Google Scholar] [CrossRef]
- Roman, L.J.; Eggleston, A.K.; Kowalczykowski, S.C. Processivity of the DNA Helicase Activity of Escherichia coli recBCD Enzyme. J. Biol. Chem. 1992, 267, 4207–4214. [Google Scholar] [CrossRef]
- Liu, B.; Baskin, R.J.; Kowalczykowski, S.C. DNA Unwinding Heterogeneity by RecBCD Results from Static Molecules Able to Equilibrate. Nature 2013, 500, 482–485. [Google Scholar] [CrossRef] [PubMed]
- Singleton, M.R.; Dillingham, M.S.; Gaudier, M.; Kowalczykowski, S.C.; Wigley, D.B. Crystal Structure of RecBCD Enzyme Reveals a Machine for Processing DNA Breaks. Nature 2004, 432, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Pavankumar, T.L.; Wong, C.J.; Wong, Y.K.; Spies, M.; Kowalczykowski, S.C. Trans-Complementation by the RecB Nuclease Domain of RecBCD Enzyme Reveals New Insight into RecA Loading upon χ Recognition. Nucleic Acids Res. 2024, 52, 2578–2589. [Google Scholar] [CrossRef]
- Fazio, N.T.; Mersch, K.N.; Hao, L.; Lohman, T.M. E. coli RecB Nuclease Domain Regulates RecBCD Helicase Activity but Not Single Stranded DNA Translocase Activity. J. Mol. Biol. 2024, 436, 168381. [Google Scholar] [CrossRef]
- Cheng, K.; Wilkinson, M.; Chaban, Y.; Wigley, D.B. A Conformational Switch in Response to Chi Converts RecBCD from Phage Destruction to DNA Repair. Nat. Struct. Mol. Biol. 2020, 27, 71–77. [Google Scholar] [CrossRef]
- Pavankumar, T.L.; Sinha, A.K.; Ray, M.K. Biochemical Characterization of RecBCD Enzyme from an Antarctic Pseudomonas Species and Identification of Its Cognate Chi (Chi) Sequence. PLoS ONE 2018, 13, e0197476. [Google Scholar] [CrossRef]
- Barria, C.; Malecki, M.; Arraiano, C.M. Bacterial Adaptation to Cold. Microbiology 2013, 159, 2437–2443. [Google Scholar] [CrossRef] [PubMed]
- Becskei, A.; Rahaman, S. The Life and Death of RNA across Temperatures. Comput. Struct. Biotechnol. J. 2022, 20, 4325–4336. [Google Scholar] [CrossRef]
- Schindelin, H.; Jiang, W.; Inouye, M.; Heinemann, U. Crystal Structure of CspA, the Major Cold Shock Protein of Escherichia coli. Proc. Natl. Acad. Sci. USA 1994, 91, 5119–5123. [Google Scholar] [CrossRef]
- Goldstein, J.; Pollitt, N.S.; Inouye, M. Major Cold Shock Protein of Escherichia coli. Proc. Natl. Acad. Sci. USA 1990, 87, 283–287. [Google Scholar] [CrossRef]
- Giuliodori, A.M.; Di Pietro, F.; Marzi, S.; Masquida, B.; Wagner, R.; Romby, P.; Gualerzi, C.O.; Pon, C.L. The cspA mRNA Is a Thermosensor That Modulates Translation of the Cold-Shock Protein CspA. Mol. Cell 2010, 37, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Phadtare, S.; Inouye, M.; Severinov, K. The Nucleic Acid Melting Activity of Escherichia coli CspE Is Critical for Transcription Antitermination and Cold Acclimation of Cells. J. Biol. Chem. 2002, 277, 7239–7245. [Google Scholar] [CrossRef]
- Zhang, Y.; Gross, C.A. Cold Shock Response in Bacteria. Annu. Rev. Genet. 2021, 55, 377–400. [Google Scholar] [CrossRef]
- Linder, P.; Lasko, P.F.; Ashburner, M.; Leroy, P.; Nielsen, P.J.; Nishi, K.; Schnier, J.; Slonimski, P.P. Birth of the D-E-A-D Box. Nature 1989, 337, 121–122. [Google Scholar] [CrossRef]
