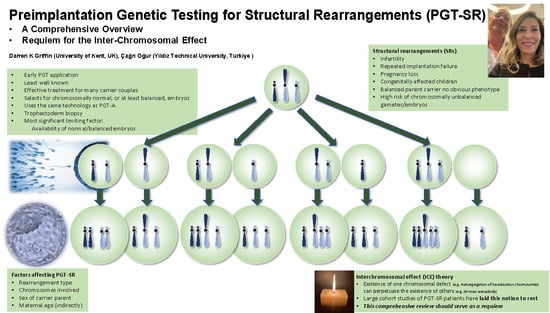
PGT-SR: A Comprehensive Overview and a Requiem for the Interchromosomal Effect
Abstract
1. Introduction
2. Balanced SRs in Overview
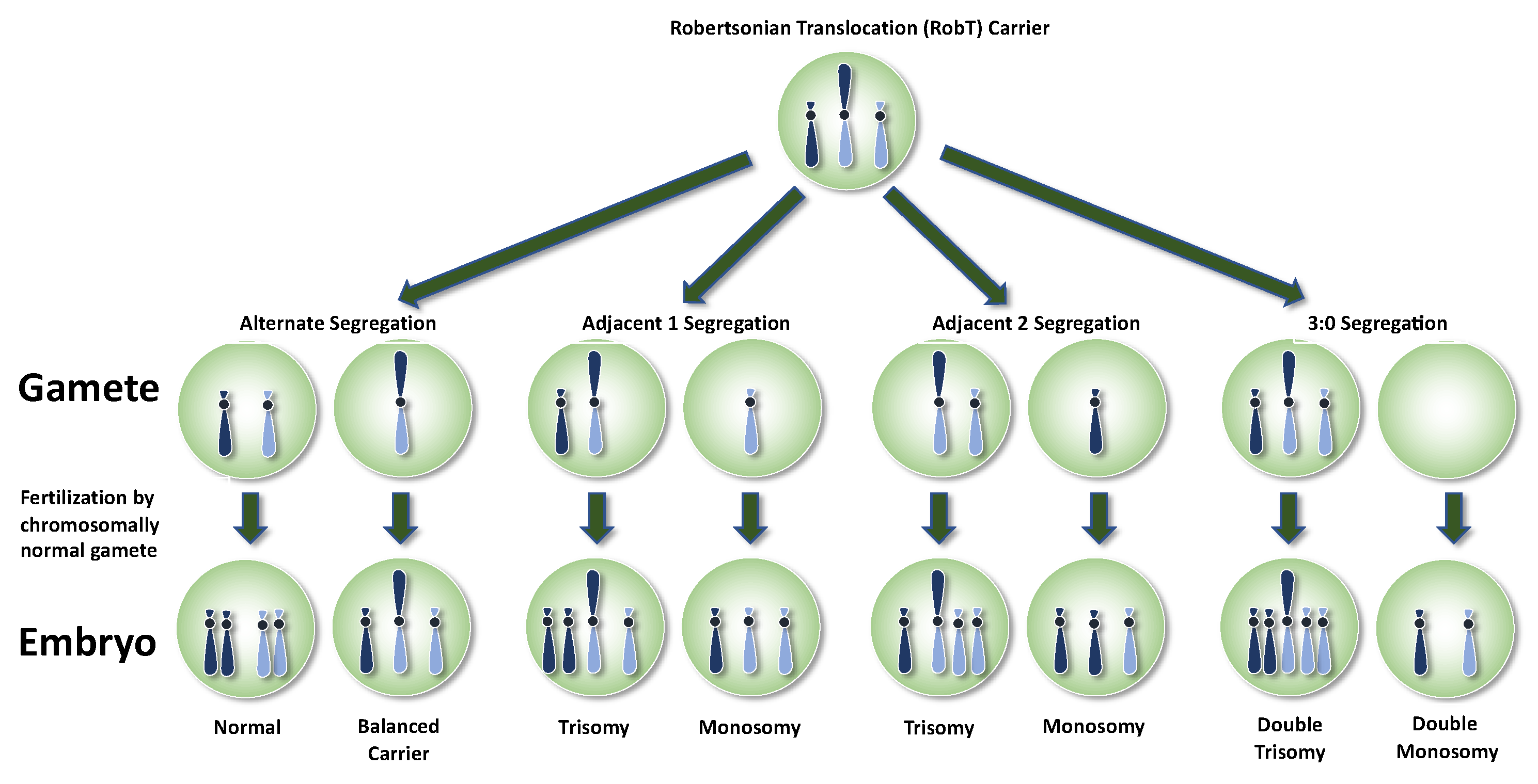
3. Robertsonian Translocations
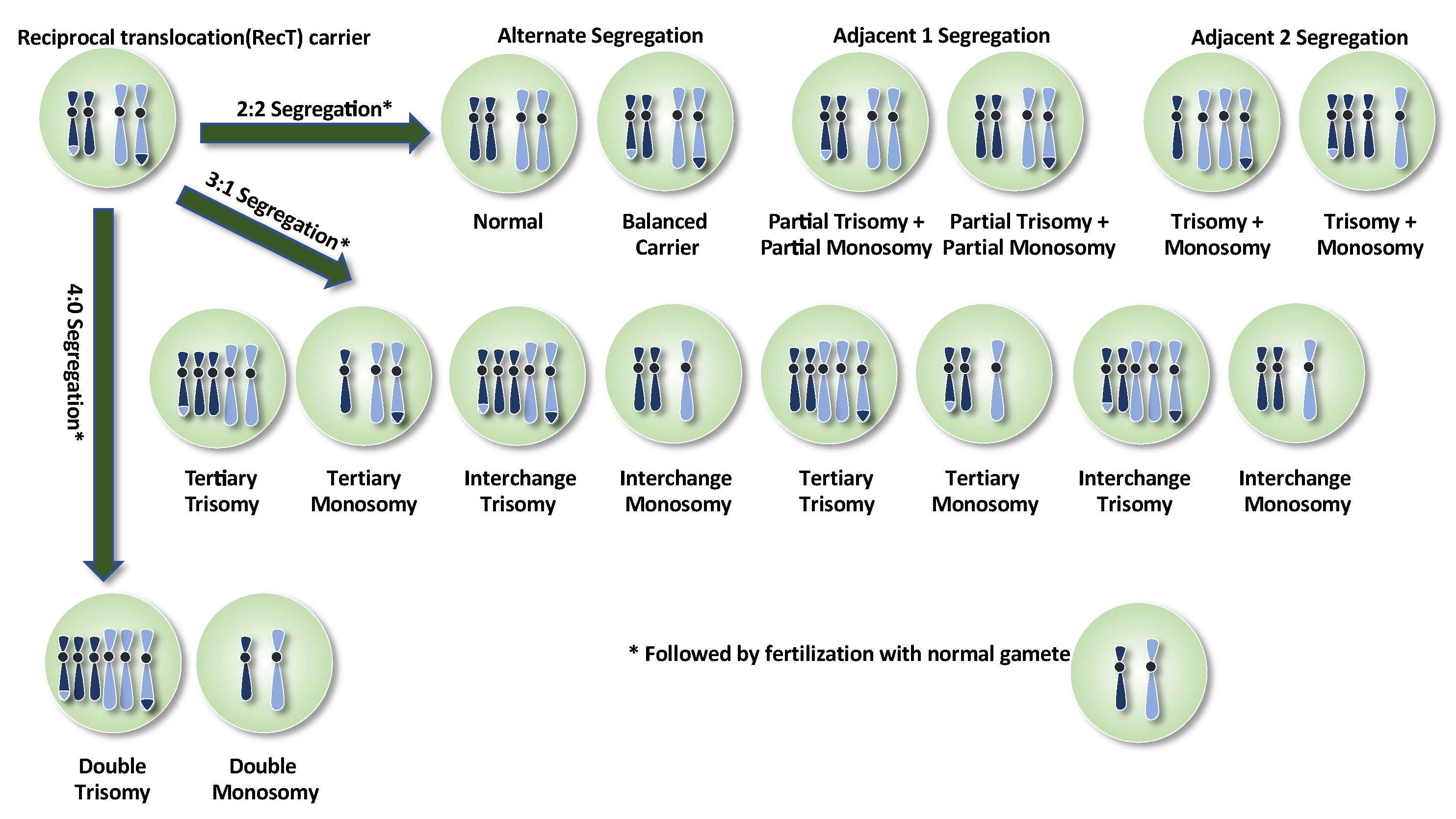
4. Reciprocal Translocations
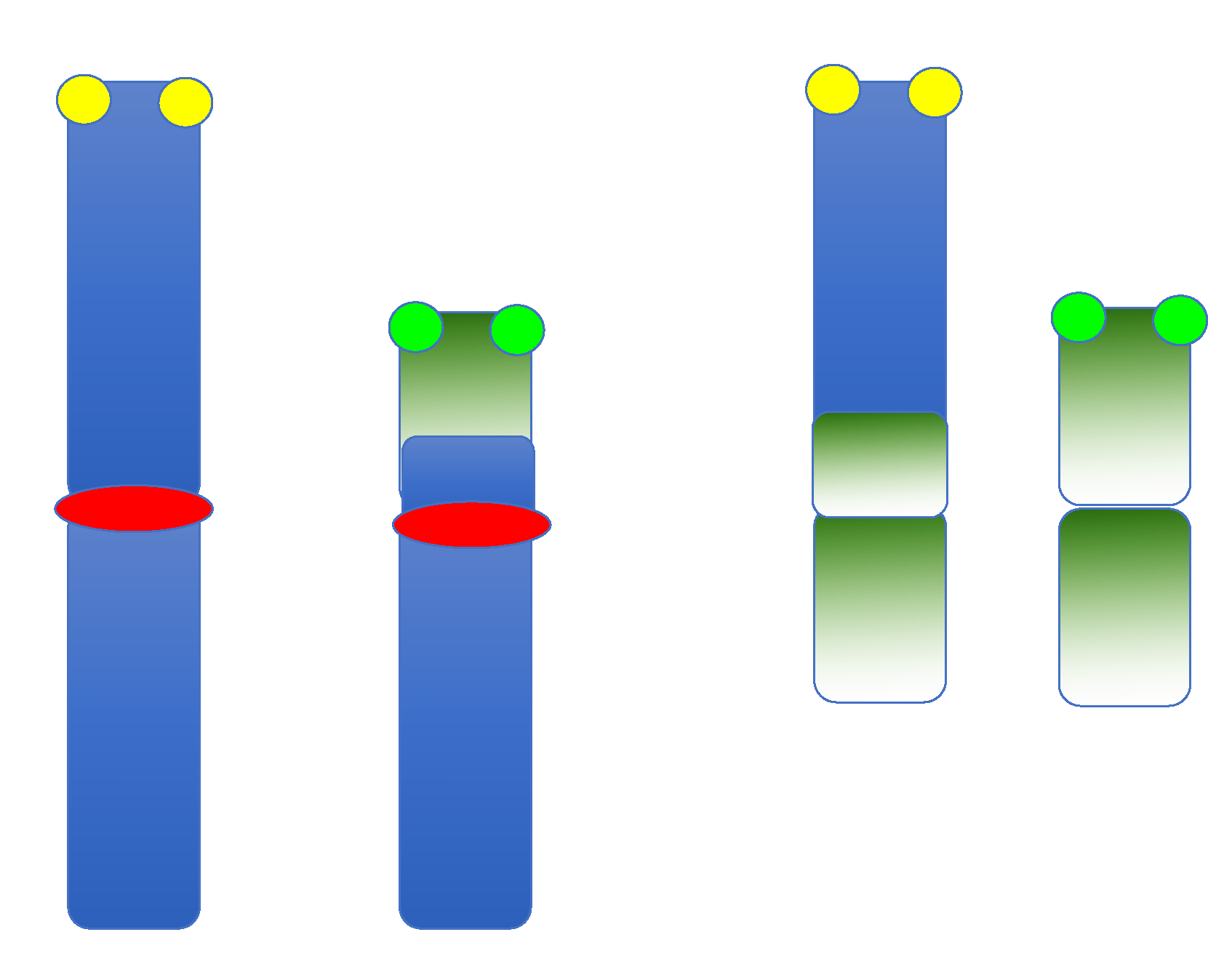
5. Inversions
6. Insertional Translocations and Complex Chromosomal Rearrangements
7. Practicing PGT-SR
8. Invasive Sampling Methods for PGT-SR
9. Minimally Invasive Sampling Methods for PGT-SR
10. FISH for PGT-SR
11. STR-Typing for PGT-SR
12. Comparative Genomic Hybridization (CGH) and Array Comparative Genomic Hybridization (aCGH)
13. Karyomapping and SNP Arrays for PGT-SR
14. Next-Generation Sequencing (NGS) for PGT-SR
15. Towards Universal PGT
16. Should We Deselect or De-Prioritize Carrier Embryos?
17. Developmental Characteristics of the Embryo and PGT-SR
18. Ovarian Response and Its Relevance to PGT-SR
19. Meiotic Segregation Patterns and the Number of Chromosomally Balanced Embryos Available for PGT-SR
20. The Time of Biopsy and Maternal Age: No Effect on Chromosome Segregation, but Relevant to PGT-SR Nonetheless
21. Aneuploidy and Chromosomal Mosaicism
22. Requiem for the Interchromosomal Effect (ICE)
23. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Weckselblatt, B.; Hermetz, K.E.; Rudd, M.K. Unbalanced Translocations Arise from Diverse Mutational Mechanisms Including Chromothripsis. Genome Res. 2015, 25, 937–947. [Google Scholar] [CrossRef]
- De Braekeleer, M.; Dao, T.N. Cytogenetic Studies in Couples Experiencing Repeated Pregnancy Losses. Hum. Reprod. 1990, 5, 519–528. [Google Scholar] [CrossRef] [PubMed]
- De Braekeleer, M.; Dao, T.N. Cytogenetic Studies in Male Infertility: A Review. Hum. Reprod. 1991, 6, 245–250. [Google Scholar] [CrossRef]
- Shah, K.; Sivapalan, G.; Gibbons, N.; Tempest, H.; Griffin, D.K. The Genetic Basis of Infertility. Reproduction 2003, 126, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Lejeune, J. Autosomal Disorders. Pediatrics 1963, 32, 326–337. [Google Scholar] [CrossRef]
- Griffin, D.K. Fluorescent Molecular Cytogenetics: Preimplantation Diagnosis, Colorectal Cancer and Mapping Chromosome 9. Ph.D. Thesis, University College London, London, UK, 1992. [Google Scholar]
- Harper, J.C.; Wilton, L.; Traeger-Synodinos, J.; Goossens, V.; Moutou, C.; SenGupta, S.B.; Pehlivan Budak, T.; Renwick, P.; De Rycke, M.; Geraedts, J.P.M.; et al. The ESHRE PGD Consortium: 10 Years of Data Collection. Hum. Reprod. Update 2012, 18, 234–247. [Google Scholar] [CrossRef] [PubMed]
- Griffin, D.K.; Ogur, C. Chromosomal Analysis in IVF: Just How Useful Is It? Reproduction 2018, 156, F29–F50. [Google Scholar] [CrossRef] [PubMed]
- Ogur, C.; Griffin, D.K. Preimplantation Genetic Testing for Structural Rearrangements. In Preimplantation Genetic Testing. Recent Advances in Reproductive Medicine; Griffin, D.K., Harton, G.L., Eds.; CRC Press: Boca Raton, UK, 2020; pp. 49–76. [Google Scholar] [CrossRef]
- Lieber, M.R. The Mechanism of Double-Strand DNA Break Repair by the Nonhomologous DNA End-Joining Pathway. Annu. Rev. Biochem. 2010, 79, 181–211. [Google Scholar] [CrossRef]
- Hurles, M.E.; Lupski, J.R. Recombination hotspots in nonallelic homologous recombination. In Genomic Disorders; Lupski, J.R., Stankiewicz, P., Eds.; Humana Press: Totowa, NJ, USA, 2006; pp. 341–355. [Google Scholar] [CrossRef]
- Ou, Z.Z.; Kochmar, S.; Yatsenko, S.A.; Woerner, A.C.; Acquaro, R.; Ortiz, D.; Surti, U.; Hu, J. Partial 5p Deletion and Partial 5q Duplication in a Patient with Multiple Congenital Anomalies: A Two-Step Mechanism in Chromosomal Rearrangement Mediated by Non-Allelic Homologous Recombination. Cytogenet. Genome Res. 2018, 156, 65–70. [Google Scholar] [CrossRef]
- Pellestor, F.; Anahory, T.; Lefort, G.; Puechberty, J.; Liehr, T.; Hedon, B.; Sarda, P. Complex Chromosomal Rearrangements: Origin and Meiotic Behavior. Hum. Reprod. Update 2011, 17, 476–494. [Google Scholar] [CrossRef]
- Lee, J.A.; Carvalho, C.M.B.; Lupski, J.R. A DNA Replication Mechanism for Generating Nonrecurrent Rearrangements Associated with Genomic Disorders. Cell 2007, 131, 1235–1247. [Google Scholar] [CrossRef]
- Tucker, J. Low-Dose Ionizing Radiation and Chromosome Translocations: A Review of the Major Considerations for Human Biological Dosimetry. Mutat. Res./Rev. Mutat. Res. 2008, 659, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Hook, E.B.; Schreinemachers, D.M.; Willey, A.M.; Cross, P.K. Inherited Structural Cytogenetic Abnormalities Detected Incidentally in Fetuses Diagnosed Prenatally: Frequency, Parental-Age Associations, Sex-Ratio Trends, and Comparisons with Rates of Mutants. Am. J. Hum. Genet. 1984, 36, 422–443. [Google Scholar] [PubMed]
- Beyazyurek, C.; Ekmekci, C.G.; Sağlam, Y.; Cinar, C.; Kahraman, S. Preimplantation Genetic Diagnosis (PGD) for Extremes—Successful Birth after PGD for a Consanguineous Couple Carrying an Identical Balanced Reciprocal Translocation. Fertil. Steril. 2010, 93, e1–e2413. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, P.A.; Browne, C.; Gregson, N.; Joyce, C.; White, H. Estimates of the Frequency of Chromosome Abnormalities Detectable in Unselected Newborns Using Moderate Levels of Banding. J. Med. Genet. 1992, 29, 103–108. [Google Scholar] [CrossRef]
