How Chromatin Motor Complexes Influence the Nuclear Architecture: A Review of Chromatin Organization, Cohesins, and Condensins with a Focus on C. elegans
Abstract
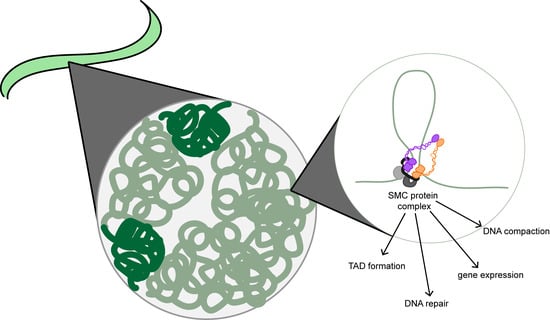
1. Introduction: Chromatin Organization
2. Structural Maintenance of Chromosome Proteins Are the Backbone of Chromatin Motor Complexes
3. The Unique Chromatin Architecture of C. elegans and the Role of Chromatin Motor Complexes in Cell Division
4. Chromatin Motor Complexes Shape Chromatin Architecture during Interphase
5. TADs in C. elegans Are a Result of Dosage Compensation
6. C. elegans Dosage Compensation as a Paradigm for Chromosome-Wide Gene Regulation
Author Contributions
Funding
Conflicts of Interest
References
- Paweletz, N. Walther Flemming: Pioneer of mitosis research. Nat. Rev. Mol. Cell Biol. 2001, 2, 72–75. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Orr-Weaver, T.L. Chromatin. In Encyclopedia of Genetics; Brenner, S., Miller, J.H., Eds.; Academic Press: New York, NY, USA, 2001; pp. 340–343. [Google Scholar]
- Richmond, T.J.; Finch, J.T.; Rushton, B.; Rhodes, D.; Klug, A. Structure of the nucleosome core particle at 7 Å resolution. Nature 1984, 311, 532–537. [Google Scholar] [CrossRef] [PubMed]
- Kornberg, R.D. Chromatin Structure: A Repeating Unit of Histones and DNA. Science 1974, 184, 868–871. [Google Scholar] [CrossRef] [PubMed]
- Antonin, W.; Neumann, H. Chromosome condensation and decondensation during mitosis. Curr. Opin. Cell Biol. 2016, 40, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Merkenschlager, M.; Nora, E.P. CTCF and Cohesin in Genome Folding and Transcriptional Gene Regulation. Annu. Rev. Genom. Hum. Genet. 2016, 17, 17–43. [Google Scholar] [CrossRef]
- Han, H.J.; Russo, J.; Kohwi, Y.; Kohwi-Shigematsu, T. SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis. Nature 2008, 452, 187–193. [Google Scholar] [CrossRef]
- Tang, Z.; Luo, O.J.; Li, X.; Zheng, M.; Zhu, J.J.; Szalaj, P.; Trzaskoma, P.; Magalska, A.; Wlodarczyk, J.; Ruszczycki, B.; et al. CTCF-Mediated Human 3D Genome Architecture Reveals Chromatin Topology for Transcription. Cell 2015, 163, 1611–1627. [Google Scholar] [CrossRef]
- Phillips-Cremins, J.E.; Sauria, M.E.G.; Sanyal, A.; Gerasimova, T.I.; Lajoie, B.R.; Bell, J.S.K.; Ong, C.-T.; Hookway, T.A.; Guo, C.; Sun, Y.; et al. Architectural Protein Subclasses Shape 3D Organization of Genomes during Lineage Commitment. Cell 2013, 153, 1281–1295. [Google Scholar] [CrossRef]
- Spitz, F.; Furlong, E.E.M. Transcription factors: From enhancer binding to developmental control. Nat. Rev. Genet. 2012, 13, 613–626. [Google Scholar] [CrossRef]
- Furlong, E.E.M.; Levine, M. Developmental enhancers and chromosome topology. Science 2018, 361, 1341–1345. [Google Scholar] [CrossRef]
- Renfro, Z.; White, B.E.; Stephens, K.E. CCAAT enhancer binding protein gamma (C/EBP-γ): An understudied transcription factor. Adv. Biol. Regul. 2022, 84, 100861. [Google Scholar] [CrossRef]
- Heslop, J.A.; Pournasr, B.; Duncan, S.A. Chromatin remodeling is restricted by transient GATA6 binding during iPSC differentiation to definitive endoderm. iScience 2022, 25, 104300. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xiao, Q.; Xiao, J.; Niu, C.; Li, Y.; Zhang, X.; Zhou, Z.; Shu, G.; Yin, G. Wnt/β-catenin signalling: Function, biological mechanisms, and therapeutic opportunities. Signal Transduct. Target. Ther. 2022, 7, 3. [Google Scholar] [CrossRef]
- Swinstead, E.E.; Paakinaho, V.; Presman, D.M.; Hager, G.L. Pioneer factors and ATP-dependent chromatin remodeling factors interact dynamically: A new perspective: Multiple transcription factors can effect chromatin pioneer functions through dynamic interactions with ATP-dependent chromatin remodeling factors. Bioessays 2016, 38, 1150–1157. [Google Scholar] [CrossRef] [PubMed]
- Radman-Livaja, M.; Rando, O.J. Nucleosome positioning: How is it established, and why does it matter? Dev. Biol. 2010, 339, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Ricci, M.A.; Manzo, C.; García-Parajo, M.F.; Lakadamyali, M.; Cosma, M.P. Chromatin fibers are formed by heterogeneous groups of nucleosomes in vivo. Cell 2015, 160, 1145–1158. [Google Scholar] [CrossRef]
- Portillo-Ledesma, S.; Tsao, L.H.; Wagley, M.; Lakadamyali, M.; Cosma, M.P.; Schlick, T. Nucleosome Clutches are Regulated by Chromatin Internal Parameters. J. Mol. Biol. 2021, 433, 166701. [Google Scholar] [CrossRef]
- Zentgraf, H.; Franke, W.W. Differences of supranucleosomal organization in different kinds of chromatin: Cell type-specific globular subunits containing different numbers of nucleosomes. J. Cell Biol. 1984, 99, 272–286. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Dixon, J.R.; Selvaraj, S.; Yue, F.; Kim, A.; Li, Y.; Shen, Y.; Hu, M.; Liu, J.S.; Ren, B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012, 485, 376–380. [Google Scholar] [CrossRef]
- Nora, E.P.; Lajoie, B.R.; Schulz, E.G.; Giorgetti, L.; Okamoto, I.; Servant, N.; Piolot, T.; van Berkum, N.L.; Meisig, J.; Sedat, J.; et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 2012, 485, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Rowley, M.J.; Nichols, M.H.; Lyu, X.; Ando-Kuri, M.; Rivera, I.S.M.; Hermetz, K.; Wang, P.; Ruan, Y.; Corces, V.G. Evolutionarily Conserved Principles Predict 3D Chromatin Organization. Mol. Cell 2017, 67, 837–852.e837. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.; Seow, W.Q.; Appert, A.; Dong, Y.; Stempor, P.; Ahringer, J. Accessible Region Conformation Capture (ARC-C) gives high-resolution insights into genome architecture and regulation. Genome Res. 2022, 32, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Ghavi-Helm, Y.; Klein, F.A.; Pakozdi, T.; Ciglar, L.; Noordermeer, D.; Huber, W.; Furlong, E.E.M. Enhancer loops appear stable during development and are associated with paused polymerase. Nature 2014, 512, 96–100. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Xu, Q.; Canzio, D.; Shou, J.; Li, J.; Gorkin, D.U.; Jung, I.; Wu, H.; Zhai, Y.; Tang, Y.; et al. CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell 2015, 162, 900–910. [Google Scholar] [CrossRef] [PubMed]
