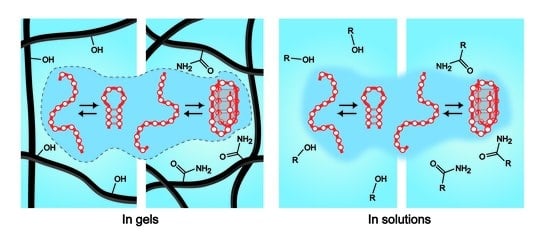
Evaluation of Thermal Stability of DNA Oligonucleotide Structures Embedded in Hydrogels
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of Buffer Solutions and Oligonucleotides
2.2. Preparation of Agarose Gels and Measurement of Their Phase Transition Temperatures
2.3. Measurement of the Stability of Oligonucleotide Structures in Agarose Gels
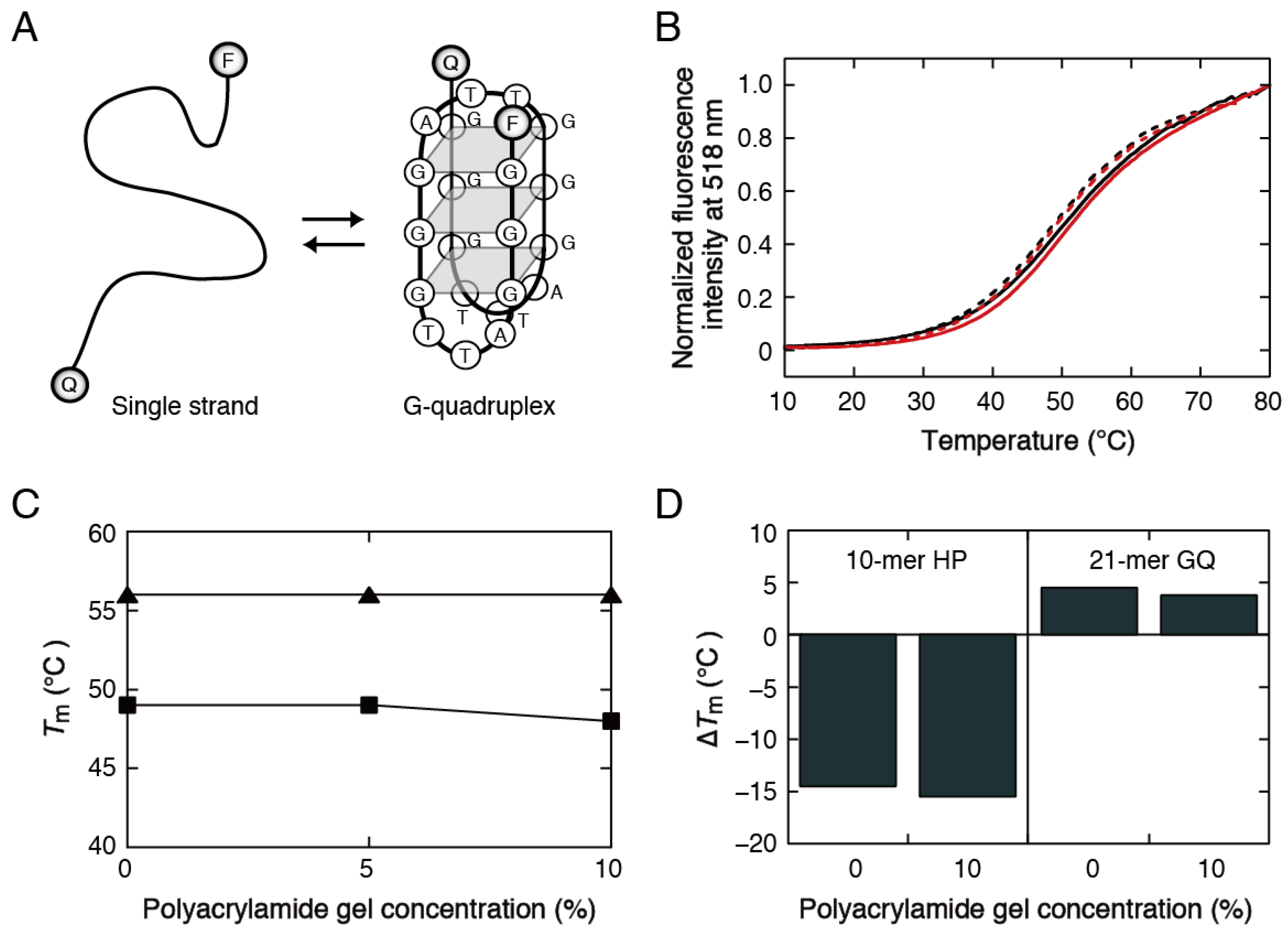
2.4. Preparation of Polyacrylamide Gels and Measurement of the Stability of Oligonucleotide Structures in the Gels
3. Results
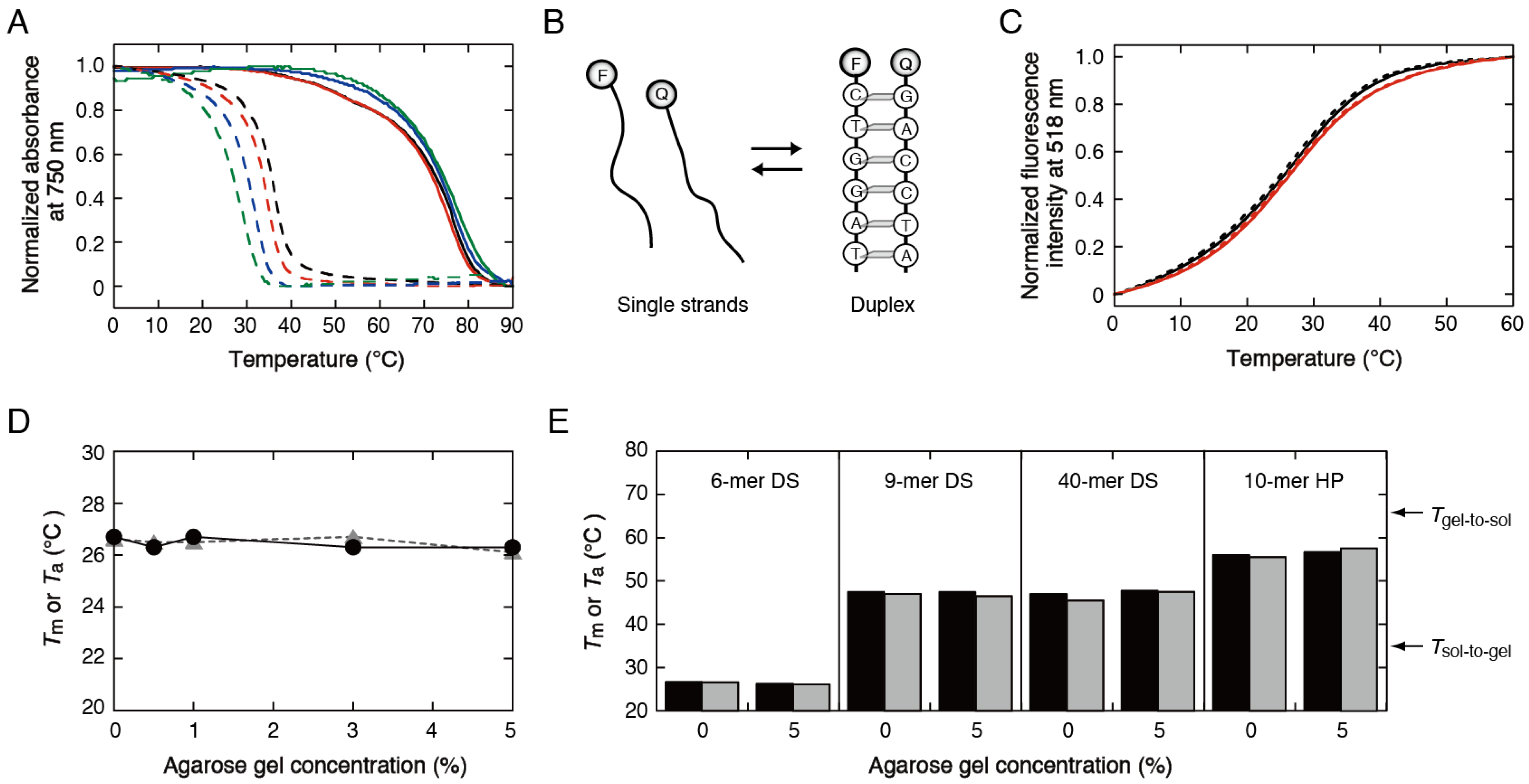
3.1. Stability Analysis of DNA Structures in Agarose Gels
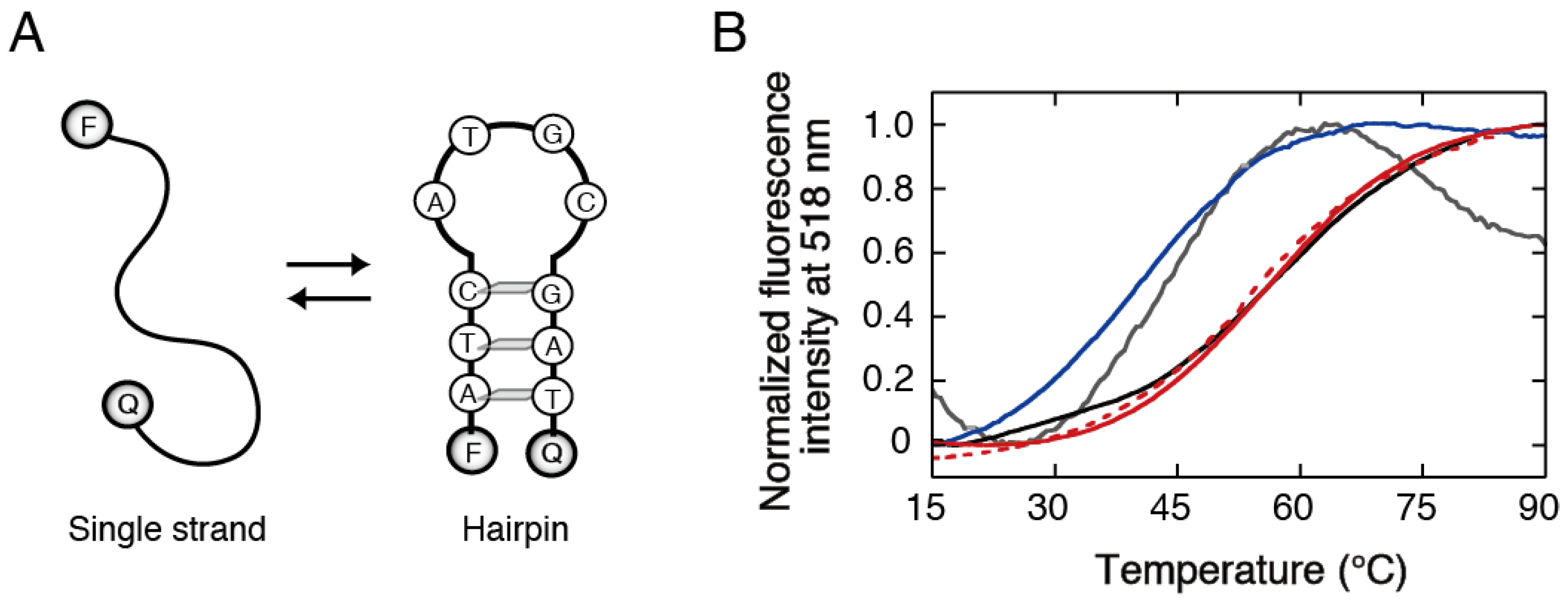
3.2. Stability Analysis of DNA Structures in Polyacrylamide Gels
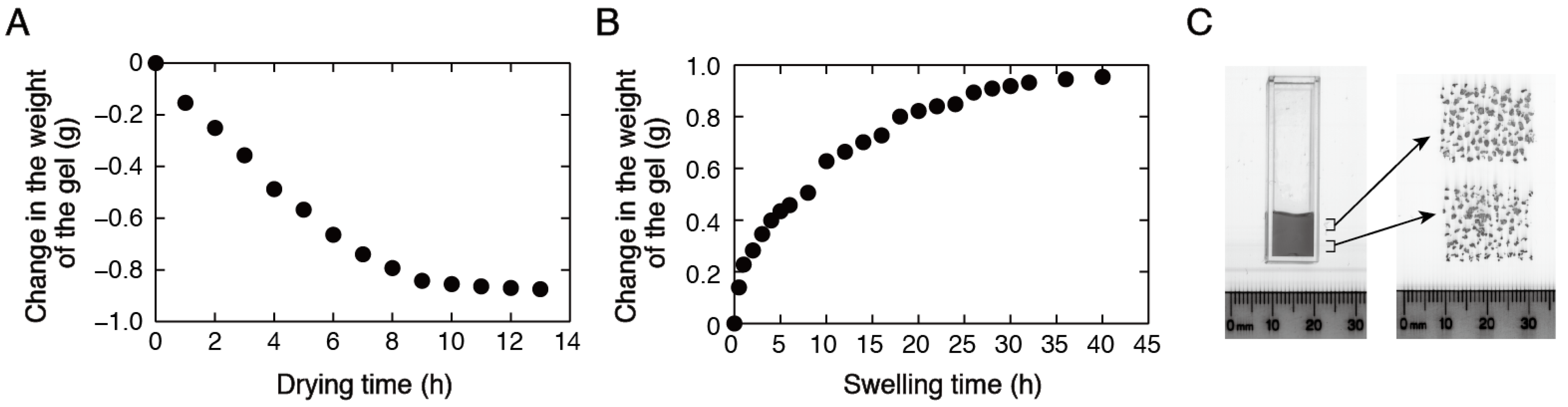
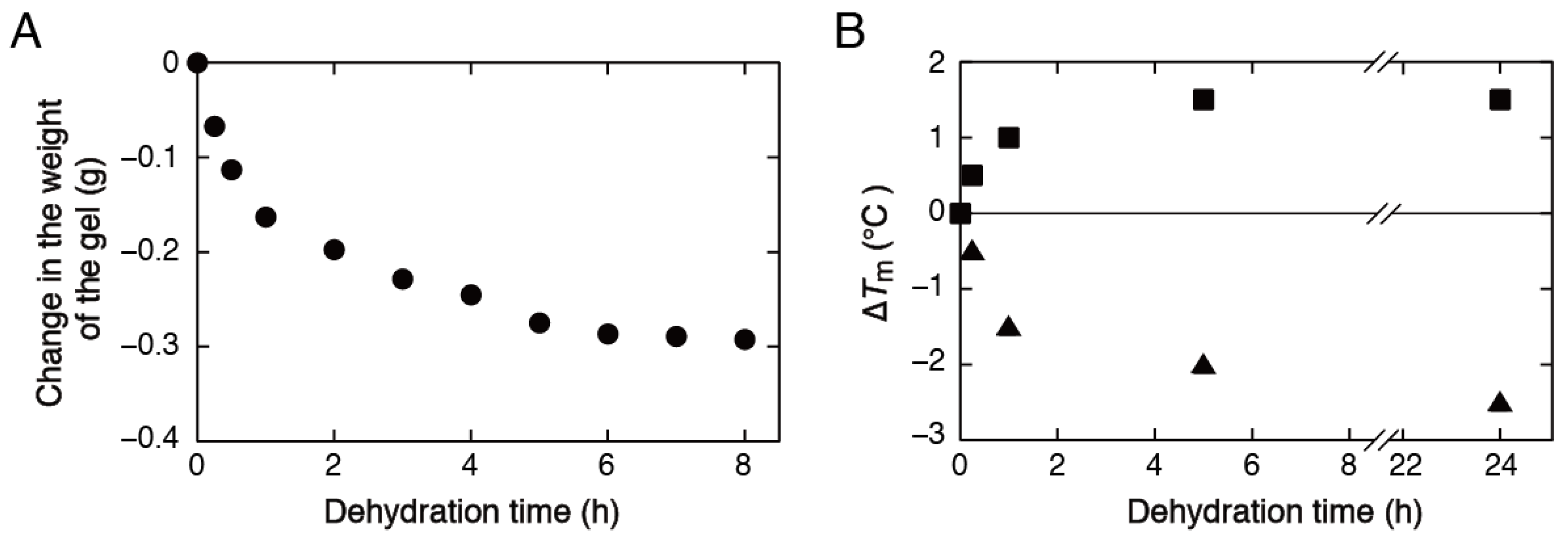
3.3. Stability Analysis of DNA Structures in Low-Water-Content Gels
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Turner, D.H. Conformational Changes. In Nucleic Acids: Structures, Properties and Functions; Bloomfield, V.A., Crothers, D.M., Tinoco, J.I., Eds.; University Science Books Press: Sausalito, CA, USA, 2000; pp. 259–334. [Google Scholar]
- Zhou, H.X.; Rivas, G.; Minton, A.P. Macromolecular crowding and confinement: Biochemical, biophysical, and potential physiological consequences. Annu. Rev. Biophys. 2008, 37, 375–397. [Google Scholar] [CrossRef] [PubMed]
- Mika, J.T.; Poolman, B. Macromolecule diffusion and confinement in prokaryotic cells. Curr. Opin. Biotechnol. 2011, 22, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Castronovo, M.; Stopar, A.; Coral, L.; Redhu, S.K.; Vidonis, M.; Kumar, V.; Ben, F.D.; Grassi, M.; Nicholson, A.W. Effects of nanoscale confinement on the functionality of nucleic acids: Implications for nanomedicine. Curr. Med. Chem. 2013, 20, 3539–3557. [Google Scholar] [CrossRef] [PubMed]