- Scriven, P.N.; Handyside, A.H.; Ogilvie, C.M. Chromosome Translocations: Segregation Modes and Strategies for Preimplantation Genetic Diagnosis. Prenat. Diagn. 1998, 18, 1437–1449. [Google Scholar] [CrossRef]
- Martin, R.H. A Detailed Method for Obtaining Preparations of Human Sperm Chromosomes. Cytogenet. Cell Genet. 1983, 35, 252–256. [Google Scholar] [CrossRef]
- Lamotte, A.; Martinez, G.; Devillard, F.; Hograindleur, J.-P.; Satre, V.; Coutton, C.; Harbuz, R.; Amblard, F.; Lespinasse, J.; Benchaib, M.; et al. Is Sperm FISH Analysis Still Useful for Robertsonian Translocations? Meiotic Analysis for 23 Patients and Review of the Literature. Basic Clin. Androl. 2018, 28, 5. [Google Scholar] [CrossRef]
- Patassini, C.; Garolla, A.; Bottacin, A.; Menegazzo, M.; Speltra, E.; Foresta, C.; Ferlin, A. Molecular Karyotyping of Human Single Sperm by Array-Comparative Genomic Hybridization. PLoS ONE 2013, 8, e60922. [Google Scholar] [CrossRef]
- Martin, R.H. Cytogenetic Determinants of Male Fertility. Hum. Reprod. Update 2008, 14, 379–390. [Google Scholar] [CrossRef]
- Goldman, A.S.; Hultén, M.A. Analysis of Chiasma Frequency and First Meiotic Segregation in a Human Male Reciprocal Translocation Heterozygote, t(1;11)(P36.3;Q13.1), Using Fluorescence in Situ Hybridisation. Cytogenet. Cell Genet. 1993, 63, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Escudero, T.; Abdelhadi, I.; Sandalinas, M.; Munné, S. Predictive Value of Sperm Fluorescence in Situ Hybridization Analysis on the Outcome of Preimplantation Genetic Diagnosis for Translocations. Fertil. Steril. 2003, 79, 1528–1534. [Google Scholar] [CrossRef] [PubMed]
- Pettenati, M.J.; Rao, P.N.; Phelan, M.C.; Grass, F.; Rao, K.W.; Cosper, P.; Carroll, A.J.; Elder, F.; Smith, J.L.; Higgins, M.D. Paracentric Inversions in Humans: A Review of 446 Paracentric Inversions with Presentation of 120 New Cases. Am. J. Med. Genet. 1995, 55, 171–187. [Google Scholar] [CrossRef] [PubMed]
- Jaarola, M.; Martin, R.H.; Ashley, T. Direct Evidence for Suppression of Recombination within Two Pericentric Inversions in Humans: A New Sperm-FISH Technique. Am. J. Hum. Genet. 1998, 63, 218–224. [Google Scholar] [CrossRef]
- Martin, R.H. Sperm Chromosome Analysis in a Man Heterozygous for a Paracentric Inversion of Chromosome 14 (Q24.1q32.1). Am. J. Hum. Genet. 1999, 64, 1480–1484. [Google Scholar] [CrossRef][Green Version]
- Bhatt, S.; Moradkhani, K.; Mrasek, K.; Puechberty, J.; Manvelyan, M.; Hunstig, F.; Lefort, G.; Weise, A.; Lespinasse, J.; Sarda, P.; et al. Breakpoint Mapping and Complete Analysis of Meiotic Segregation Patterns in Three Men Heterozygous for Paracentric Inversions. Eur. J. Hum. Genet. 2009, 17, 44–50. [Google Scholar] [CrossRef][Green Version]
- Yapan, C.; Beyazyurek, C.; Ekmekci, C.; Kahraman, S. The Largest Paracentric Inversion, the Highest Rate of Recombinant Spermatozoa. Case Report: 46,XY, Inv(2)(Q21.2q37.3) and Literature Review. Balk. J. Med. Genet. 2014, 17, 55–62. [Google Scholar] [CrossRef]
- Morel, F.; Laudier, B.; Guérif, F.; Couet, M.L.; Royère, D.; Roux, C.; Bresson, J.L.; Amice, V.; De Braekeleer, M.; Douet-Guilbert, N. Meiotic Segregation Analysis in Spermatozoa of Pericentric Inversion Carriers Using Fluorescence In-Situ Hybridization. Hum. Reprod. 2007, 22, 136–141. [Google Scholar] [CrossRef]
- Abdi, A. Prevalence of Chromosome Inversions (Pericentric and Paracentric) in Patients with Recurrent Abortions. SJRM 2017, 2, 45–50. [Google Scholar] [CrossRef]
- Anton, E.; Blanco, J.; Egozcue, J.; Vidal, F. Sperm Studies in Heterozygote Inversion Carriers: A Review. Cytogenet. Genome Res. 2005, 111, 297–304. [Google Scholar] [CrossRef]
- Salaun, G.; Tchirkov, A.; Francannet, C.; Pons, H.; Brugnon, F.; Pebrel-Richard, C.; Gouas, L.; Eymard-Pierre, E.; Vago, P.; Goumy, C. Sperm Meiotic Segregation of a Balanced Interchromosomal Reciprocal Insertion Resulting in Recurrent Spontaneous Miscarriage. Reprod. BioMed. Online 2018, 37, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Melotte, C.; Debrock, S.; D’Hooghe, T.; Fryns, J.P.; Vermeesch, J.R. Preimplantation Genetic Diagnosis for an Insertional Translocation Carrier. Hum. Reprod. 2004, 19, 2777–2783. [Google Scholar] [CrossRef] [PubMed]
- Xanthopoulou, L.; Mantzouratou, A.; Mania, A.; Cawood, S.; Doshi, A.; Ranieri, D.M.; Delhanty, J.D. Male and Female Meiotic Behaviour of an Intrachromosomal Insertion Determined by Preimplantation Genetic Diagnosis. Mol. Cytogenet. 2010, 3, 2. [Google Scholar] [CrossRef] [PubMed]
- Scriven, P.N.; Bint, S.M.; Davies, A.F.; Ogilvie, C.M. Meiotic Outcomes of Three-Way Translocations Ascertained in Cleavage-Stage Embryos: Refinement of Reproductive Risks and Implications for PGD. Eur. J. Hum. Genet. 2014, 22, 748–753. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pujol, A.; Durban, M.; Benet, J.; Boiso, I.; Calafell, J.M.; Egozcue, J.; Navarro, J. Multiple Aneuploidies in the Oocytes of Balanced Translocation Carriers: A Preimplantation Genetic Diagnosis Study Using First Polar Body. Reproduction 2003, 126, 701–711. [Google Scholar] [CrossRef]