- Lupiáñez, D.G.; Kraft, K.; Heinrich, V.; Krawitz, P.; Brancati, F.; Klopocki, E.; Horn, D.; Kayserili, H.; Opitz, J.M.; Laxova, R.; et al. Disruptions of Topological Chromatin Domains Cause Pathogenic Rewiring of Gene-Enhancer Interactions. Cell 2015, 161, 1012–1025. [Google Scholar] [CrossRef]
- Franke, M.; Ibrahim, D.M.; Andrey, G.; Schwarzer, W.; Heinrich, V.; Schöpflin, R.; Kraft, K.; Kempfer, R.; Jerković, I.; Chan, W.-L.; et al. Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature 2016, 538, 265–269. [Google Scholar] [CrossRef]
- Yu, M.; Ren, B. The Three-Dimensional Organization of Mammalian Genomes. Annu. Rev. Cell Dev. Biol. 2017, 33, 265–289. [Google Scholar] [CrossRef]
- Li, D.; He, M.; Tang, Q.; Tian, S.; Zhang, J.; Li, Y.; Wang, D.; Jin, L.; Ning, C.; Zhu, W.; et al. Comparative 3D genome architecture in vertebrates. BMC Biol. 2022, 20, 99. [Google Scholar] [CrossRef]
- Wang, C.; Liu, C.; Roqueiro, D.; Grimm, D.; Schwab, R.; Becker, C.; Lanz, C.; Weigel, D. Genome-wide analysis of local chromatin packing in Arabidopsis thaliana. Genome Res. 2015, 25, 246–256. [Google Scholar] [CrossRef]
- Rao, S.S.P.; Huang, S.C.; Glenn St Hilaire, B.; Engreitz, J.M.; Perez, E.M.; Kieffer-Kwon, K.R.; Sanborn, A.L.; Johnstone, S.E.; Bascom, G.D.; Bochkov, I.D.; et al. Cohesin Loss Eliminates All Loop Domains. Cell 2017, 171, 305–320.e324. [Google Scholar] [CrossRef]
- Nora, E.P.; Goloborodko, A.; Valton, A.L.; Gibcus, J.H.; Uebersohn, A.; Abdennur, N.; Dekker, J.; Mirny, L.A.; Bruneau, B.G. Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell 2017, 169, 930–944.e922. [Google Scholar] [CrossRef]
- Lieberman-Aiden, E.; van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science 2009, 326, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Sexton, T.; Yaffe, E.; Kenigsberg, E.; Bantignies, F.; Leblanc, B.; Hoichman, M.; Parrinello, H.; Tanay, A.; Cavalli, G. Three-Dimensional Folding and Functional Organization Principles of the Drosophila Genome. Cell 2012, 148, 458–472. [Google Scholar] [CrossRef] [PubMed]
- Krietenstein, N.; Abraham, S.; Venev, S.V.; Abdennur, N.; Gibcus, J.; Hsieh, T.-H.S.; Parsi, K.M.; Yang, L.; Maehr, R.; Mirny, L.A.; et al. Ultrastructural Details of Mammalian Chromosome Architecture. Mol. Cell 2020, 78, 554–565.e557. [Google Scholar] [CrossRef]
- Hsieh, T.-H.S.; Cattoglio, C.; Slobodyanyuk, E.; Hansen, A.S.; Rando, O.J.; Tjian, R.; Darzacq, X. Resolving the 3D Landscape of Transcription-Linked Mammalian Chromatin Folding. Mol. Cell 2020, 78, 539–553.e538. [Google Scholar] [CrossRef]
- Harris, H.L.; Gu, H.; Olshansky, M.; Wang, A.; Farabella, I.; Eliaz, Y.; Kalluchi, A.; Krishna, A.; Jacobs, M.; Cauer, G.; et al. Chromatin alternates between A and B compartments at kilobase scale for subgenic organization. Nat. Commun. 2023, 14, 3303. [Google Scholar] [CrossRef]
- Cremer, T.; Cremer, M. Chromosome territories. Cold Spring Harb. Perspect. Biol. 2010, 2, a003889. [Google Scholar] [CrossRef]
- Branco, M.R.; Pombo, A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol. 2006, 4, e138. [Google Scholar] [CrossRef] [PubMed]
- Pecinka, A.; Schubert, V.; Meister, A.; Kreth, G.; Klatte, M.; Lysak, M.A.; Fuchs, J.; Schubert, I. Chromosome territory arrangement and homologous pairing in nuclei of Arabidopsis thaliana are predominantly random except for NOR-bearing chromosomes. Chromosoma 2004, 113, 258–269. [Google Scholar] [CrossRef]
- Zink, D.; Cremer, T.; Saffrich, R.; Fischer, R.; Trendelenburg, M.F.; Ansorge, W.; Stelzer, E.H. Structure and dynamics of human interphase chromosome territories in vivo. Hum. Genet. 1998, 102, 241–251. [Google Scholar] [CrossRef]
- Croft, J.A.; Bridger, J.M.; Boyle, S.; Perry, P.; Teague, P.; Bickmore, W.A. Differences in the localization and morphology of chromosomes in the human nucleus. J. Cell Biol. 1999, 145, 1119–1131. [Google Scholar] [CrossRef]
- Fritz, A.J.; Sehgal, N.; Pliss, A.; Xu, J.; Berezney, R. Chromosome territories and the global regulation of the genome. Genes Chromosomes Cancer 2019, 58, 407–426. [Google Scholar] [CrossRef]
- Harvey, S.H.; Krien, M.J.; O’Connell, M.J. Structural maintenance of chromosomes (SMC) proteins, a family of conserved ATPases. Genome Biol. 2002, 3, Reviews3003. [Google Scholar] [CrossRef]
- Lammens, A.; Schele, A.; Hopfner, K.P. Structural biochemistry of ATP-driven dimerization and DNA-stimulated activation of SMC ATPases. Curr. Biol. 2004, 14, 1778–1782. [Google Scholar] [CrossRef]
- Palou, R.; Dhanaraman, T.; Marrakchi, R.; Pascariu, M.; Tyers, M.; D’Amours, D. Condensin ATPase motifs contribute differentially to the maintenance of chromosome morphology and genome stability. PLoS Biol. 2018, 16, e2003980. [Google Scholar] [CrossRef]
- Abdul, F.; Diman, A.; Baechler, B.; Ramakrishnan, D.; Kornyeyev, D.; Beran, R.K.; Fletcher, S.P.; Strubin, M. Smc5/6 silences episomal transcription by a three-step function. Nat. Struct. Mol. Biol. 2022, 29, 922–931. [Google Scholar] [CrossRef]
- Haering, C.H.; Lowe, J.; Hochwagen, A.; Nasmyth, K. Molecular Architecture of SMC Proteins and the Yeast Cohesin Complex. Mol. Cell 2002, 9, 773–788. [Google Scholar] [CrossRef] [PubMed]