- Lorsch, J.R.; Herschlag, D. The DEAD Box Protein eIF4A. 2. A Cycle of Nucleotide and RNA-Dependent Conformational Changes. Biochemistry 1998, 37, 2194–2206. [Google Scholar] [CrossRef] [PubMed]
- Cartier, G.; Lorieux, F.; Allemand, F.; Dreyfus, M.; Bizebard, T. Cold Adaptation in DEAD-Box Proteins. Biochemistry 2010, 49, 2636–2646. [Google Scholar] [CrossRef]
- Charollais, J.; Dreyfus, M.; Iost, I. CsdA, a Cold-Shock RNA Helicase from Escherichia coli, Is Involved in the Biogenesis of 50S Ribosomal Subunit. Nucleic Acids Res. 2004, 32, 2751–2759. [Google Scholar] [CrossRef] [PubMed]
- Charollais, J.; Pflieger, D.; Vinh, J.; Dreyfus, M.; Iost, I. The DEAD-Box RNA Helicase SrmB Is Involved in the Assembly of 50S Ribosomal Subunits in Escherichia coli. Mol. Microbiol. 2003, 48, 1253–1265. [Google Scholar] [CrossRef]
- Awano, N.; Xu, C.; Ke, H.; Inoue, K.; Inouye, M.; Phadtare, S. Complementation Analysis of the Cold-Sensitive Phenotype of the Escherichia coli csdA Deletion Strain. J. Bacteriol. 2007, 189, 5808–5815. [Google Scholar] [CrossRef]
- Hussain, A.; Ray, M.K. Role of DEAD-Box RNA Helicases in Low-Temperature Adapted Growth of Antarctic Pseudomonas syringae Lz4W. Microbiol. Spectr. 2024, 12, e0433522. [Google Scholar] [CrossRef]
- Jiang, X.; Keto-Timonen, R.; Skurnik, M.; Korkeala, H. Role of DEAD-Box RNA Helicase Genes in the Growth of Yersinia pseudotuberculosis IP32953 under Cold, pH, Osmotic, Ethanol and Oxidative Stresses. PLoS ONE 2019, 14, e0219422. [Google Scholar] [CrossRef] [PubMed]
- Lehnik-Habrink, M.; Rempeters, L.; Kovács, Á.T.; Wrede, C.; Baierlein, C.; Krebber, H.; Kuipers, O.P.; Stülke, J. DEAD-Box RNA Helicases in Bacillus subtilis Have Multiple Functions and Act Independently from Each Other. J. Bacteriol. 2013, 195, 534–544. [Google Scholar] [CrossRef] [PubMed]
- Pandiani, F.; Brillard, J.; Bornard, I.; Michaud, C.; Chamot, S.; Nguyen-the, C.; Broussolle, V. Differential Involvement of the Five RNA Helicases in Adaptation of Bacillus cereus ATCC 14579 to Low Growth Temperatures. Appl. Environ. Microbiol. 2010, 76, 6692–6697. [Google Scholar] [CrossRef]
- Carpousis, A.J. The RNA Degradosome of Escherichia coli: An mRNA-Degrading Machine Assembled on RNase E. Annu. Rev. Microbiol. 2007, 61, 71–87. [Google Scholar] [CrossRef] [PubMed]
- Prud’homme-Genereux, A.; Beran, R.K.; Iost, I.; Ramey, C.S.; Mackie, G.A.; Simons, R.W. Physical and Functional Interactions among RNase E, Polynucleotide Phosphorylase and the Cold-Shock Protein, CsdA: Evidence for a “Cold Shock Degradosome”. Mol. Microbiol. 2004, 54, 1409–1421. [Google Scholar] [CrossRef]
- Aguirre, A.A.; Vicente, A.M.; Hardwick, S.W.; Alvelos, D.M.; Mazzon, R.R.; Luisi, B.F.; Marques, M.V. Association of the Cold Shock DEAD-Box RNA Helicase RhlE to the RNA Degradosome in Caulobacter Crescentus. J. Bacteriol. 2017, 199, e00135-17. [Google Scholar] [CrossRef]
- Purusharth, R.I.; Klein, F.; Sulthana, S.; Jager, S.; Jagannadham, M.V.; Evguenieva-Hackenberg, E.; Ray, M.K.; Klug, G. Exoribonuclease R Interacts with Endoribonuclease E and an RNA Helicase in the Psychrotrophic Bacterium Pseudomonas syringae Lz4W. J. Biol. Chem. 2005, 280, 14572–14578. [Google Scholar] [CrossRef]