- Molina Gomes, D.; Hammoud, I.; Bailly, M.; Bergere, M.; Wainer, R.; Selva, J.; Vialard, F. Preconceptional Diagnosis for Robertsonian Translocation as an Alternative to Preimplantation Genetic Diagnosis in Two Situations: A Pilot Study. J. Assist. Reprod. Genet. 2009, 26, 113–117. [Google Scholar] [CrossRef][Green Version]
- Scott, R.T.; Upham, K.M.; Forman, E.J.; Zhao, T.; Treff, N.R. Cleavage-Stage Biopsy Significantly Impairs Human Embryonic Implantation Potential While Blastocyst Biopsy Does Not: A Randomized and Paired Clinical Trial. Fertil. Steril. 2013, 100, 624–630. [Google Scholar] [CrossRef]
- Chatzimeletiou, K.; Petrogiannis, N.; Sioga, A.; Emmanouil-Nikoloussi, E.N.; Panagiotidis, Y.; Prapa, M.; Patrikiou, A.; Filippa, M.; Zervakakou, G.; Papanikolaou, K.; et al. The human embryo following biopsy on day 5 versus day 3: Viability, ultrastructure and spindle/chromosome configurations. Reprod. Biomed. Online 2022, 45, 219–233. [Google Scholar] [CrossRef]
- Magli, M.C.; Pomante, A.; Cafueri, G.; Valerio, M.; Crippa, A.; Ferraretti, A.P.; Gianaroli, L. Preimplantation Genetic Testing: Polar Bodies, Blastomeres, Trophectoderm Cells, or Blastocoelic Fluid? Fertil. Steril. 2016, 105, 676–683.e5. [Google Scholar] [CrossRef]
- Liu, W.; Liu, J.; Du, H.; Ling, J.; Sun, X.; Chen, D. Non-Invasive Pre-Implantation Aneuploidy Screening and Diagnosis of Beta Thalassemia IVSII654 Mutation Using Spent Embryo Culture Medium. Ann. Med. 2017, 49, 319–328. [Google Scholar] [CrossRef]
- Xu, J.; Fang, R.; Chen, L.; Chen, D.; Xiao, J.-P.; Yang, W.; Wang, H.; Song, X.; Ma, T.; Bo, S.; et al. Noninvasive Chromosome Screening of Human Embryos by Genome Sequencing of Embryo Culture Medium for in Vitro Fertilization. Proc. Natl. Acad. Sci. USA 2016, 113, 11907–11912. [Google Scholar] [CrossRef] [PubMed]
- Jiao, J.; Shi, B.; Sagnelli, M.; Yang, D.; Yao, Y.; Li, W.; Shao, L.; Lu, S.; Li, D.; Wang, X. Minimally Invasive Preimplantation Genetic Testing Using Blastocyst Culture Medium. Hum. Reprod. 2019, 34, 1369–1379. [Google Scholar] [CrossRef] [PubMed]
- Griffin, D.K.; Handyside, A.H.; Penketh, R.J.; Winston, R.M.; Delhanty, J.D. Fluorescent In-Situ Hybridization to Interphase Nuclei of Human Preimplantation Embryos with X and Y Chromosome Specific Probes. Hum. Reprod. 1991, 6, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Schrurs, B.M.; Winston, R.M.; Handyside, A.H. Preimplantation Diagnosis of Aneuploidy Using Fluorescent In-Situ Hybridization: Evaluation Using a Chromosome 18-Specific Probe. Hum. Reprod. 1993, 8, 296–301. [Google Scholar] [CrossRef] [PubMed]
- Cassel, M.J.; Munné, S.; Fung, J.; Weier, H.U. Carrier-Specific Breakpoint-Spanning DNA Probes: An Approach to Preimplantation Genetic Diagnosis in Interphase Cells. Hum. Reprod. 1997, 12, 2019–2027. [Google Scholar] [CrossRef]
- Munne, S.; Sandalinas, M.; Escudero, T.; Fung, J.; Gianaroli, L.; Cohen, J. Outcome of Preimplantation Genetic Diagnosis of Translocations. Fertil. Steril. 2000, 73, 1209–1218. [Google Scholar] [CrossRef]
- Munné, S.; Escudero, T.; Fischer, J.; Chen, S.; Hill, J.; Stelling, J.R.; Anna, E. Negligible Interchromosomal Effect in Embryos of Robertsonian Translocation Carriers. Reprod. BioMed. Online 2005, 10, 363–369. [Google Scholar] [CrossRef]
- Velilla, E.; Escudero, T.; Munné, S. Blastomere Fixation Techniques and Risk of Misdiagnosis for Preimplantation Genetic Diagnosis of Aneuploidy. Reprod. BioMed. Online 2002, 4, 210–217. [Google Scholar] [CrossRef]
- Ghevaria, H.; SenGupta, S.; Shmitova, N.; Serhal, P.; Delhanty, J. The Origin and Significance of Additional Aneuploidy Events in Couples Undergoing Preimplantation Genetic Diagnosis for Translocations by Array Comparative Genomic Hybridization. Reprod. BioMed. Online 2016, 32, 178–189. [Google Scholar] [CrossRef][Green Version]
- Verlinsky, Y.; Cieslak, J.; Evsikov, S.; Galat, V.; Kuliev, A. Nuclear Transfer for Full Karyotyping and Preimplantation Diagnosis for Translocations. Reprod. BioMed. Online 2002, 5, 300–305. [Google Scholar] [CrossRef]
- Kuliev, A.; Janzen, J.C.; Zlatopolsky, Z.; Kirillova, I.; Ilkevitch, Y.; Verlinsky, Y. Conversion and Non-Conversion Approach to Preimplantation Diagnosis for Chromosomal Rearrangements in 475 Cycles. Reprod. BioMed. Online 2010, 21, 93–99. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lim, C.K.; Cho, J.W.; Song, I.O.; Kang, I.S.; Yoon, Y.-D.; Jun, J.H. Estimation of Chromosomal Imbalances in Preimplantation Embryos from Preimplantation Genetic Diagnosis Cycles of Reciprocal Translocations with or without Acrocentric Chromosomes. Fertil. Steril. 2008, 90, 2144–2151. [Google Scholar] [CrossRef] [PubMed]
- Ko, D.S.; Cho, J.W.; Park, S.Y.; Kim, J.Y.; Koong, M.K.; Song, I.O.; Kang, I.S.; Lim, C.K. Clinical Outcomes of Preimplantation Genetic Diagnosis (PGD) and Analysis of Meiotic Segregation Modes in Reciprocal Translocation Carriers. Am. J. Med. Genet. 2010, 1428–1433. [Google Scholar] [CrossRef] [PubMed]
- Gianaroli, L. Possible Interchromosomal Effect in Embryos Generated by Gametes from Translocation Carriers. Hum. Reprod. 2002, 17, 3201–3207. [Google Scholar] [CrossRef] [PubMed]
- Pujol, A.; Benet, J.; Staessen, C.; Van Assche, E.; Campillo, M.; Egozcue, J.; Navarro, J. The Importance of Aneuploidy Screening in Reciprocal Translocation Carriers. Reproduction 2006, 131, 1025–1035. [Google Scholar] [CrossRef] [PubMed]