- Jessberger, R. The many functions of smc proteins in chromosome dynamics. Nat. Rev. Mol. Cell Biol. 2002, 3, 767–778. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.; Yang, Y.; Chen, Y.H.; Arenz, J.; Rangi, G.K.; Zhao, X.; Ye, H. Architecture of the Smc5/6 Complex of Saccharomyces cerevisiae Reveals a Unique Interaction between the Nse5-6 Subcomplex and the Hinge Regions of Smc5 and Smc6. J. Biol. Chem. 2009, 284, 8507–8515. [Google Scholar] [CrossRef] [PubMed]
- Hirano, T.; Kobayashi, R.; Hirano, M. Condensins, Chromosome Condensation Protein Complexes Containing XCAP-C, XCAP-E and a Xenopus Homolog of the Drosophila Barren Protein. Cell 1997, 89, 511–521. [Google Scholar] [CrossRef]
- Hassler, M.; Shaltiel, I.A.; Kschonsak, M.; Simon, B.; Merkel, F.; Tharichen, L.; Bailey, H.J.; Macosek, J.; Bravo, S.; Metz, J.; et al. Structural Basis of an Asymmetric Condensin ATPase Cycle. Mol. Cell 2019, 74, 1175–1188.e9. [Google Scholar] [CrossRef]
- Hirano, M.; Hirano, T. Opening closed arms: Long-distance activation of SMC ATPase by hinge-DNA interactions. Mol. Cell 2006, 21, 175–186. [Google Scholar] [CrossRef]
- Pradhan, B.; Kanno, T.; Umeda Igarashi, M.; Loke, M.S.; Baaske, M.D.; Wong, J.S.K.; Jeppsson, K.; Björkegren, C.; Kim, E. The Smc5/6 complex is a DNA loop-extruding motor. Nature 2023, 616, 843–848. [Google Scholar] [CrossRef]
- Cheng, H.; Zhang, N.; Pati, D. Cohesin subunit RAD21: From biology to disease. Gene 2020, 758, 144966. [Google Scholar] [CrossRef]
- Schleiffer, A.; Kaitna, S.; Maurer-Stroh, S.; Glotzer, M.; Nasmyth, K.; Eisenhaber, F. Kleisins: A Superfamily of Bacterial and Eukaryotic SMC Protein Partners. Mol. Cell 2003, 11, 571–575. [Google Scholar] [CrossRef]
- Mito, Y.; Sugimoto, A.; Yamamoto, M. Distinct developmental function of two Caenorhabditis elegans homologs of the cohesin subunit Scc1/Rad21. Mol. Biol. Cell 2003, 14, 2399–2409. [Google Scholar] [CrossRef] [PubMed]
- Severson, A.F.; Ling, L.; van Zuylen, V.; Meyer, B.J. The axial element protein HTP-3 promotes cohesin loading and meiotic axis assembly in C. elegans to implement the meiotic program of chromosome segregation. Genes. Dev. 2009, 23, 1763–1778. [Google Scholar] [CrossRef]
- Severson, A.F.; Meyer, B.J. Divergent kleisin subunits of cohesin specify mechanisms to tether and release meiotic chromosomes. eLife 2014, 3, e03467. [Google Scholar] [CrossRef] [PubMed]
- Brooker, A.S.; Berkowitz, K.M. The roles of cohesins in mitosis, meiosis, and human health and disease. Methods Mol. Biol. 2014, 1170, 229–266. [Google Scholar] [CrossRef] [PubMed]
- Cutts, E.E.; Vannini, A. Condensin complexes: Understanding loop extrusion one conformational change at a time. Biochem. Soc. Trans. 2020, 48, 2089–2100. [Google Scholar] [CrossRef] [PubMed]
- Tane, S.; Shintomi, K.; Kinoshita, K.; Tsubota, Y.; Yoshida, M.M.; Nishiyama, T.; Hirano, T. Cell cycle-specific loading of condensin I is regulated by the N-terminal tail of its kleisin subunit. eLife 2022, 11, e84694. [Google Scholar] [CrossRef] [PubMed]
- Shaltiel, I.A.; Datta, S.; Lecomte, L.; Hassler, M.; Kschonsak, M.; Bravo, S.; Stober, C.; Ormanns, J.; Eustermann, S.; Haering, C.H. A hold-and-feed mechanism drives directional DNA loop extrusion by condensin. Science 2022, 376, 1087–1094. [Google Scholar] [CrossRef]
- Palecek, J.J.; Gruber, S. Kite Proteins: A Superfamily of SMC/Kleisin Partners Conserved Across Bacteria, Archaea, and Eukaryotes. Structure 2015, 23, 2183–2190. [Google Scholar] [CrossRef] [PubMed]
- Wells, J.N.; Gligoris, T.G.; Nasmyth, K.A.; Marsh, J.A. Evolution of condensin and cohesin complexes driven by replacement of Kite by Hawk proteins. Curr. Biol. 2017, 27, R17–R18. [Google Scholar] [CrossRef] [PubMed]
- Ono, T.; Losada, A.; Hirano, M.; Myers, M.P.; Neuwald, A.F.; Hirano, T. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell 2003, 115, 109–121. [Google Scholar] [CrossRef]
- Yoshida, M.M.; Kinoshita, K.; Shintomi, K.; Aizawa, Y.; Hirano, T. Regulation of condensin II by self-suppression and release mechanisms. Mol. Biol. Cell 2023, 35, ar21. [Google Scholar] [CrossRef]
- Csankovszki, G.; Collette, K.; Spahl, K.; Carey, J.; Snyder, M.; Petty, E.; Patel, U.; Tabuchi, T.; Liu, H.; McLeod, I.; et al. Three distinct condensin complexes control C. elegans chromosome dynamics. Curr. Biol. 2009, 19, 9–19. [Google Scholar] [CrossRef]
- Solé-Soler, R.; Torres-Rosell, J. Smc5/6, an atypical SMC complex with two RING-type subunits. Biochem. Soc. Trans. 2020, 48, 2159–2171. [Google Scholar] [CrossRef] [PubMed]
- Pebernard, S.; Wohlschlegel, J.; McDonald, W.H.; Yates, J.R., 3rd; Boddy, M.N. The Nse5-Nse6 dimer mediates DNA repair roles of the Smc5-Smc6 complex. Mol. Cell Biol. 2006, 26, 1617–1630. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.P.; Zhao, X. The multi-functional Smc5/6 complex in genome protection and disease. Nat. Struct. Mol. Biol. 2023, 30, 724–734. [Google Scholar] [CrossRef]
- Odiba, A.S.; Ezechukwu, C.S.; Liao, G.; Li, S.; Chen, Z.; Liu, X.; Fang, W.; Jin, C.; Wang, B. Loss of NSE-4 Perturbs Genome Stability and DNA Repair in Caenorhabditis elegans. Int. J. Mol. Sci. 2022, 23, 7202. [Google Scholar] [CrossRef]
- Hong, Y.; Sonneville, R.; Agostinho, A.; Meier, B.; Wang, B.; Blow, J.J.; Gartner, A. The SMC-5/6 Complex and the HIM-6 (BLM) Helicase Synergistically Promote Meiotic Recombination Intermediate Processing and Chromosome Maturation during Caenorhabditis elegans Meiosis. PLoS Genet. 2016, 12, e1005872. [Google Scholar] [CrossRef]
- Barra, V.; Fachinetti, D. The dark side of centromeres: Types, causes and consequences of structural abnormalities implicating centromeric DNA. Nat. Commun. 2018, 9, 4340. [Google Scholar] [CrossRef]
- Kyriacou, E.; Heun, P. Centromere structure and function: Lessons from Drosophila. Genetics 2023, 225, iyad170. [Google Scholar] [CrossRef]