- Elder, R.M.; Pfaendtner, J.; Jayaraman, A. Effect of hydrophobic and hydrophilic surfaces on the stability of double-stranded DNA. Biomacromolecules 2015, 16, 1862–1869. [Google Scholar] [CrossRef]
- Gao, M.; Gnutt, D.; Orban, A.; Appel, B.; Righetti, F.; Winter, R.; Narberhaus, F.; Muller, S.; Ebbinghaus, S. RNA hairpin folding in the crowded cell. Angew. Chem. Int. Ed. Engl. 2016, 55, 3224–3228. [Google Scholar] [CrossRef] [PubMed]
- Ganser, L.R.; Kelly, M.L.; Herschlag, D.; Al-Hashimi, H.M. The roles of structural dynamics in the cellular functions of RNAs. Nat. Rev. Mol. Cell. Biol. 2019, 20, 474–489. [Google Scholar] [CrossRef] [PubMed]
- Nakano, S.; Miyoshi, D.; Sugimoto, N. Effects of molecular crowding on the structures, interactions, and functions of nucleic acids. Chem. Rev. 2014, 114, 2733–2758. [Google Scholar] [CrossRef]
- Riback, J.A.; Katanski, C.D.; Kear-Scott, J.L.; Pilipenko, E.V.; Rojek, A.E.; Sosnick, T.R.; Drummond, D.A. Stress-triggered phase separation Is an adaptive, evolutionarily tuned response. Cell 2017, 168, 1028–1040.e19. [Google Scholar] [CrossRef] [PubMed]
- Jun, S.; Mulder, B. Entropy-driven spatial organization of highly confined polymers: Lessons for the bacterial chromosome. Proc. Natl. Acad. Sci. USA 2006, 103, 12388–12393. [Google Scholar] [CrossRef]
- Buenemann, M.; Lenz, P. A geometrical model for DNA organization in bacteria. PLoS ONE 2010, 5, e13806. [Google Scholar] [CrossRef]
- Yanagisawa, M.; Sakaue, T.; Yoshikawa, K. Characteristic behavior of crowding macromolecules confined in cell-sized droplets. Int. Rev. Cell Mol. Biol. 2014, 307, 175–204. [Google Scholar]