- Pavankumar, T.L. RNase R vs. PNPase: Selecting the Best-Suited Exoribonuclease for Environmental Adaptation. Extremophiles 2024, 28, 35. [Google Scholar] [CrossRef]
- Awano, N.; Rajagopal, V.; Arbing, M.; Patel, S.; Hunt, J.; Inouye, M.; Phadtare, S. Escherichia coli RNase R Has Dual Activities, Helicase and RNase. J. Bacteriol. 2010, 192, 1344–1352. [Google Scholar] [CrossRef]
- Mittal, P.; Sipani, R.; Pandiyan, A.; Sulthana, S.; Sinha, A.K.; Hussain, A.; Ray, M.K.; Pavankumar, T.L. Exoribonuclease RNase R Protects Antarctic Pseudomonas syringae Lz4W from DNA Damage and Oxidative Stress. Appl. Environ. Microbiol. 2023, 89, e0116823. [Google Scholar] [CrossRef]
- Sulthana, S.; Deutscher, M.P. Multiple Exoribonucleases Catalyze Maturation of the 3’ Terminus of 16S Ribosomal RNA (rRNA). J. Biol. Chem. 2013, 288, 12574–12579. [Google Scholar] [CrossRef] [PubMed]
- Carpousis, A.J.; Van Houwe, G.; Ehretsmann, C.; Krisch, H.M. Copurification of E. coli RNAase E and PNPase: Evidence for a Specific Association between Two Enzymes Important in RNA Processing and Degradation. Cell 1994, 76, 889–900. [Google Scholar] [CrossRef] [PubMed]
- Luttinger, A.; Hahn, J.; Dubnau, D. Polynucleotide Phosphorylase Is Necessary for Competence Development in Bacillus subtilis. Mol. Microbiol. 1996, 19, 343–356. [Google Scholar] [CrossRef] [PubMed]
- Piazza, F.; Zappone, M.; Sana, M.; Briani, F.; Deho, G. Polynucleotide Phosphorylase of Escherichia coli Is Required for the Establishment of Bacteriophage P4 Immunity. J. Bacteriol. 1996, 178, 5513–5521. [Google Scholar] [CrossRef]
- Carzaniga, T.; Sbarufatti, G.; Briani, F.; Deho, G. Polynucleotide Phosphorylase Is Implicated in Homologous Recombination and DNA Repair in Escherichia coli. BMC Microbiol. 2017, 17, 81. [Google Scholar] [CrossRef]
- Wu, J.; Jiang, Z.; Liu, M.; Gong, X.; Wu, S.; Burns, C.M.; Li, Z. Polynucleotide Phosphorylase Protects Escherichia coli against Oxidative Stress. Biochemistry 2009, 48, 2012–2020. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Y.; Deutscher, M.P. Exoribonuclease Superfamilies: Structural Analysis and Phylogenetic Distribution. Nucleic Acids Res. 2001, 29, 1017–1026. [Google Scholar] [CrossRef]
- Cheng, Z.F.; Zuo, Y.; Li, Z.; Rudd, K.E.; Deutscher, M.P. The vacB Gene Required for Virulence in Shigella Flexneri and Escherichia coli Encodes the Exoribonuclease RNase R. J. Biol. Chem. 1998, 273, 14077–14080. [Google Scholar] [CrossRef]
- Donovan, W.P.; Kushner, S.R. Polynucleotide Phosphorylase and Ribonuclease II Are Required for Cell Viability and mRNA Turnover in Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 1986, 83, 120–124. [Google Scholar] [CrossRef]
- Spickler, C.; Mackie, G.A. Action of RNase II and Polynucleotide Phosphorylase against RNAs Containing Stem-Loops of Defined Structure. J. Bacteriol. 2000, 182, 2422–2427. [Google Scholar] [CrossRef]
- Cheng, Z.F.; Deutscher, M.P. An Important Role for RNase R in mRNA Decay. Mol. Cell 2005, 17, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Hossain, S.T.; Malhotra, A.; Deutscher, M.P. How RNase R Degrades Structured RNA: Role of the Helicase Activity and the S1 Domain. J. Biol. Chem. 2016, 291, 7877–7887. [Google Scholar] [CrossRef] [PubMed]