- Keymolen, K.; Staessen, C.; Verpoest, W.; Liebaers, I.; Bonduelle, M. Preimplantation Genetic Diagnosis in Female and Male Carriers of Reciprocal Translocations: Clinical Outcome until Delivery of 312 Cycles. Eur. J. Hum. Genet. 2012, 20, 376–380. [Google Scholar] [CrossRef]
- Franssen, M.T.M.; Musters, A.M.; van der Veen, F.; Repping, S.; Leschot, N.J.; Bossuyt, P.M.M.; Goddijn, M.; Korevaar, J.C. Reproductive Outcome after PGD in Couples with Recurrent Miscarriage Carrying a Structural Chromosome Abnormality: A Systematic Review. Hum. Reprod. Update 2011, 17, 467–475. [Google Scholar] [CrossRef]
- Iews, M.; Tan, J.; Taskin, O.; Alfaraj, S.; AbdelHafez, F.F.; Abdellah, A.H.; Bedaiwy, M.A. Does Preimplantation Genetic Diagnosis Improve Reproductive Outcome in Couples with Recurrent Pregnancy Loss Owing to Structural Chromosomal Rearrangement? A Systematic Review. Reprod. BioMed. Online 2018, 36, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Traversa, M.V.; Carey, L.; Leigh, D. A Molecular Strategy for Routine Preimplantation Genetic Diagnosis in Both Reciprocal and Robertsonian Translocation Carriers. Mol. Hum. Reprod. 2010, 16, 329–337. [Google Scholar] [CrossRef][Green Version]
- Fiorentino, F.; Kokkali, G.; Biricik, A.; Stavrou, D.; Ismailoglu, B.; De Palma, R.; Arizzi, L.; Harton, G.; Sessa, M.; Pantos, K. Polymerase Chain Reaction-Based Detection of Chromosomal Imbalances on Embryos: The Evolution of Preimplantation Genetic Diagnosis for Chromosomal Translocations. Fertil. Steril. 2010, 94, 2001–2011.e6. [Google Scholar] [CrossRef]
- Malmgren, H.; Sahlén, S.; Inzunza, J.; Aho, M.; Rosenlund, B.; Fridström, M.; Hovatta, O.; Ahrlund-Richter, L.; Nordenskjöld, M.; Blennow, E. Single Cell CGH Analysis Reveals a High Degree of Mosaicism in Human Embryos from Patients with Balanced Structural Chromosome Aberrations. Mol. Hum. Reprod. 2002, 8, 502–510. [Google Scholar] [CrossRef] [PubMed]
- Le Caignec, C.; Spits, C.; Sermon, K.; De Rycke, M.; Thienpont, B.; Debrock, S.; Staessen, C.; Moreau, Y.; Fryns, J.-P.; Van Steirteghem, A.; et al. Single-Cell Chromosomal Imbalances Detection by Array CGH. Nucleic Acids Res. 2006, 34, e68. [Google Scholar] [CrossRef] [PubMed]
- Kallioniemi, A.; Kallioniemi, O.P.; Sudar, D.; Rutovitz, D.; Gray, J.W.; Waldman, F.; Pinkel, D. Comparative Genomic Hybridization for Molecular Cytogenetic Analysis of Solid Tumors. Science 1992, 258, 818–821. [Google Scholar] [CrossRef] [PubMed]
- Wilton, L. Preimplantation Genetic Diagnosis and Chromosome Analysis of Blastomeres Using Comparative Genomic Hybridization. Hum. Reprod. Update 2005, 11, 33–41. [Google Scholar] [CrossRef]
- Rius, M.; Obradors, A.; Daina, G.; Ramos, L.; Pujol, A.; Martínez-Passarell, O.; Marquès, L.; Oliver-Bonet, M.; Benet, J.; Navarro, J. Detection of Unbalanced Chromosome Segregations in Preimplantation Genetic Diagnosis of Translocations by Short Comparative Genomic Hibridization. Fertil. Steril. 2011, 96, 134–142. [Google Scholar] [CrossRef]
- Hu, L.; Cheng, D.; Gong, F.; Lu, C.; Tan, Y.; Luo, K.; Wu, X.; He, W.; Xie, P.; Feng, T.; et al. Reciprocal Translocation Carrier Diagnosis in Preimplantation Human Embryos. EBioMedicine 2016, 14, 139–147. [Google Scholar] [CrossRef]
- Fiorentino, F.; Spizzichino, L.; Bono, S.; Biricik, A.; Kokkali, G.; Rienzi, L.; Ubaldi, F.M.; Iammarrone, E.; Gordon, A.; Pantos, K. PGD for Reciprocal and Robertsonian Translocations Using Array Comparative Genomic Hybridization. Hum. Reprod. 2011, 26, 1925–1935. [Google Scholar] [CrossRef]
- Ramos, L.; del Rey, J.; Daina, G.; García-Aragonés, M.; Armengol, L.; Fernandez-Encinas, A.; Parriego, M.; Boada, M.; Martinez-Passarell, O.; Martorell, M.R.; et al. Oligonucleotide Arrays vs. Metaphase-Comparative Genomic Hybridisation and BAC Arrays for Single-Cell Analysis: First Applications to Preimplantation Genetic Diagnosis for Robertsonian Translocation Carriers. PLoS ONE 2014, 9, e113223. [Google Scholar] [CrossRef]
- Christodoulou, C.; Dheedene, A.; Heindryckx, B.; van Nieuwerburgh, F.; Deforce, D.; De Sutter, P.; Menten, B.; Van den Abbeel, E. Preimplantation Genetic Diagnosis for Chromosomal Rearrangements with the Use of Array Comparative Genomic Hybridization at the Blastocyst Stage. Fertil. Steril. 2017, 107, 212–219.e3. [Google Scholar] [CrossRef][Green Version]
- Alfarawati, S.; Fragouli, E.; Colls, P.; Wells, D. First Births after Preimplantation Genetic Diagnosis of Structural Chromosome Abnormalities Using Comparative Genomic Hybridization and Microarray Analysis. Hum. Reprod. 2011, 26, 1560–1574. [Google Scholar] [CrossRef]
- Fodina, V.; Dudorova, A.; Alksere, B.; Dzalbs, A.; Vedmedovska, N.; Andersone, S.; Una, C.; Erenpreiss, J.; Dace, B. The application of PGT-A for carriers of balanced structural chromosomal rearrangements. Gynecol. Endocrinol. 2019, 35 (Suppl. S1), 18–23. [Google Scholar] [CrossRef] [PubMed]
- Handyside, A.H.; Harton, G.L.; Mariani, B.; Thornhill, A.R.; Affara, N.; Shaw, M.-A.; Griffin, D.K. Karyomapping: A Universal Method for Genome Wide Analysis of Genetic Disease Based on Mapping Crossovers between Parental Haplotypes. J. Med. Genet. 2010, 47, 651–658. [Google Scholar] [CrossRef]