- Gursoy, G.; Xu, Y.; Liang, J. Spatial organization of the budding yeast genome in the cell nucleus and identification of specific chromatin interactions from multi-chromosome constrained chromatin model. PLoS Comput. Biol. 2017, 13, e1005658. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.C.; Hsu, C.Y.; Chen, J.Y.; Fang, Z.S.; Chen, H.W.; Yao, B.Y.; Shiau, G.H.M.; Tsai, J.S.; Gu, M.; Jung, M.; et al. Facile transformation of murine and human primary dendritic cells into robust and modular artificial antigen-presenting systems by intracellular hydrogelation. Adv. Mater. 2021, 33, e2101190. [Google Scholar] [CrossRef] [PubMed]
- Macdougall, L.J.; Hoffman, T.E.; Kirkpatrick, B.E.; Fairbanks, B.D.; Bowman, C.N.; Spencer, S.L.; Anseth, K.S. Intracellular crowding by bio-orthogonal hydrogel formation induces reversible molecular stasis. Adv. Mater. 2022, 34, e2202882. [Google Scholar] [CrossRef] [PubMed]
- Reisner, W.; Pedersen, J.N.; Austin, R.H. DNA confinement in nanochannels: Physics and biological applications. Rep. Prog. Phys. 2012, 75, 106601. [Google Scholar] [CrossRef]
- Hoffman, A.S. Hydrogels for biomedical applications. Adv. Drug Deliv. Rev. 2002, 54, 3–12. [Google Scholar] [CrossRef]
- Morya, V.; Walia, S.; Mandal, B.B.; Ghoroi, C.; Bhatia, D. Functional DNA based hydrogels: Development, properties and biological applications. ACS Biomater. Sci. Eng. 2020, 6, 6021–6035. [Google Scholar] [CrossRef]
- Zhu, Y.; Granick, S. Viscosity of interfacial water. Phys. Rev. Lett. 2001, 87, 096104. [Google Scholar] [CrossRef]
- Raviv, U.; Laurat, P.; Klein, J. Fluidity of water confined to subnanometre films. Nature 2001, 413, 51–54. [Google Scholar] [CrossRef]