- Cairrao, F.; Cruz, A.; Mori, H.; Arraiano, C.M. Cold Shock Induction of RNase R and Its Role in the Maturation of the Quality Control Mediator SsrA/tmRNA. Mol. Microbiol. 2003, 50, 1349–1360. [Google Scholar] [CrossRef]
- Jones, P.G.; VanBogelen, R.A.; Neidhardt, F.C. Induction of Proteins in Response to Low Temperature in Escherichia coli. J. Bacteriol. 1987, 169, 2092–2095. [Google Scholar] [CrossRef]
- Zangrossi, S.; Briani, F.; Ghisotti, D.; Regonesi, M.E.; Tortora, P.; Dehò, G. Transcriptional and Post-Transcriptional Control of Polynucleotide Phosphorylase during Cold Acclimation in Escherichia coli. Mol. Microbiol. 2000, 36, 1470–1480. [Google Scholar] [CrossRef] [PubMed]
- Purusharth, R.I.; Madhuri, B.; Ray, M.K. Exoribonuclease R in Pseudomonas syringae Is Essential for Growth at Low Temperature and Plays a Novel Role in the 3’ End Processing of 16 and 5 S Ribosomal RNA. J. Biol. Chem. 2007, 282, 16267–16277. [Google Scholar] [CrossRef]
- Mittal, P.; Sinha, A.K.; Pandiyan, A.; Kumari, L.; Ray, M.K.; Pavankumar, T.L. A Type II Toxin-Antitoxin System Is Responsible for the Cell Death at Low Temperature in Pseudomonas syringae Lz4W Lacking RNase R. J. Biol. Chem. 2024, 300, 107600. [Google Scholar] [CrossRef]
- Vincent, H.A.; Deutscher, M.P. The Roles of Individual Domains of RNase R in Substrate Binding and Exoribonuclease Activity. The Nuclease Domain Is Sufficient for Digestion of Structured RNA. J. Biol. Chem. 2009, 284, 486–494. [Google Scholar] [CrossRef]
- Hussain, A.; Ray, M.K. Functional Activity of E. Coli RNase R in the Antarctic Pseudomonas syringae Lz4W. J. Genet. Eng. Biotechnol. 2023, 21, 101. [Google Scholar] [CrossRef]
- Chu, L.Y.; Hsieh, T.J.; Golzarroshan, B.; Chen, Y.P.; Agrawal, S.; Yuan, H.S. Structural Insights into RNA Unwinding and Degradation by RNase R. Nucleic Acids Res. 2017, 45, 12015–12024. [Google Scholar] [CrossRef]
- Apel, K.; Hirt, H. Reactive Oxygen Species: Metabolism, Oxidative Stress, and Signal Transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef] [PubMed]
- Blagojević, D.P. Antioxidant Systems in Supporting Environmental and Programmed Adaptations to Low Temperatures. Cryo Lett. 2007, 28, 137–150. [Google Scholar]
- Imlay, J.A. Cellular Defenses against Superoxide and Hydrogen Peroxide. Annu. Rev. Biochem. 2008, 77, 755–776. [Google Scholar] [CrossRef]
- Smirnova, G.V.; Zakirova, O.N.; Oktiabr’skii, O.N. Role of the Antioxidant System in Response of Escherichia coli Bacteria to Cold Stress. Mikrobiologiia 2001, 70, 55–60. [Google Scholar]
- Chattopadhyay, M.K.; Raghu, G.; Sharma, Y.V.R.K.; Biju, A.R.; Rajasekharan, M.V.; Shivaji, S. Increase in Oxidative Stress at Low Temperature in an Antarctic Bacterium. Curr. Microbiol. 2011, 62, 544–546. [Google Scholar] [CrossRef]
- Arvizu-Gómez, J.L.; Hernández-Morales, A.; Llanos-Vargas, K.D.; Olmedo-Álvarez, G.; Campos-Guillén, J.; Vallejo-Cardona, A.A.; Hernández-Flores, J.L.; González-Reyes, C. Influence of the Low Temperatures (18 °C) in the Generation of Intracellular Oxidative Stress in the Phytopathogen Bacterium Pseudomonas savastanoi Pv. phaseolicolaNPS3121. J. Phytopathol. 2024, 172, e13367. [Google Scholar] [CrossRef]