- Treff, N.R.; Thompson, K.; Rafizadeh, M.; Chow, M.; Morrison, L.; Tao, X.; Garnsey, H.; Reda, C.V.; Metzgar, T.L.; Neal, S.; et al. SNP Array-Based Analyses of Unbalanced Embryos as a Reference to Distinguish between Balanced Translocation Carrier and Normal Blastocysts. J. Assist. Reprod. Genet. 2016, 33, 1115–1119. [Google Scholar] [CrossRef] [PubMed]
- Sundheimer, L.W.; Liu, L.; Buyalos, R.P.; Hubert, G.; Al-Safi, Z.; Shamonki, M. Diagnosis of Parental Balanced Reciprocal Translocations by Trophectoderm Biopsy and Comprehensive Chromosomal Screening. J. Assist. Reprod. Genet. 2018, 35, 165–169. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Lei, C.; Wu, J.; Zhou, J.; Sun, H.; Fu, J.; Sun, Y.; Sun, X.; Lu, D.; Zhang, Y. The Establishment and Application of Preimplantation Genetic Haplotyping in Embryo Diagnosis for Reciprocal and Robertsonian Translocation Carriers. BMC Med. Genom. 2017, 10, 60. [Google Scholar] [CrossRef] [PubMed]
- Zamani Esteki, M.; Dimitriadou, E.; Mateiu, L.; Melotte, C.; Van der Aa, N.; Kumar, P.; Das, R.; Theunis, K.; Cheng, J.; Legius, E.; et al. Concurrent Whole-Genome Haplotyping and Copy-Number Profiling of Single Cells. Am. J. Hum. Genet. 2015, 96, 894–912. [Google Scholar] [CrossRef] [PubMed]
- Treff, N.R.; Northrop, L.E.; Kasabwala, K.; Su, J.; Levy, B.; Scott, R.T. Single Nucleotide Polymorphism Microarray-Based Concurrent Screening of 24-Chromosome Aneuploidy and Unbalanced Translocations in Preimplantation Human Embryos. Fertil. Steril. 2011, 95, e1–e2. [Google Scholar] [CrossRef]
- Van Uum, C.M.; Stevens, S.J.; Dreesen, J.C.; Drüsedau, M.; Smeets, H.J.; Hollanders-Crombach, B.; de Die-Smulders, C.E.; Geraedts, J.P.; Engelen, J.J.; Coonen, E. SNP Array-Based Copy Number and Genotype Analyses for Preimplantation Genetic Diagnosis of Human Unbalanced Translocations. Eur. J. Hum. Genet. 2012, 20, 938–944. [Google Scholar] [CrossRef][Green Version]
- Tan, Y.-Q.; Tan, K.; Zhang, S.-P.; Gong, F.; Cheng, D.-H.; Xiong, B.; Lu, C.-F.; Tang, X.-C.; Luo, K.-L.; Lin, G.; et al. Single-Nucleotide Polymorphism Microarray-Based Preimplantation Genetic Diagnosis Is Likely to Improve the Clinical Outcome for Translocation Carriers. Hum. Reprod. 2013, 28, 2581–2592. [Google Scholar] [CrossRef]
- Tobler, K.J.; Brezina, P.R.; Benner, A.T.; Du, L.; Xu, X.; Kearns, W.G. Two Different Microarray Technologies for Preimplantation Genetic Diagnosis and Screening, Due to Reciprocal Translocation Imbalances, Demonstrate Equivalent Euploidy and Clinical Pregnancy Rates. J. Assist. Reprod. Genet. 2014, 31, 843–850. [Google Scholar] [CrossRef]
- Xiong, B.; Tan, K.; Tan, Y.Q.; Gong, F.; Zhang, S.P.; Lu, C.F.; Luo, K.L.; Lu, G.X.; Lin, G. Using SNP Array to Identify Aneuploidy and Segmental Imbalance in Translocation Carriers. Genom. Data 2014, 2, 92–95. [Google Scholar] [CrossRef] [PubMed]
- Idowu, D.; Merrion, K.; Wemmer, N.; Mash, J.G.; Pettersen, B.; Kijacic, D.; Lathi, R.B. Pregnancy Outcomes Following 24-Chromosome Preimplantation Genetic Diagnosis in Couples with Balanced Reciprocal or Robertsonian Translocations. Fertil. Steril. 2015, 103, 1037–1042. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ding, C.; Wang, J.; Zeng, Y.; Zhou, W.; Li, R.; Zhou, C.; Deng, M.-F.; Xu, Y. Number of Blastocysts Biopsied as a Predictive Indicator to Obtain at Least One Normal/Balanced Embryo Following Preimplantation Genetic Diagnosis with Single Nucleotide Polymorphism Microarray in Translocation Cases. J. Assist. Reprod. Genet. 2017, 34, 51–59. [Google Scholar] [CrossRef][Green Version]
- Zhang, S.; Zhao, D.; Zhang, J.; Mao, Y.; Kong, L.; Zhang, Y.; Liang, B.; Sun, X.; Xu, C. BasePhasing: A highly efficient approach for preimplantation genetic haplotyping in clinical application of balanced translocation carriers. BMC Med. Genom. 2019, 12, 52. [Google Scholar] [CrossRef] [PubMed]
- Beyer, C.E.; Willats, E. Natural Selection between Day 3 and Day 5/6 PGD Embryos in Couples with Reciprocal or Robertsonian Translocations. J. Assist. Reprod. Genet. 2017, 34, 1483–1492. [Google Scholar] [CrossRef]
- Wells, D.; Kaur, K.; Grifo, J.; Glassner, M.; Taylor, J.C.; Fragouli, E.; Munne, S. Clinical Utilisation of a Rapid Low-Pass Whole Genome Sequencing Technique for the Diagnosis of Aneuploidy in Human Embryos Prior to Implantation. J. Med. Genet. 2014, 51, 553–562. [Google Scholar] [CrossRef]
- Maxwell, S.M.; Colls, P.; Hodes-Wertz, B.; McCulloh, D.H.; McCaffrey, C.; Wells, D.; Munné, S.; Grifo, J.A. Why Do Euploid Embryos Miscarry? A Case-Control Study Comparing the Rate of Aneuploidy within Presumed Euploid Embryos That Resulted in Miscarriage or Live Birth Using next-Generation Sequencing. Fertil. Steril. 2016, 106, 1414–1419.e5. [Google Scholar] [CrossRef]
- Biricik, A.; Cotroneo, E.; Minasi, M.G.; Greco, P.F.; Bono, S.; Surdo, M.; Lecciso, F.; Sessa, M.; Fiorentino, F.; Spinella, F.; et al. Cross-Validation of Next-Generation Sequencing Technologies for Diagnosis of Chromosomal Mosaicism and Segmental Aneuploidies in Preimplantation Embryos Model. Life 2021, 11, 340. [Google Scholar] [CrossRef]
- Cuman, C.; Beyer, C.E.; Brodie, D.; Fullston, T.; Lin, J.I.; Willats, E.; Zander-Fox, D.; Mullen, J. Defining the Limits of Detection for Chromosome Rearrangements in the Preimplantation Embryo Using next Generation Sequencing. Hum. Reprod. 2018, 33, 1566–1576. [Google Scholar] [CrossRef]