- Perez-Hernandez, N.; Luong, T.Q.; Perez, C.; Martin, J.D.; Havenith, M. Pore size dependent dynamics of confined water probed by FIR spectroscopy. Phys. Chem. Chem. Phys. 2010, 12, 6928–6932. [Google Scholar] [CrossRef]
- Zhou, H.X.; Dill, K.A. Stabilization of proteins in confined spaces. Biochemistry 2001, 40, 11289–11293. [Google Scholar] [CrossRef] [PubMed]
- Mittal, J.; Best, R.B. Thermodynamics and kinetics of protein folding under confinement. Proc. Natl. Acad. Sci. USA 2008, 105, 20233–20238. [Google Scholar] [CrossRef] [PubMed]
- Eggers, D.K.; Valentine, J.S. Crowding and hydration effects on protein conformation: A study with sol-gel encapsulated proteins. J. Mol. Biol. 2001, 314, 911–922. [Google Scholar] [PubMed]
- Cheung, M.S.; Thirumalai, D. Nanopore-protein interactions dramatically alter stability and yield of the native state in restricted spaces. J. Mol. Biol. 2006, 357, 632–643. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.J.; Chen, S.J. Ion-mediated RNA structural collapse: Effect of spatial confinement. Biophys. J. 2012, 103, 827–836. [Google Scholar] [CrossRef][Green Version]
- Feng, C.; Tan, Y.L.; Cheng, Y.X.; Shi, Y.Z.; Tan, Z.J. Salt-dependent RNA pseudoknot stability: Effect of spatial confinement. Front. Mol. Biosci. 2021, 8, 666369. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, Z.; Li, N.; He, X.; Liang, H. Denaturation and renaturation behaviors of short DNA in a confined space. J. Chem. Phys. 2014, 141, 044911. [Google Scholar] [CrossRef]
- Reiter-Schad, M.; Werner, E.; Tegenfeldt, J.O.; Mehlig, B.; Ambjornsson, T. How nanochannel confinement affects the DNA melting transition within the Poland-Scheraga model. J. Chem. Phys. 2015, 143, 115101. [Google Scholar] [CrossRef]
- Maity, A.; Singh, N. Melting of DNA in confined geometries. Eur. Biophys. J. 2020, 49, 561–569. [Google Scholar] [CrossRef]