- García-Ríos, E.; Ramos-Alonso, L.; Guillamón, J.M. Correlation between Low Temperature Adaptation and Oxidative Stress in Saccharomyces cerevisiae. Front. Microbiol. 2016, 7, 1199. [Google Scholar] [CrossRef]
- Suzuki, N.; Mittler, R. Reactive Oxygen Species and Temperature Stresses: A Delicate Balance between Signaling and Destruction. Physiol. Plant. 2006, 126, 45–51. [Google Scholar] [CrossRef]
- O’Kane, D.; Gill, V.; Boyd, P.; Burdon, R. Chilling, Oxidative Stress and Antioxidant Responses in Arabidopsis thaliana callus. Planta 1996, 198, 371–377. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.; Zhu, J.-K. Cold Stress Regulation of Gene Expression in Plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef]
- Guo, Z.; Cai, L.; Liu, C.; Chen, Z.; Guan, S.; Ma, W.; Pan, G. Low-Temperature Stress Affects Reactive Oxygen Species, Osmotic Adjustment Substances, and Antioxidants in Rice (Oryza sativa L.) at the Reproductive Stage. Sci. Rep. 2022, 12, 6224. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Liu, H.; Zhang, H.; Wang, X.; Song, F. OsBIRH1, a DEAD-Box RNA Helicase with Functions in Modulating Defence Responses against Pathogen Infection and Oxidative Stress. J. Exp. Bot. 2008, 59, 2133–2146. [Google Scholar] [CrossRef] [PubMed]
- Xing, W.; Yin, M.; Lv, Q.; Hu, Y.; Liu, C.; Zhang, J. 1-Oxygen Solubility, Diffusion Coefficient, and Solution Viscosity. In Rotating Electrode Methods and Oxygen Reduction Electrocatalysts; Xing, W., Yin, G., Zhang, J., Eds.; Elsevier: Amsterdam, The Netherlands, 2014; pp. 1–31. ISBN 978-0-444-63278-4. [Google Scholar]
- Li, X.; Li, C.; Zhu, J.; Zhong, S.; Zhu, H.; Zhang, X. Functions and Mechanisms of RNA Helicases in Plants. J. Exp. Bot. 2023, 74, 2295–2310. [Google Scholar] [CrossRef]
- Yadav, S.; Tuteja, N. Chapter 4—Evolution of RNA Helicases in Plants: Molecular and Functional Insights. In Helicases from All Domains of Life; Tuteja, R., Ed.; Academic Press: Cambridge, MA, USA, 2019; pp. 53–75. ISBN 978-0-12-814685-9. [Google Scholar]
- Nidumukkala, S.; Tayi, L.; Chittela, R.K.; Vudem, D.R.; Khareedu, V.R. DEAD Box Helicases as Promising Molecular Tools for Engineering Abiotic Stress Tolerance in Plants. Crit. Rev. Biotechnol. 2019, 39, 395–407. [Google Scholar] [CrossRef]
- Liu, Y.; Tabata, D.; Imai, R. A Cold-Inducible DEAD-Box RNA Helicase from Arabidopsis thaliana Regulates Plant Growth and Development under Low Temperature. PLoS ONE 2016, 11, e0154040. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Kim, K.A.; Oh, T.R.; Park, C.M.; Kang, H. Functional Characterization of DEAD-Box RNA Helicases in Arabidopsis thaliana under Abiotic Stress Conditions. Plant Cell Physiol. 2008, 49, 1563–1571. [Google Scholar] [CrossRef]
- Guan, Q.; Wu, J.; Zhang, Y.; Jiang, C.; Liu, R.; Chai, C.; Zhu, J. A DEAD Box RNA Helicase Is Critical for Pre-mRNA Splicing, Cold-Responsive Gene Regulation, and Cold Tolerance in Arabidopsis. Plant Cell 2013, 25, 342–356. [Google Scholar] [CrossRef]