- Simon, L.; Voisin, M.; Tatout, C.; Probst, A.V. Structure and Function of Centromeric and Pericentromeric Heterochromatin in Arabidopsis thaliana. Front. Plant Sci. 2015, 6, 1049. [Google Scholar] [CrossRef]
- Xu, R.; Pan, Z.; Nakagawa, T. Gross Chromosomal Rearrangement at Centromeres. Biomolecules 2023, 14, 28. [Google Scholar] [CrossRef]
- Melters, D.P.; Paliulis, L.V.; Korf, I.F.; Chan, S.W.L. Holocentric chromosomes: Convergent evolution, meiotic adaptations, and genomic analysis. Chromosome Res. 2012, 20, 579–593. [Google Scholar] [CrossRef]
- Emerson, F.J.; Lee, S.S. Chromatin: The old and young of it. Front. Mol. Biosci. 2023, 10, 1270285. [Google Scholar] [CrossRef]
- Márquez-Corro, J.I.; Escudero, M.; Luceño, M. Do holocentric chromosomes represent an evolutionary advantage? A study of paired analyses of diversification rates of lineages with holocentric chromosomes and their monocentric closest relatives. Chromosome Res. 2018, 26, 139–152. [Google Scholar] [CrossRef]
- Nabeshima, K.; Villeneuve, A.M.; Colaiácovo, M.P. Crossing over is coupled to late meiotic prophase bivalent differentiation through asymmetric disassembly of the SC. J. Cell Biol. 2005, 168, 683–689. [Google Scholar] [CrossRef]
- Chan, R.C.; Severson, A.F.; Meyer, B.J. Condensin restructures chromosomes in preparation for meiotic divisions. J. Cell Biol. 2004, 167, 613–625. [Google Scholar] [CrossRef]
- Hillers, K.J.; Villeneuve, A.M. Chromosome-Wide Control of Meiotic Crossing over in C. elegans. Curr. Biol. 2003, 13, 1641–1647. [Google Scholar] [CrossRef]
- Schvarzstein, M.; Wignall, S.M.; Villeneuve, A.M. Coordinating cohesion, co-orientation, and congression during meiosis: Lessons from holocentric chromosomes. Genes. Dev. 2010, 24, 219–228. [Google Scholar] [CrossRef]
- Ganji, M.; Shaltiel, I.A.; Bisht, S.; Kim, E.; Kalichava, A.; Haering, C.H.; Dekker, C. Real-time imaging ofDNA loop extrusion by condensin. Science 2018, 360, 102–105. [Google Scholar] [CrossRef]
- Kim, Y.; Shi, Z.; Zhang, H.; Finkelstein, I.J.; Yu, H. Human cohesin compacts DNA by loop extrusion. Science 2019, 366, 1345–1349. [Google Scholar] [CrossRef]
- Higashi, T.L.; Pobegalov, G.; Tang, M.; Molodtsov, M.I.; Uhlmann, F. A Brownian ratchet model for DNA loop extrusion by the cohesin complex. eLife 2021, 10, e67530. [Google Scholar] [CrossRef]
- Marko, J.F.; De Los Rios, P.; Barducci, A.; Gruber, S. DNA-segment-capture model for loop extrusion by structural maintenance of chromosome (SMC) protein complexes. Nucleic Acids Res. 2019, 47, 6956–6972. [Google Scholar] [CrossRef]
- Nomidis, S.K.; Carlon, E.; Gruber, S.; Marko, J.F. DNA tension-modulated translocation and loop extrusion by SMC complexes revealed by molecular dynamics simulations. Nucleic Acids Res. 2022, 50, 4974–4987. [Google Scholar] [CrossRef]
- Dekker, C.; Haering, C.H.; Peters, J.-M.; Rowland, B.D. How do molecular motors fold the genome? Science 2023, 382, 646–648. [Google Scholar] [CrossRef]
- Hagstrom, K.A.; Holmes, V.F.; Cozzarelli, N.R.; Meyer, B.J. C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis. Genes. Dev. 2002, 16, 729–742. [Google Scholar] [CrossRef]
- Uhlmann, F.; Nasmyth, K. Cohesion between sister chromatids must be established during DNA replication. Curr. Biol. 1998, 8, 1095–1102. [Google Scholar] [CrossRef] [PubMed]
- Chan, R.C.; Chan, A.; Jeon, M.; Wu, T.F.; Pasqualone, D.; Rougvie, A.E.; Meyer, B.J. Chromosome cohesion is regulated by a clock gene paralogue TIM-1. Nature 2003, 423, 1002–1009. [Google Scholar] [CrossRef]
- Pasierbek, P.; Födermayr, M.; Jantsch, V.; Jantsch, M.; Schweizer, D.; Loidl, J. The Caenorhabditis elegans SCC-3 homologue is required for meiotic synapsis and for proper chromosome disjunction in mitosis and meiosis. Exp. Cell Res. 2003, 289, 245–255. [Google Scholar] [CrossRef]
- Siomos, M.F.; Badrinath, A.; Pasierbek, P.; Livingstone, D.; White, J.; Glotzer, M.; Nasmyth, K. Separase is required for chromosome segregation during meiosis I in Caenorhabditis elegans. Curr. Biol. 2001, 11, 1825–1835. [Google Scholar] [CrossRef]
- Luo, S.; Tong, L. Structure and Function of the Separase-Securin Complex. Subcell. Biochem. 2021, 96, 217–232. [Google Scholar] [CrossRef]
- Michaelis, C.; Ciosk, R.; Nasmyth, K. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell 1997, 91, 35–45. [Google Scholar] [CrossRef]
- Ishiguro, K.I. The cohesin complex in mammalian meiosis. Genes. Cells 2019, 24, 6–30. [Google Scholar] [CrossRef]
- Pasierbek, P.; Jantsch, M.; Melcher, M.; Schleiffer, A.; Schweizer, D.; Loidl, J. A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes. Dev. 2001, 15, 1349–1360. [Google Scholar] [CrossRef]
- Castellano-Pozo, M.; Sioutas, G.; Barroso, C.; Prince, J.P.; Lopez-Jimenez, P.; Davy, J.; Jaso-Tamame, A.-L.; Crawley, O.; Shao, N.; Page, J.; et al. The kleisin subunit controls the function of C. elegans meiotic cohesins by determining the mode of DNA binding and differential regulation by SCC-2 and WAPL-1. eLife 2023, 12, e84138. [Google Scholar] [CrossRef]
- Klein, F.; Mahr, P.; Galova, M.; Buonomo, S.B.; Michaelis, C.; Nairz, K.; Nasmyth, K. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 1999, 98, 91–103. [Google Scholar] [CrossRef]
- Kitajima, T.S.; Yokobayashi, S.; Yamamoto, M.; Watanabe, Y. Distinct cohesin complexes organize meiotic chromosome domains. Science 2003, 300, 1152–1155. [Google Scholar] [CrossRef]
- Ono, T.; Fang, Y.; Spector, D.L.; Hirano, T. Spatial and temporal regulation of Condensins I and II in mitotic chromosome assembly in human cells. Mol. Biol. Cell 2004, 15, 3296–3308. [Google Scholar] [CrossRef] [PubMed]