- Pal, S.; Paul, S. Theoretical investigation of conformational deviation of the human parallel telomeric G-quadruplex DNA in the presence of different salt concentrations and temperatures under confinement. Phys. Chem. Chem. Phys. 2021, 23, 14372–14382. [Google Scholar] [CrossRef]
- Chen, I.A.; Salehi-Ashtiani, K.; Szostak, J.W. RNA catalysis in model protocell vesicles. J. Am. Chem. Soc. 2005, 127, 13213–13219. [Google Scholar] [CrossRef] [PubMed]
- Pramanik, S.; Nagatoishi, S.; Sugimoto, N. DNA tetraplex structure formation from human telomeric repeat motif (TTAGGG):(CCCTAA) in nanocavity water pools of reverse micelles. Chem. Commun. 2012, 48, 4815–4817. [Google Scholar] [CrossRef] [PubMed]
- Strulson, C.A.; Molden, R.C.; Keating, C.D.; Bevilacqua, P.C. RNA catalysis through compartmentalization. Nat. Chem. 2012, 4, 941–946. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, P.; Jonchhe, S.; Emura, T.; Hidaka, K.; Endo, M.; Sugiyama, H.; Mao, H. Confined space facilitates G-quadruplex formation. Nat. Nanotechnol. 2017, 12, 582–588. [Google Scholar] [CrossRef]
- Jonchhe, S.; Pandey, S.; Emura, T.; Hidaka, K.; Hossain, M.A.; Shrestha, P.; Sugiyama, H.; Endo, M.; Mao, H. Decreased water activity in nanoconfinement contributes to the folding of G-quadruplex and i-motif structures. Proc. Natl. Acad. Sci. USA 2018, 115, 9539–9544. [Google Scholar] [CrossRef]
- Downs, A.M.; McCallum, C.; Pennathur, S. Confinement effects on DNA hybridization in electrokinetic micro- and nanofluidic systems. Electrophoresis 2019, 40, 792–798. [Google Scholar] [CrossRef]
- Jonchhe, S.; Pandey, S.; Karna, D.; Pokhrel, P.; Cui, Y.; Mishra, S.; Sugiyama, H.; Endo, M.; Mao, H. Duplex DNA is weakened in nanoconfinement. J. Am. Chem. Soc. 2020, 142, 10042–10049. [Google Scholar] [CrossRef]