- Yin, X.; Tan, K.; Vajta, G.; Jiang, H.; Tan, Y.; Zhang, C.; Chen, F.; Chen, S.; Zhang, C.; Pan, X.; et al. Massively Parallel Sequencing for Chromosomal Abnormality Testing in Trophectoderm Cells of Human Blastocysts. Biol. Reprod. 2013, 88, 69. [Google Scholar] [CrossRef]
- Tan, Y.; Yin, X.; Zhang, S.; Jiang, H.; Tan, K.; Li, J.; Xiong, B.; Gong, F.; Zhang, C.; Pan, X.; et al. Clinical Outcome of Preimplantation Genetic Diagnosis and Screening Using next Generation Sequencing. Gigascience 2014, 3, 30. [Google Scholar] [CrossRef] [PubMed]
- Gui, B.; Yao, Z.; Li, Y.; Liu, D.; Liu, N.; Xia, Y.; Huang, Y.; Mei, L.; Ma, R.; Lu, S.; et al. Chromosomal Analysis of Blastocysts from Balanced Chromosomal Rearrangement Carriers. Reproduction 2016, 151, 455–464. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chow, J.F.C.; Yeung, W.S.B.; Lee, V.C.Y.; Lau, E.Y.L.; Ng, E.H.Y. Evaluation of Preimplantation Genetic Testing for Chromosomal Structural Rearrangement by a Commonly Used next Generation Sequencing Workflow. Eur. J. Obs. Gynecol. Reprod. Biol. 2018, 224, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Shen, J.; Cram, D.S.; Ma, M.; Wang, H.; Zhang, W.; Fan, J.; Gao, Z.; Zhang, L.; Li, Z.; et al. Preferential Selection and Transfer of Euploid Noncarrier Embryos in Preimplantation Genetic Diagnosis Cycles for Reciprocal Translocations. Fertil. Steril. 2017, 108, 620–627.e4. [Google Scholar] [CrossRef] [PubMed]
- Nakano, T.; Ammae, M.; Satoh, M.; Mizuno, S.; Nakaoka, Y.; Morimoto, Y. Analysis of Clinical Outcomes and Meiotic Segregation Modes Following Preimplantation Genetic Testing for Structural Rearrangements Using aCGH/NGS in Couples with Balanced Chromosome Rearrangement. Reprod Med. Biol. 2022, 21, e12476. [Google Scholar] [CrossRef]
- Chen, S.; Yin, X.; Zhang, S.; Xia, J.; Liu, P.; Xie, P.; Yan, H.; Liang, X.; Zhang, J.; Chen, Y.; et al. Comprehensive Preimplantation Genetic Testing by Massively Parallel Sequencing. Hum. Reprod. 2021, 36, 236–247. [Google Scholar] [CrossRef]
- Boynukalin, F.K.; Gultomruk, M.; Turgut, N.E.; Rubio, C.; Rodrigo, L.; Yarkiner, Z.; Ecemis, S.; Karlikaya, G.; Findikli, N.; Bahceci, M. The Impact of Patient, Embryo, and Translocation Characteristics on the Ploidy Status of Young Couples Undergoing Preimplantation Genetic Testing for Structural Rearrangements (PGT-SR) by next Generation Sequencing (NGS). J Assist Reprod. Genet. 2021, 38, 387–396. [Google Scholar] [CrossRef]
- Walters-Sen, L.; Neitzel, D.; Wilkinson, J.; Poll, S.; Faulkner, N.; Aradhya, S. EP472: Deriving Risk Estimates for Balanced Rearrangement Carriers Utilizing PGT-SR Data. Genet. Med. 2022, 24, S299–S300. [Google Scholar] [CrossRef]
- Yuan, P.; Zheng, L.; Ou, S.; Zhao, H.; Li, R.; Luo, H.; Tan, X.; Zhang, Q.; Wang, W. Evaluation of Chromosomal Abnormalities from Preimplantation Genetic Testing to the Reproductive Outcomes: A Comparison between Three Different Structural Rearrangements Based on next-Generation Sequencing. J. Assist. Reprod. Genet. 2021, 38, 709–718. [Google Scholar] [CrossRef]
- Tong, J.; Jiang, J.; Niu, Y.; Zhang, T. Do Chromosomal Inversion Carriers Really Need Preimplantation Genetic Testing? J. Assist. Reprod. Genet. 2022, 39, 2573–2579. [Google Scholar] [CrossRef]
- Zheng, W.; Ren, B.; Mu, M.; Liu, Y.; Liu, X.; Yang, C.; Yang, S.; Yang, R.; Li, J.; Zu, R.; et al. Perinatal Outcomes of Singleton Live Births Following Preimplantation Genetic Testing for Chromosomal Structural Rearrangements in Single Frozen-Thawed Blastocyst Transfer Cycles: A Retrospective Cohort Study. Reprod. Sci. 2022, 29, 3039–3046. [Google Scholar] [CrossRef] [PubMed]
- Zeevi, D.A.; Backenroth, D.; Hakam-Spector, E.; Renbaum, P.; Mann, T.; Zahdeh, F.; Segel, R.; Zeligson, S.; Eldar-Geva, T.; Ben-Ami, I.; et al. Expanded clinical validation of Haploseek for comprehensive preimplantation genetic testing. Genet. Med. Off. J. Am. Coll. Med. Genet. 2021, 23, 1334–1340. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Ullmann, R.; Langnick, C.; Menzel, C.; Wotschofsky, Z.; Hu, H.; Döring, A.; Hu, Y.; Kang, H.; Tzschach, A.; et al. Breakpoint Analysis of Balanced Chromosome Rearrangements by Next-Generation Paired-End Sequencing. Eur. J. Hum. Genet. 2010, 18, 539–543. [Google Scholar] [CrossRef]
- Zhai, F.; Wang, Y.; Li, H.; Wang, Y.; Zhu, X.; Kuo, Y.; Guan, S.; Li, J.; Song, S.; He, Q.; et al. Preimplantation Genetic Testing for Structural Rearrangement Based on Low-Coverage next-Generation Sequencing Accurately Discriminates between Normal and Carrier Embryos for Patients with Translocations. Reprod. BioMed. Online 2022, 45, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Yin, B.; Zhu, Y.; Li, G.; Ye, L.; Liang, D.; Zeng, Y. First Report on an X-Linked Hypohidrotic Ectodermal Dysplasia Family with X Chromosome Inversion: Breakpoint Mapping Reveals the Pathogenic Mechanism and Preimplantation Genetics Diagnosis Achieves an Unaffected Birth. Clin. Chim. Acta 2017, 475, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Findikli, N.; Kahraman, S.; Kumtepe, Y.; Donmez, E.; Benkhalifa, M.; Biricik, A.; Sertyel, S.; Berkil, H.; Oncu, N. Assessment of DNA Fragmentation and Aneuploidy on Poor Quality Human Embryos. Reprod. Biomed. Online 2004, 8, 196–206. [Google Scholar] [CrossRef]