- Collette, K.S.; Petty, E.L.; Golenberg, N.; Bembenek, J.N.; Csankovszki, G. Different roles for Aurora B in condensin targeting during mitosis and meiosis. J. Cell Sci. 2011, 124, 3684–3694. [Google Scholar] [CrossRef]
- Shintomi, K.; Hirano, T. The relative ratio of condensin I to II determines chromosome shapes. Genes. Dev. 2011, 25, 1464–1469. [Google Scholar] [CrossRef]
- Bembenek, J.N.; Verbrugghe, K.J.; Khanikar, J.; Csankovszki, G.; Chan, R.C. Condensin and the spindle midzone prevent cytokinesis failure induced by chromatin bridges in C. elegans embryos. Curr. Biol. 2013, 23, 937–946. [Google Scholar] [CrossRef]
- Golfier, S.; Quail, T.; Kimura, H.; Brugues, J. Cohesin and condensin extrude DNA loops in a cell cycle-dependent manner. Elife 2020, 9, e53885. [Google Scholar] [CrossRef]
- Lee, J.; Ogushi, S.; Saitou, M.; Hirano, T. Condensins I and II are essential for construction of bivalent chromosomes in mouse oocytes. Mol. Biol. Cell 2011, 22, 3465–3477. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.J.; Osman, K.; Franklin, F.C. The condensin complexes play distinct roles to ensure normal chromosome morphogenesis during meiotic division in Arabidopsis. Plant J. 2014, 80, 255–268. [Google Scholar] [CrossRef] [PubMed]
- Resnick, T.D.; Dej, K.J.; Xiang, Y.; Hawley, R.S.; Ahn, C.; Orr-Weaver, T.L. Mutations in the chromosomal passenger complex and the condensin complex differentially affect synaptonemal complex disassembly and metaphase I configuration in Drosophila female meiosis. Genetics 2009, 181, 875–887. [Google Scholar] [CrossRef]
- Hernandez, M.R.; Davis, M.B.; Jiang, J.; Brouhard, E.A.; Severson, A.F.; Csankovszki, G. Condensin I protects meiotic cohesin from WAPL-1 mediated removal. PLoS Genet. 2018, 14, e1007382. [Google Scholar] [CrossRef]
- Dixon, J.R.; Gorkin, D.U.; Ren, B. Chromatin Domains: The Unit of Chromosome Organization. Mol. Cell 2016, 62, 668–680. [Google Scholar] [CrossRef]
- Dekker, J.; Heard, E. Structural and functional diversity of Topologically Associating Domains. FEBS Lett. 2015, 589, 2877–2884. [Google Scholar] [CrossRef]
- Makrantoni, V.; Marston, A.L. Cohesin and chromosome segregation. Curr. Biol. 2018, 28, R688–R693. [Google Scholar] [CrossRef]
- Holwerda, S.J.; de Laat, W. CTCF: The protein, the binding partners, the binding sites and their chromatin loops. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368, 20120369. [Google Scholar] [CrossRef] [PubMed]
- Ong, C.-T.; Corces, V.G. CTCF: An architectural protein bridging genome topology and function. Nat. Rev. Genet. 2014, 15, 234–246. [Google Scholar] [CrossRef] [PubMed]
- Crane, E.; Bian, Q.; McCord, R.P.; Lajoie, B.R.; Wheeler, B.S.; Ralston, E.J.; Uzawa, S.; Dekker, J.; Meyer, B.J. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature 2015, 523, 240–244. [Google Scholar] [CrossRef] [PubMed]
- Ikegami, K.; Egelhofer, T.A.; Strome, S.; Lieb, J.D. Caenorhabditis elegans chromosome arms are anchored to the nuclear membrane via discontinuous association with LEM-2. Genome Biol. 2010, 11, R120. [Google Scholar] [CrossRef]
- Cabianca, D.S.; Muñoz-Jiménez, C.; Kalck, V.; Gaidatzis, D.; Padeken, J.; Seeber, A.; Askjaer, P.; Gasser, S.M. Active chromatin marks drive spatial sequestration of heterochromatin in C. elegans nuclei. Nature 2019, 569, 734–739. [Google Scholar] [CrossRef]
- Towbin, B.D.; González-Aguilera, C.; Sack, R.; Gaidatzis, D.; Kalck, V.; Meister, P.; Askjaer, P.; Gasser, S.M. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell 2012, 150, 934–947. [Google Scholar] [CrossRef]
- Gonzalez-Sandoval, A.; Towbin, B.D.; Kalck, V.; Cabianca, D.S.; Gaidatzis, D.; Hauer, M.H.; Geng, L.; Wang, L.; Yang, T.; Wang, X.; et al. Perinuclear Anchoring of H3K9-Methylated Chromatin Stabilizes Induced Cell Fate in C. elegans Embryos. Cell 2015, 163, 1333–1347. [Google Scholar] [CrossRef]
- Alagna, N.S.; Thomas, T.I.; Wilson, K.L.; Reddy, K.L. Choreography of lamina-associated domains: Structure meets dynamics. FEBS Lett. 2023, 597, 2806–2822. [Google Scholar] [CrossRef]
- Wang, N.; Wang, Z.; Tzourtzou, S.; Wang, X.; Bi, X.; Leimeister, J.; Xu, L.; Sakamoto, T.; Matsunaga, S.; Schaller, A.; et al. The plant nuclear lamina disassembles to regulate genome folding in stress conditions. Nat. Plants 2023, 9, 1081–1093. [Google Scholar] [CrossRef]
- Liu, T.; Rechtsteiner, A.; Egelhofer, T.A.; Vielle, A.; Latorre, I.; Cheung, M.S.; Ercan, S.; Ikegami, K.; Jensen, M.; Kolasinska-Zwierz, P.; et al. Broad chromosomal domains of histone modification patterns in C. elegans. Genome Res. 2011, 21, 227–236. [Google Scholar] [CrossRef]
- Hassan, A.; Rodriguez, P.A.; Heidmann, S.K.; Walmsley, E.L.; Aughey, G.N.; Southall, T.D. Condensin I subunit Cap-G is essential for proper gene expression during the maturation of post-mitotic neurons. eLife 2020, 9, e55159. [Google Scholar] [CrossRef]
- Bauer, C.R.; Hartl, T.A.; Bosco, G. Condensin II promotes the formation of chromosome territories by inducing axial compaction of polyploid interphase chromosomes. PLoS Genet. 2012, 8, e1002873. [Google Scholar] [CrossRef]
- Iwasaki, O.; Tanizawa, H.; Kim, K.D.; Kossenkov, A.; Nacarelli, T.; Tashiro, S.; Majumdar, S.; Showe, L.C.; Zhang, R.; Noma, K.I. Involvement of condensin in cellular senescence through gene regulation and compartmental reorganization. Nat. Commun. 2019, 10, 5688. [Google Scholar] [CrossRef]
- Hoencamp, C.; Dudchenko, O.; Elbatsh, A.M.O.; Brahmachari, S.; Raaijmakers, J.A.; van Schaik, T.; Sedeño Cacciatore, Á.; Contessoto, V.G.; van Heesbeen, R.; van den Broek, B.; et al. 3D genomics across the tree of life reveals condensin II as a determinant of architecture type. Science 2021, 372, 984–989. [Google Scholar] [CrossRef]