- Nakano, S.; Yamaguchi, D.; Sugimoto, N. Thermal stability and conformation of DNA and proteins under the confined condition in the matrix of hydrogels. Mol. Biol. Rep. 2018, 45, 403–411. [Google Scholar] [CrossRef]
- Puglisi, J.D.; Tinoco, I., Jr. Absorbance melting curves of RNA. Methods. Enzymol. 1989, 180, 304–325. [Google Scholar]
- Knowles, D.B.; LaCroix, A.S.; Deines, N.F.; Shkel, I.; Record, M.T., Jr. Separation of preferential interaction and excluded volume effects on DNA duplex and hairpin stability. Proc. Natl. Acad. Sci. USA 2011, 108, 12699–12704. [Google Scholar] [CrossRef]
- Nakano, S.; Yamaguchi, D.; Tateishi-Karimata, H.; Miyoshi, D.; Sugimoto, N. Hydration changes upon DNA folding studied by osmotic stress experiments. Biophys. J. 2012, 102, 2808–2817. [Google Scholar] [CrossRef] [PubMed]
- Blake, R.D.; Delcourt, S.G. Thermodynamic effects of formamide on DNA stability. Nucleic Acids Res. 1996, 24, 2095–2103. [Google Scholar] [CrossRef] [PubMed]
- Nordstrom, L.J.; Clark, C.A.; Andersen, B.; Champlin, S.M.; Schwinefus, J.J. Effect of ethylene glycol, urea, and N-methylated glycines on DNA thermal stability: The role of DNA base pair composition and hydration. Biochemistry 2006, 45, 9604–9614. [Google Scholar] [CrossRef] [PubMed]
- Williamson, J.R. G-quartet structures in telomeric DNA. Annu. Rev. Biophys. Biomol. Struct. 1994, 23, 703–730. [Google Scholar] [CrossRef] [PubMed]
- Nakano, S.; Wu, L.; Oka, H.; Karimata, H.T.; Kirihata, T.; Sato, Y.; Fujii, S.; Sakai, H.; Kuwahara, M.; Sawai, H.; et al. Conformation and the sodium ion condensation on DNA and RNA structures in the presence of a neutral cosolute as a mimic of the intracellular media. Mol. BioSyst. 2008, 4, 579–588. [Google Scholar] [CrossRef] [PubMed]