- Evsikov, S.; Cieslak, J.; Verlinsky, Y. Effect of Chromosomal Translocations on the Development of Preimplantation Human Embryos in Vitro. Fertil. Steril. 2000, 74, 672–677. [Google Scholar] [CrossRef]
- Chang, E.M.; Han, J.E.; Kwak, I.P.; Lee, W.S.; Yoon, T.K.; Shim, S.H. Preimplantation Genetic Diagnosis for Couples with a Robertsonian Translocation: Practical Information for Genetic Counseling. J. Assist. Reprod. Genet. 2012, 29, 67–75. [Google Scholar] [CrossRef]
- Findikli, N.; Kahraman, S.; Kumtepe, Y.; Donmez, E.; Biricik, A.; Sertyel, S.; Berkil, H.; Melil, S. Embryo Development Characteristics in Robertsonian and Reciprocal Translocations: A Comparison of Results with Non-Translocation Cases. Reprod. BioMed. Online 2003, 7, 563–571. [Google Scholar] [CrossRef]
- Campbell, A.; Fishel, S.; Bowman, N.; Duffy, S.; Sedler, M.; Hickman, C.F.L. Modelling a Risk Classification of Aneuploidy in Human Embryos Using Non-Invasive Morphokinetics. Reprod. BioMed. Online 2013, 26, 477–485. [Google Scholar] [CrossRef]
- Meseguer, M.; Herrero, J.; Tejera, A.; Hilligsoe, K.M.; Ramsing, N.B.; Remohi, J. The Use of Morphokinetics as a Predictor of Embryo Implantation. Hum. Reprod. 2011, 26, 2658–2671. [Google Scholar] [CrossRef] [PubMed]
- Amir, H.; Barbash-Hazan, S.; Kalma, Y.; Frumkin, T.; Malcov, M.; Samara, N.; Hasson, J.; Reches, A.; Azem, F.; Ben-Yosef, D. Time-Lapse Imaging Reveals Delayed Development of Embryos Carrying Unbalanced Chromosomal Translocations. J. Assist. Reprod. Genet. 2019, 36, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Lammers, J.; Reignier, A.; Splingart, C.; Moradkhani, K.; Barrière, P.; Fréour, T. Morphokinetic Parameters in Chromosomal Translocation Carriers Undergoing Preimplantation Genetic Testing. Reprod. BioMed. Online 2019, 38, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Insogna, I.G.; Lanes, A.; Dobson, L.; Ginsburg, E.S.; Racowsky, C.; Yanushpolsky, E. Blastocyst. Conversion Rate and Ploidy in Patients with Structural Rearrangements. J. Assist. Reprod. Genet. 2021, 38, 1143–1151. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.H.; Escudero, T.; Cekleniak, N.A.; Sable, D.B.; Garrisi, M.G.; Munne, S. Patterns of Ovarian Response to Gonadotropin Stimulation in Female Carriers of Balanced Translocation. Fertil. Steril. 2005, 83, 1504–1509. [Google Scholar] [CrossRef]
- Dechanet, C.; Castelli, C.; Reyftmann, L.; Hamamah, S.; Hedon, B.; Dechaud, H.; Anahory, T. Do Female Translocations Influence the Ovarian Response Pattern to Controlled Ovarian Stimulation in Preimplantation Genetic Diagnosis? Hum. Reprod. 2011, 26, 1232–1240. [Google Scholar] [CrossRef]
- Lledo, B.; Ortiz, J.A.; Morales, R.; Ten, J.; de la Fuente, P.E.; Garcia-Ochoa, C.; Bernabeu, R. The Paternal Effect of Chromosome Translocation Carriers Observed from Meiotic Segregation in Embryos. Hum. Reprod. 2010, 25, 1843–1848. [Google Scholar] [CrossRef]
- Ye, Y.; Qian, Y.; Xu, C.; Jin, F. Meiotic Segregation Analysis of Embryos from Reciprocal Translocation Carriers in PGD Cycles. Reprod. BioMed. Online 2012, 24, 83–90. [Google Scholar] [CrossRef]
- Zhang, L.; Jiang, W.; Zhu, Y.; Chen, H.; Yan, J.; Chen, Z.-J. Effects of a Carrier’s Sex and Age on the Segregation Patterns of the Trivalent of Robertsonian Translocations. J. Assist. Reprod. Genet. 2019, 36, 1963–1969. [Google Scholar] [CrossRef]
- Song, H.; Shi, H.; Yang, E.-T.; Bu, Z.-Q.; Jin, Z.-Q.; Huo, M.-Z.; Zhang, Y. Effects of Gender of Reciprocal Chromosomal Translocation on Blastocyst Formation and Pregnancy Outcome in Preimplantation Genetic Testing. Front. Endocrinol. 2021, 12, 704299. [Google Scholar] [CrossRef]
- LeMaire-Adkins, R.; Radke, K.; Hunt, P.A. Lack of Checkpoint Control at the Metaphase/Anaphase Transition: A Mechanism of Meiotic Nondisjunction in Mammalian Females. J. Cell Biol. 1997, 139, 1611–1619. [Google Scholar] [CrossRef]
- Xie, P.; Hu, L.; Peng, Y.; Tan, Y.-Q.; Luo, K.; Gong, F.; Lu, G.; Lin, G. Risk Factors Affecting Alternate Segregation in Blastocysts From Preimplantation Genetic Testing Cycles of Autosomal Reciprocal Translocations. Front. Genet. 2022, 13, 880208. [Google Scholar] [CrossRef] [PubMed]
- Benkhalifa, M.; Kasakyan, S.; Clement, P.; Baldi, M.; Tachdjian, G.; Demirol, A.; Gurgan, T.; Fiorentino, F.; Mohammed, M.; Qumsiyeh, M.B. Array comparative genomic hybridization profiling of first-trimester spontaneous abortions that fail to grow in vitro. Prenat. Diagn. 2005, 25, 894–900. [Google Scholar] [CrossRef] [PubMed]
- Franasiak, J.M.; Forman, E.J.; Hong, K.H.; Werner, M.D.; Upham, K.M.; Treff, N.R.; Scott, R.T., Jr. The nature of aneuploidy with increasing age of the female partner: A review of 15,169 consecutive trophectoderm biopsies evaluated with comprehensive chromosomal screening. Fertil. Steril. 2014, 101, 656–663.e1. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Diez, C.; FitzHarris, G. Causes and consequences of chromosome segregation error in preimplantation embryos. Reproduction 2018, 155, R63–R76. [Google Scholar] [CrossRef]