- Jin, Q.W.; Fuchs, J.; Loidl, J. Centromere clustering is a major determinant of yeast interphase nuclear organization. J. Cell Sci. 2000, 113, 1903–1912. [Google Scholar] [CrossRef]
- Sakamoto, T.; Sugiyama, T.; Yamashita, T.; Matsunaga, S. Plant condensin II is required for the correct spatial relationship between centromeres and rDNA arrays. Nucleus 2019, 10, 116–125. [Google Scholar] [CrossRef]
- Schuster, A.T.; Sarvepalli, K.; Murphy, E.A.; Longworth, M.S. Condensin II subunit dCAP-D3 restricts retrotransposon mobilization in Drosophila somatic cells. PLoS Genet. 2013, 9, e1003879. [Google Scholar] [CrossRef]
- Kranz, A.L.; Jiao, C.Y.; Winterkorn, L.H.; Albritton, S.E.; Kramer, M.; Ercan, S. Genome-wide analysis of condensin binding in Caenorhabditis elegans. Genome Biol. 2013, 14, R112. [Google Scholar] [CrossRef] [PubMed]
- Madl, J.E.; Herman, R.K. Polyploids and Sex Determination in Caenorhabditis elegans. Genetics 1979, 93, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Brockdorff, N.; Turner, B.M. Dosage compensation in mammals. Cold Spring Harb. Perspect. Biol. 2015, 7, a019406. [Google Scholar] [CrossRef] [PubMed]
- Lucchesi, J.C.; Kuroda, M.I. Dosage compensation in Drosophila. Cold Spring Harb. Perspect. Biol. 2015, 7, a019398. [Google Scholar] [CrossRef]
- Lau, A.C.; Csankovszki, G. Balancing up and downregulation of the C. elegans X chromosomes. Curr. Opin. Genet. Dev. 2015, 31, 50–56. [Google Scholar] [CrossRef][Green Version]
- Sanderson, B.J.; Feng, G.; Hu, N.; Carlson, C.H.; Smart, L.B.; Keefover-Ring, K.; Yin, T.; Ma, T.; Liu, J.; DiFazio, S.P.; et al. Sex determination through X-Y heterogamety in Salix nigra. Heredity 2021, 126, 630–639. [Google Scholar] [CrossRef] [PubMed]
- Hayman, E.S.; Fairgrieve, W.T.; Luckenbach, J.A. Molecular and morphological sex differentiation in sablefish (Anoplopoma fimbria), a marine teleost with XX/XY sex determination. Gene 2021, 764, 145093. [Google Scholar] [CrossRef]
- Nakamura, M. Sex determination in amphibians. Semin. Cell Dev. Biol. 2009, 20, 271–282. [Google Scholar] [CrossRef]
- Irwin, D.E. Sex chromosomes and speciation in birds and other ZW systems. Mol. Ecol. 2018, 27, 3831–3851. [Google Scholar] [CrossRef]
- Chandler, C.H. When and why does sex chromosome dosage compensation evolve? Ann. N. Y. Acad. Sci. 2017, 1389, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Lau, A.C.; Nabeshima, K.; Csankovszki, G. The C. elegans dosage compensation complex mediates interphase X chromosome compaction. Epigenetics Chromatin 2014, 7, 31. [Google Scholar] [CrossRef] [PubMed]
- Meyer, B.J. Mechanisms of sex determination and X-chromosome dosage compensation. Genetics 2022, 220, iyab197. [Google Scholar] [CrossRef] [PubMed]
- Chuang, P.-T.; Lieb, J.D.; Meyer, B.J. Sex-Specific Assembly of a Dosage Compensation Complex on the Nematode X Chromosome. Science 1996, 274, 1736–1739. [Google Scholar] [CrossRef] [PubMed]
- Lieb, J.D.; Albrecht, M.R.; Chuang, P.T.; Meyer, B.J. MIX-1: An essential component of the C. elegans mitotic machinery executes X chromosome dosage compensation. Cell 1998, 92, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Lieb, J.D.; Capowski, E.E.; Meneely, P.; Meyer, B.J. DPY-26, a link between dosage compensation and meiotic chromosome segregation in the nematode. Science 1996, 274, 1732–1736. [Google Scholar] [CrossRef] [PubMed]
- Nusbaum, C.; Meyer, B.J. The Caenorhabditis elegans gene sdc-2 controls sex determination and dosage compensation in XX animals. Genetics 1989, 122, 579–593. [Google Scholar] [CrossRef] [PubMed]
- Villeneuve, A.M.; Meyer, B.J. sdc-1: A link between sex determination and dosage compensation in C. elegans. Cell 1987, 48, 25–37. [Google Scholar] [CrossRef]
- DeLong, L.; Plenefisch, J.D.; Klein, R.D.; Meyer, B.J. Feedback control of sex determination by dosage compensation revealed through Caenorhabditis elegans sdc-3 mutations. Genetics 1993, 133, 875–896. [Google Scholar] [CrossRef]
- Yonker, S.A.; Meyer, B.J. Recruitment of C. elegans dosage compensation proteins for gene-specific versus chromosome-wide repression. Development 2003, 130, 6519–6532. [Google Scholar] [CrossRef]
- Brejc, K.; Bian, Q.; Uzawa, S.; Wheeler, B.S.; Anderson, E.C.; King, D.S.; Kranzusch, P.J.; Preston, C.G.; Meyer, B.J. Dynamic Control of X Chromosome Conformation and Repression by a Histone H4K20 Demethylase. Cell 2017, 171, 85–102.e23. [Google Scholar] [CrossRef]
- Pferdehirt, R.R.; Kruesi, W.S.; Meyer, B.J. An MLL/COMPASS subunit functions in the C. elegans dosage compensation complex to target X chromosomes for transcriptional regulation of gene expression. Genes. Dev. 2011, 25, 499–515. [Google Scholar] [CrossRef]
- Dong, X.; Peng, Y.; Peng, Y.; Xu, F.; He, X.; Wang, F.; Peng, X.; Qiang, B.; Yuan, J.; Rao, Z. Characterization and crystallization of human DPY-30-like protein, an essential component of dosage compensation complex. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2005, 1753, 257–262. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, V.; Zhang, P.; Chaturvedi, C.P.; Thornton, J.; Brunzelle, J.S.; Skiniotis, G.; Shilatifard, A.; Brand, M.; Couture, J.F. Molecular basis for DPY-30 association to COMPASS-like and NURF complexes. Structure 2014, 22, 1821–1830. [Google Scholar] [CrossRef] [PubMed]
- Petty, E.; Laughlin, E.; Csankovszki, G. Regulation of DCC localization by HTZ-1/H2A.Z and DPY-30 does not correlate with H3K4 methylation levels. PLoS ONE 2011, 6, e25973. [Google Scholar] [CrossRef]
- Custer, L.M.; Snyder, M.J.; Flegel, K.; Csankovszki, G. The onset of C. elegans dosage compensation is linked to the loss of developmental plasticity. Dev. Biol. 2014, 385, 279–290. [Google Scholar] [CrossRef]