- Maaloum, M.; Pernodet, N.; Tinland, B. Agarose gel structure using atomic force microscopy: Gel concentration and ionic strength effects. Electrophoresis 1998, 19, 1606–1610. [Google Scholar] [CrossRef]
- Xiong, J.Y.; Narayanan, J.; Liu, X.Y.; Chong, T.K.; Chen, S.B.; Chung, T.S. Topology evolution and gelation mechanism of agarose gel. J. Phys. Chem. B 2005, 109, 5638–5643. [Google Scholar] [CrossRef]
- Zhou, J.; Zhou, M.; Caruso, R.A. Agarose template for the fabrication of macroporous metal oxide structures. Langmuir 2006, 22, 3332–3336. [Google Scholar] [CrossRef]
- Haggerty, L.; Sugarman, J.H.; Prud’homme, R.K. Diffusion of polymers through polyacrylamide gels. Polymer 1988, 29, 1058–1063. [Google Scholar] [CrossRef]
- Holmes, D.L.; Stellwagen, N.C. Estimation of polyacrylamide gel pore size from Ferguson plots of linear DNA fragments. II. Comparison of gels with different crosslinker concentrations, added agarose and added linear polyacrylamide. Electrophoresis 1991, 12, 612–619. [Google Scholar] [CrossRef]
- Sim, A.Y.; Lipfert, J.; Herschlag, D.; Doniach, S. Salt dependence of the radius of gyration and flexibility of single-stranded DNA in solution probed by small-angle x-ray scattering. Phys. Rev. E 2012, 86, 021901. [Google Scholar] [CrossRef] [PubMed]
- Plumridge, A.; Meisburger, S.P.; Pollack, L. Visualizing single-stranded nucleic acids in solution. Nucleic Acids Res. 2017, 45, e66. [Google Scholar] [CrossRef] [PubMed]
- Rao, J.S.; Cruz, L. Effects of confinement on the structure and dynamics of an intrinsically disordered peptide: A molecular-dynamics study. J. Phys. Chem. B 2013, 117, 3707–3719. [Google Scholar] [CrossRef] [PubMed]