- Lu, C.-A.; Huang, C.-K.; Huang, W.-S.; Huang, T.-S.; Liu, H.-Y.; Chen, Y.-F. DEAD-Box RNA Helicase 42 Plays a Critical Role in Pre-mRNA Splicing under Cold Stress. Plant Physiol. 2020, 182, 255–271. [Google Scholar] [CrossRef]
- Gu, L.; Xu, T.; Lee, K.; Lee, K.H.; Kang, H. A Chloroplast-Localized DEAD-Box RNA helicaseAtRH3 Is Essential for Intron Splicing and Plays an Important Role in the Growth and Stress Response in Arabidopsis thaliana. Plant Physiol. Biochem. 2014, 82, 309–318. [Google Scholar] [CrossRef]
- Asakura, Y.; Galarneau, E.; Watkins, K.P.; Barkan, A.; van Wijk, K.J. Chloroplast RH3 DEAD Box RNA Helicases in Maize and Arabidopsis Function in Splicing of Specific Group II Introns and Affect Chloroplast Ribosome Biogenesis. Plant Physiol. 2012, 159, 961–974. [Google Scholar] [CrossRef]
- Xiaomei, W.; Rongrong, K.; Ting, Z.; Yuanyuan, G.; Jianlong, X.; Zhongze, P.; Gangseob, L.; Dongzhi, L.; Yanjun, D. A DEAD-Box RNA Helicase TCD33 That Confers Chloroplast Development in Rice at Seedling Stage under Cold Stress. J. Plant Physiol. 2020, 248, 153138. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhao, X.; Feng, C.; Liu, N.; Feng, H.; Wang, X.; Mu, X.; Huang, L.; Kang, Z. The Cloning and Characterization of a DEAD-Box RNA Helicase from Stress-Responsive Wheat. Physiol. Mol. Plant Pathol. 2014, 88, 36–42. [Google Scholar] [CrossRef]
- Chung, E.; Cho, C.-W.; Yun, B.-H.; Choi, H.-K.; So, H.-A.; Lee, S.-W.; Lee, J.-H. Molecular Cloning and Characterization of the Soybean DEAD-Box RNA Helicase Gene Induced by Low Temperature and High Salinity Stress. Gene 2009, 443, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Wigley, D.B. Bacterial DNA Repair: Recent Insights into the Mechanism of RecBCD, AddAB and AdnAB. Nat. Rev. Microbiol. 2013, 11, 9–13. [Google Scholar] [CrossRef]
- Pavankumar, T.L.; Exell, J.C.; Kowalczykowski, S.C. Direct Fluorescent Imaging of Translocation and Unwinding by Individual DNA Helicases; Academic Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Miranda, A.; Kuzminov, A. Chromosomal Lesion Suppression and Removal in Escherichia coli via Linear DNA Degradation. Genetics 2003, 163, 1255–1271. [Google Scholar] [CrossRef]
- Dermić, D. Functions of Multiple Exonucleases Are Essential for Cell Viability, DNA Repair and Homologous Recombination in recD Mutants of Escherichia coli. Genetics 2006, 172, 2057–2069. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pavankumar, T.L.; Rai, N.; Pandey, P.K.; Vincent, N. Helicases at Work: The Importance of Nucleic Acids Unwinding Under Cold Stress. DNA 2024, 4, 455-472. https://doi.org/10.3390/dna4040031
Pavankumar TL, Rai N, Pandey PK, Vincent N. Helicases at Work: The Importance of Nucleic Acids Unwinding Under Cold Stress. DNA. 2024; 4(4):455-472. https://doi.org/10.3390/dna4040031
Chicago/Turabian StylePavankumar, Theetha L., Navneet Rai, Pramod K. Pandey, and Nishanth Vincent. 2024. "Helicases at Work: The Importance of Nucleic Acids Unwinding Under Cold Stress" DNA 4, no. 4: 455-472. https://doi.org/10.3390/dna4040031
APA StylePavankumar, T. L., Rai, N., Pandey, P. K., & Vincent, N. (2024). Helicases at Work: The Importance of Nucleic Acids Unwinding Under Cold Stress. DNA, 4(4), 455-472. https://doi.org/10.3390/dna4040031