- Dawes, H.E.; Berlin, D.S.; Lapidus, D.M.; Nusbaum, C.; Davis, T.L.; Meyer, B.J. Dosage Compensation Proteins Targeted to X Chromosomes by a Determinant of Hermaphrodite Fate. Science 1999, 284, 1800–1804. [Google Scholar] [CrossRef]
- McDonel, P.; Jans, J.; Peterson, B.K.; Meyer, B.J. Clustered DNA motifs mark X chromosomes for repression by a dosage compensation complex. Nature 2006, 444, 614–618. [Google Scholar] [CrossRef]
- Jans, J.; Gladden, J.M.; Ralston, E.J.; Pickle, C.S.; Michel, A.H.; Pferdehirt, R.R.; Eisen, M.B.; Meyer, B.J. A condensin-like dosage compensation complex acts at a distance to control expression throughout the genome. Genes. Dev. 2009, 23, 602–618. [Google Scholar] [CrossRef] [PubMed]
- Fuda, N.J.; Brejc, K.; Kruesi, W.S.; Ralston, E.J.; Bigley, R.; Shin, A.; Okada, M.; Meyer, B.J. Combinatorial clustering of distinct DNA motifs directs synergistic binding of Caenorhabditis elegans dosage compensation complex to X chromosomes. Proc. Natl. Acad. Sci. USA 2022, 119, e2211642119. [Google Scholar] [CrossRef]
- Albritton, S.E.; Kranz, A.L.; Winterkorn, L.H.; Street, L.A.; Ercan, S. Cooperation between a hierarchical set of recruitment sites targets the X chromosome for dosage compensation. eLife 2017, 6, e23645. [Google Scholar] [CrossRef] [PubMed]
- Ercan, S.; Giresi, P.G.; Whittle, C.M.; Zhang, X.; Green, R.D.; Lieb, J.D. X chromosome repression by localization of the C. elegans dosage compensation machinery to sites of transcription initiation. Nat. Genet. 2007, 39, 403–408. [Google Scholar] [CrossRef]
- Kim, J.; Jimenez, D.S.; Ragipani, B.; Zhang, B.; Street, L.A.; Kramer, M.; Albritton, S.E.; Winterkorn, L.; Morao, A.; Ercan, S. Condensin DC loads and spreads from recruitment sites to create loop- anchored TADs in C. elegans. eLife 2022, 11, e68745. [Google Scholar] [CrossRef]
- Csankovszki, G.; McDonel, P.; Meyer, B.J. Recruitment and spreading of the C. elegans dosage compensation complex along X chromosomes. Science 2004, 303, 1182–1185. [Google Scholar] [CrossRef]
- Anderson, E.C.; Frankino, P.A.; Higuchi-Sanabria, R.; Yang, Q.; Bian, Q.; Podshivalova, K.; Shin, A.; Kenyon, C.; Dillin, A.; Meyer, B.J. X Chromosome Domain Architecture Regulates Caenorhabditis elegans Lifespan but Not Dosage Compensation. Dev. Cell 2019, 51, 192–207.e196. [Google Scholar] [CrossRef]
- Rowley, M.J.; Poulet, A.; Nichols, M.H.; Bixler, B.J.; Sanborn, A.L.; Brouhard, E.A.; Hermetz, K.; Linsenbaum, H.; Csankovszki, G.; Lieberman Aiden, E.; et al. Analysis of Hi-C data using SIP effectively identifies loops in organisms from C. elegans to mammals. Genome Res. 2020, 30, 447–458. [Google Scholar] [CrossRef] [PubMed]
- Petit, F.; Sears, K.E.; Ahituv, N. Limb development: A paradigm of gene regulation. Nat. Rev. Genet. 2017, 18, 245–258. [Google Scholar] [CrossRef] [PubMed]
- Arvanitis, M.; Tayeb, K.; Strober, B.J.; Battle, A. Redefining tissue specificity of genetic regulation of gene expression in the presence of allelic heterogeneity. Am. J. Hum. Genet. 2022, 109, 223–239. [Google Scholar] [CrossRef]
- Chang, Y.N.; Zhu, C.; Jiang, J.; Zhang, H.; Zhu, J.K.; Duan, C.G. Epigenetic regulation in plant abiotic stress responses. J. Integr. Plant Biol. 2020, 62, 563–580. [Google Scholar] [CrossRef]
- Kruesi, W.S.; Core, L.J.; Waters, C.T.; Lis, J.T.; Meyer, B.J. Condensin controls recruitment of RNA polymerase II to achieve nematode X-chromosome dosage compensation. eLife 2013, 2, e00808. [Google Scholar] [CrossRef]
- Tzatsos, A.; Paskaleva, P.; Ferrari, F.; Deshpande, V.; Stoykova, S.; Contino, G.; Wong, K.K.; Lan, F.; Trojer, P.; Park, P.J.; et al. KDM2B promotes pancreatic cancer via Polycomb-dependent and -independent transcriptional programs. J. Clin. Investig. 2013, 123, 727–739. [Google Scholar] [CrossRef]
- Ward, J.R.; Vasu, K.; Deutschman, E.; Halawani, D.; Larson, P.A.; Zhang, D.; Willard, B.; Fox, P.L.; Moran, J.V.; Longworth, M.S. Condensin II and GAIT complexes cooperate to restrict LINE-1 retrotransposition in epithelial cells. PLoS Genet. 2017, 13, e1007051. [Google Scholar] [CrossRef] [PubMed]
- Vielle, A.; Lang, J.; Dong, Y.; Ercan, S.; Kotwaliwale, C.; Rechtsteiner, A.; Appert, A.; Chen, Q.B.; Dose, A.; Egelhofer, T.; et al. H4K20me1 contributes to downregulation of X-linked genes for C. elegans dosage compensation. PLoS Genet. 2012, 8, e1002933. [Google Scholar] [CrossRef] [PubMed]
- Wells, M.B.; Snyder, M.J.; Custer, L.M.; Csankovszki, G. Caenorhabditis elegans dosage compensation regulates histone H4 chromatin state on X chromosomes. Mol. Cell Biol. 2012, 32, 1710–1719. [Google Scholar] [CrossRef] [PubMed]
- Budhavarapu, V.N.; Chavez, M.; Tyler, J.K. How is epigenetic information maintained through DNA replication? Epigenetics Chromatin 2013, 6, 32. [Google Scholar] [CrossRef] [PubMed]
- Corvalan, A.Z.; Coller, H.A. Methylation of histone 4’s lysine 20: A critical analysis of the state of the field. Physiol. Genom. 2021, 53, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Oda, H.; Okamoto, I.; Murphy, N.; Chu, J.; Price, S.M.; Shen, M.M.; Torres-Padilla, M.E.; Heard, E.; Reinberg, D. Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Mol. Cell Biol. 2009, 29, 2278–2295. [Google Scholar] [CrossRef] [PubMed]
- Snyder, M.J.; Lau, A.C.; Brouhard, E.A.; Davis, M.B.; Jiang, J.; Sifuentes, M.H.; Csankovszki, G. Anchoring of Heterochromatin to the Nuclear Lamina Reinforces Dosage Compensation-Mediated Gene Repression. PLoS Genet. 2016, 12, e1006341. [Google Scholar] [CrossRef] [PubMed]
- Poleshko, A.; Katz, R.A. Specifying peripheral heterochromatin during nuclear lamina reassembly. Nucleus 2014, 5, 32–39. [Google Scholar] [CrossRef]