- Rao, J.S.; Smith, M.D.; Cruz, L. The stability of a beta-hairpin is altered by surface-water interactions under confinement. J. Phys. Chem. B 2014, 118, 3517–3523. [Google Scholar] [CrossRef]
- Miyoshi, D.; Karimata, H.; Sugimoto, N. Hydration regulates thermodynamics of G-quadruplex formation under molecular crowding conditions. J. Am. Chem. Soc. 2006, 128, 7957–7963. [Google Scholar] [CrossRef]
- Buscaglia, R.; Miller, M.C.; Dean, W.L.; Gray, R.D.; Lane, A.N.; Trent, J.O.; Chaires, J.B. Polyethylene glycol binding alters human telomere G-quadruplex structure by conformational selection. Nucleic Acids Res. 2013, 41, 7934–7946. [Google Scholar] [CrossRef]
- Nakano, M.; Tateishi-Karimata, H.; Tanaka, S.; Tama, F.; Miyashita, O.; Nakano, S.; Sugimoto, N. Local thermodynamics of the water molecules around single- and double-stranded DNA studied by grid inhomogeneous solvation theory. Chem. Phys. Lett. 2016, 660, 250–255. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamaguchi, D.; Yoshida, M.; Nakano, S.-i. Evaluation of Thermal Stability of DNA Oligonucleotide Structures Embedded in Hydrogels. DNA 2022, 2, 302-313. https://doi.org/10.3390/dna2040021
Yamaguchi D, Yoshida M, Nakano S-i. Evaluation of Thermal Stability of DNA Oligonucleotide Structures Embedded in Hydrogels. DNA. 2022; 2(4):302-313. https://doi.org/10.3390/dna2040021
Chicago/Turabian StyleYamaguchi, Daisuke, Masatoshi Yoshida, and Shu-ichi Nakano. 2022. "Evaluation of Thermal Stability of DNA Oligonucleotide Structures Embedded in Hydrogels" DNA 2, no. 4: 302-313. https://doi.org/10.3390/dna2040021
APA StyleYamaguchi, D., Yoshida, M., & Nakano, S.-i. (2022). Evaluation of Thermal Stability of DNA Oligonucleotide Structures Embedded in Hydrogels. DNA, 2(4), 302-313. https://doi.org/10.3390/dna2040021