- Taylor, T.H.; Gitlin, S.A.; Patrick, J.L.; Crain, J.L.; Wilson, J.M.; Griffin, D.K. The origin, mechanisms, incidence and clinical consequences of chromosomal mosaicism in humans. Hum. Reprod. Update 2014, 20, 571–581. [Google Scholar] [CrossRef]
- Coonen, E.; Derhaag, J.G.; Dumoulin, J.C.; van Wissen, L.C.; Bras, M.; Janssen, M.; Evers, J.L.; Geraedts, J.P. Anaphase lagging mainly explains chromosomal mosaicism in human preimplantation embryos. Hum. Reprod. 2004, 19, 316–324. [Google Scholar] [CrossRef] [PubMed]
- Lindenbaum, R.H.; Hultén, M.; McDermott, A.; Seabright, M. The Prevalence of Translocations in Parents of Children with Regular Trisomy 21: A Possible Interchromosomal Effect? J. Med. Genet. 1985, 22, 24–28. [Google Scholar] [CrossRef] [PubMed]
- Navarro, J.; Vidal, F.; Benet, J.; Templado, C.; Marina, S.; Egozcue, J. XY-Trivalent Association and Synaptic Anomalies in a Male Carrier of a Robertsonian t(13;14) Translocation. Hum. Reprod. 1991, 6, 376–381. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.H.; Spriggs, E.L. Sperm Chromosome Complements in a Man Heterozygous for a Reciprocal Translocation 46,XY,t(9;13)(Q21.1;Q21.2) and a Review of the Literature. Clin. Genet. 1995, 47, 42–46. [Google Scholar] [CrossRef] [PubMed]
- Amiel, A.; Sardos-Albertini, F.; Fejgin, M.D.; Sharony, R.; Diukman, R.; Bartoov, B. Interchromosomal Effect Leading to an Increase in Aneuploidy in Sperm Nuclei in a Man Heterozygous for Pericentric Inversion (Inv 9) and C-Heterochromatin. J. Hum. Genet. 2001, 46, 245–250. [Google Scholar] [CrossRef]
- Machev, N.; Gosset, P.; Warter, S.; Treger, M.; Schillinger, M.; Viville, S. Fluorescence in Situ Hybridization Sperm Analysis of Six Translocation Carriers Provides Evidence of an Interchromosomal Effect. Fertil. Steril. 2005, 84, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Godo, A.; Blanco, J.; Vidal, F.; Anton, E. Accumulation of Numerical and Structural Chromosome Imbalances in Spermatozoa from Reciprocal Translocation Carriers. Hum. Reprod. 2013, 28, 840–849. [Google Scholar] [CrossRef] [PubMed]
- Scriven, P.N.; Flinter, F.A.; Braude, P.R.; Ogilvie, C.M. Robertsonian Translocations--Reproductive Risks and Indications for Preimplantation Genetic Diagnosis. Hum. Reprod. 2001, 16, 2267–2273. [Google Scholar] [CrossRef] [PubMed]
- Ferfouri, F.; Boitrelle, F.; Clément, P.; Molina Gomes, D.; Selva, J.; Vialard, F. Can One Translocation Impact the Meiotic Segregation of Another Translocation? A Sperm-FISH Analysis of a 46,XY,t(1;16)(Q21;P11.2),t(8;9) (Q24.3;P24) Patient and His 46,XY,t(8;9)(Q24.3;P24) Brother and Cousin. MHR: Basic Sci. Reprod. Med. 2013, 19, 109–117. [Google Scholar] [CrossRef][Green Version]
- Young, D.; Klepacka, D.; McGarvey, M.; Schoolcraft, W.B.; Katz-Jaffe, M.G. Infertility Patients with Chromosome Inversions Are Not Susceptible to an Inter-Chromosomal Effect. J. Assist. Reprod. Genet. 2019, 36, 509–516. [Google Scholar] [CrossRef]
- Vanneste, E.; Voet, T.; Melotte, C.; Debrock, S.; Sermon, K.; Staessen, C.; Liebaers, I.; Fryns, J.-P.; D’Hooghe, T.; Vermeesch, J.R. What next for Preimplantation Genetic Screening? High Mitotic Chromosome Instability Rate Provides the Biological Basis for the Low Success Rate. Hum. Reprod. 2009, 24, 2679–2682. [Google Scholar] [CrossRef]
- Alfarawati, S.; Fragouli, E.; Colls, P.; Wells, D. Embryos of Robertsonian Translocation Carriers Exhibit a Mitotic Interchromosomal Effect That Enhances Genetic Instability during Early Development. PLoS Genet. 2012, 8, e1003025. [Google Scholar] [CrossRef]
- Lynch, C.A.; Gordon, T.; Griffin, D.K. Retrospective analysis of 479 Pgt-Sr cycles—Analysis of chromosome information and availability of embryos for transfer. Fertil. Steril. 2021, 116, e397. [Google Scholar] [CrossRef]
- Lynch, C.A.; Whiting, O.; Sloan, J.; Cameron, E.; Xanthopoulou, L.; Gordon, T.; Griffin, D.K. The importance of retrospective data analysis in genetic counselling for Pgt-sr—maternal age is more significant than rearrangement type. Fertil. Steril. 2022, 118, e97. [Google Scholar] [CrossRef]
- Ogur, C.; Kahraman, S.; Griffin, D.K.; Cinar, C.; Tufekci, M.A.; Cetinkaya, M.; Temel, S.G.; Yilmaz, A. Preimplantation Genetic Testing for Structural Rearrangements (PGT-SR) in 300 Couples Reveals Individual Specific Risk Factors but an Inter Chromosomal Effect (ICE) Is Unlikely. Reprod. BioMed. Online 2022, S1472648322005223. [Google Scholar] [CrossRef]
- Scriven, P.N. PGT-SR: The Red-Herring and the Siren; Interchromosomal Effect and Screening for Unrelated Aneuploidy. J. Assist. Reprod. Genet. 2021, 38, 1015–1018. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Griffin, D.K.; Ogur, C. PGT-SR: A Comprehensive Overview and a Requiem for the Interchromosomal Effect. DNA 2023, 3, 41-64. https://doi.org/10.3390/dna3010004
Griffin DK, Ogur C. PGT-SR: A Comprehensive Overview and a Requiem for the Interchromosomal Effect. DNA. 2023; 3(1):41-64. https://doi.org/10.3390/dna3010004
Chicago/Turabian StyleGriffin, Darren K., and Cagri Ogur. 2023. "PGT-SR: A Comprehensive Overview and a Requiem for the Interchromosomal Effect" DNA 3, no. 1: 41-64. https://doi.org/10.3390/dna3010004
APA StyleGriffin, D. K., & Ogur, C. (2023). PGT-SR: A Comprehensive Overview and a Requiem for the Interchromosomal Effect. DNA, 3(1), 41-64. https://doi.org/10.3390/dna3010004