- Davis, M.B.; Jash, E.; Chawla, B.; Haines, R.A.; Tushman, L.E.; Troll, R.; Csankovszki, G. Dual roles for nuclear RNAi Argonautes in Caenorhabditis elegans dosage compensation. Genetics 2022, 221, iyac033. [Google Scholar] [CrossRef]
- Pal-Bhadra, M.; Leibovitch, B.A.; Gandhi, S.G.; Chikka, M.R.; Bhadra, U.; Birchler, J.A.; Elgin, S.C. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 2004, 303, 669–672. [Google Scholar] [CrossRef]
- Breimann, L.; Morao, A.K.; Kim, J.; Jimenez, D.S.; Maryn, N.; Bikkasani, K.; Carrozza, M.J.; Albritton, S.E.; Kramer, M.; Street, L.A.; et al. The histone H4 lysine 20 demethylase DPY-21 regulates thedynamics of condensin DC binding. J. Cell Sci. 2022, 135, jcs258818. [Google Scholar] [CrossRef]
- Kschonsak, M.; Merkel, F.; Bisht, S.; Metz, J.; Rybin, V.; Hassler, M.; Haering, C.H. Structural Basis for a Safety-Belt Mechanism That Anchors Condensin to Chromosomes. Cell 2017, 171, 588–600.e524. [Google Scholar] [CrossRef]
- Terakawa, T.; Bisht, S.; Eeftens, J.M.; Dekker, C.; Haering, C.H.; Greene, E.C. The condensin complex is a mechanochemical motor that translocates along DNA. Science 2017, 358, 672–676. [Google Scholar] [CrossRef] [PubMed]
- Kong, M.; Cutts, E.; Pan, D.; Beuron, F.; Kaliyappan, T.; Xue, C.; Morris, E.; Musacchio, A.; Vannini, A.; Greene, E.C. Human Condensin I and II Drive Extensive ATP-Dependent Compaction of Nucleosome-Bound DNA. Mol. Cell 2020, 79, 99–114. [Google Scholar] [CrossRef] [PubMed]
- Eeftens, J.M.; Bisht, S.; Kerssemakers, J.; Kschonsak, M.; Haering, C.H.; Dekker, C. Real-time detection of condensin-driven DNA compaction reveals a multistep binding mechanism. EMBO J. 2017, 36, 3448–3457. [Google Scholar] [CrossRef] [PubMed]
- Morao, A.K.; Kim, J.; Obaji, D.; Sun, S.; Ercan, S. Topoisomerases I and II facilitate condensin DC translocation to organize and repress X chromosomes in C. elegans. Mol. Cell 2022, 82, 4202–4217.e4205. [Google Scholar] [CrossRef] [PubMed]
- Uusküla-Reimand, L.; Hou, H.; Samavarchi-Tehrani, P.; Rudan, M.V.; Liang, M.; Medina-Rivera, A.; Mohammed, H.; Schmidt, D.; Schwalie, P.; Young, E.J.; et al. Topoisomerase II beta interacts with cohesin and CTCF at topological domain borders. Genome Biol. 2016, 17, 182. [Google Scholar] [CrossRef] [PubMed]
- Meyer, B.J.; Casson, L.P. Caenorhabditis elegans compensates for the difference in X chromosome dosage between the sexes by regulating transcript levels. Cell 1986, 47, 871–881. [Google Scholar] [CrossRef]
- Lau, A.C.; Zhu, K.P.; Brouhard, E.A.; Davis, M.B.; Csankovszki, G. An H4K16 histone acetyltransferase mediates decondensation of the X chromosome in C. elegans males. Epigenetics Chromatin 2016, 9, 44. [Google Scholar] [CrossRef]




| Organism | C. elegans | Saccharomyces cerevisiae | Saccharomyces pombe | Mammals | Drosophila melanogaster | Xenopus |
|---|---|---|---|---|---|---|
| SMC1 | SMC-1 | Smc1 | Psm1 | SMC1α, SMC1β * | DmSMC1/Smc1 | XSMC1a/Smc1a XSMC1b/Smc1b |
| SMC3 | SMC-3 | Smc3 | Psm3 | SMC3 | Cap/Smc3 | XSMC3/Smc3 |
| Kleisin | SCC-1/COH-2 COH-1 REC-8 * COH-3 * COH-4 * | Scc1/Mcd1 Rec8 * | Rad21 Rec8 * | RAD21 RAD21L * REC8 * | DRAD21/Rad21 c(2)M * | XRAD21/Rad21 XREC8/Rec8 * |
| HAWK | SCC-3 | Scc3/Irr1 | Psc3 Rec11 * | SA1/STAG1 SA2/STAG2 SA3/STAG3 * | DSA1/Sa1 SA-2/Sa2 * | XSA1/Sa1 XSA2/Sa2 |
| CTCF | - | - | - | CTCF | DmCtcf/Ctcf | XCTCF/Ctcf |
| Organism | C. elegans | S. cerevisiae | S. pombe | Mammals | D. melanogaster | Xenopus |
|---|---|---|---|---|---|---|
| SMC2 | SMC-2 | Smc2 | Cut14 | SMC2 | DmSmc2/Smc2 | XCAP-E/Smc2 |
| SMC4 | SMC-4 DPY-27 * | Smc4 | Cut3 | SMC4 | DmSmc4/Smc4 | XCAP-C/Smc4 |
| Kleisin | DPY-26 | Brn1 | Cnd2 | CAP-H | Cap-H/Barren | XCAP-H/Cap-H |
| HAWKs | DPY-28 CAP-G1 | Ysc4 Ysc1 | Cnd1 Cnd3 | CAP-D2 CAP-G | dCap-D2/Cap-D2 dcap-g/Cap-G | XCAP-D2/Cap-D2 XCAP-G/Cap-G |
| Organism | C. elegans | Mammals | D. melanogaster | Xenopus |
|---|---|---|---|---|
| SMC2 | SMC-2 | SMC2 | DmSMC2/Smc2 | XCAP-E/Smc2 |
| SMC4 | SMC-4 | SMC4 | Glu/SMC4 | XCAP-C/Smc3 |
| Kleisin | KLE-2 | CAP-H2 | dCap-H2/Cap-H2 | XCAP-H2/Cap-H2 |
| HAWKs | HCP-6 CAP-G2 | CAP-D3 CAP-G2 | dCAP-D3/Cap-D3 -1 | XCAP-D3/Cap-D3 XCAP-G2/Cap-G2 |
| Organism | C. elegans | S. cerevisiae | S. pombe | Mammals | D. melanogaster | Xenopus |
|---|---|---|---|---|---|---|
| SMC5 | SMC-5 | Smc5 | Smc5/Spr18 | SMC5 | Smc5 | XSMC5/Smc5 |
| SMC6 | SMC-6 | Smc6 | Smc6/Rad18 | SMC6 | Smc6 | XSMC6/Smc6 |
| Kleisin | NSE-4 | Nse4 | Nse4/Rad62 | NSE4/NSMCE4 | Nse4 | Nse4 |
| Kites | NSE-1 NSE-2 NSE-3 | Nse1 Nse2/Mms21 Nse3 | Nse1 Nse2 Nse3 | NSE1/NSMCE1 NSE2/NSMCE2 NSE3/NSMCE3 | Nse1 Qjt/Nse2 MAGE/Nse3 | Nse1 Nse2/Nsmce2/Mms21 Nse3 |
| Other 1 | - - | Nse5 Nse6/Kre29 | Nse5 Nse6 | SIMC1, SLF1 SLF2 | - - | Slf1 Slf2/Fam178a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chawla, B.; Csankovszki, G. How Chromatin Motor Complexes Influence the Nuclear Architecture: A Review of Chromatin Organization, Cohesins, and Condensins with a Focus on C. elegans. DNA 2024, 4, 84-103. https://doi.org/10.3390/dna4010005
Chawla B, Csankovszki G. How Chromatin Motor Complexes Influence the Nuclear Architecture: A Review of Chromatin Organization, Cohesins, and Condensins with a Focus on C. elegans. DNA. 2024; 4(1):84-103. https://doi.org/10.3390/dna4010005
Chicago/Turabian StyleChawla, Bahaar, and Györgyi Csankovszki. 2024. "How Chromatin Motor Complexes Influence the Nuclear Architecture: A Review of Chromatin Organization, Cohesins, and Condensins with a Focus on C. elegans" DNA 4, no. 1: 84-103. https://doi.org/10.3390/dna4010005
APA StyleChawla, B., & Csankovszki, G. (2024). How Chromatin Motor Complexes Influence the Nuclear Architecture: A Review of Chromatin Organization, Cohesins, and Condensins with a Focus on C. elegans. DNA, 4(1), 84-103. https://doi.org/10.3390/dna4010005