DNA G-Quadruplex-Binding Proteins: An Updated Overview
Abstract
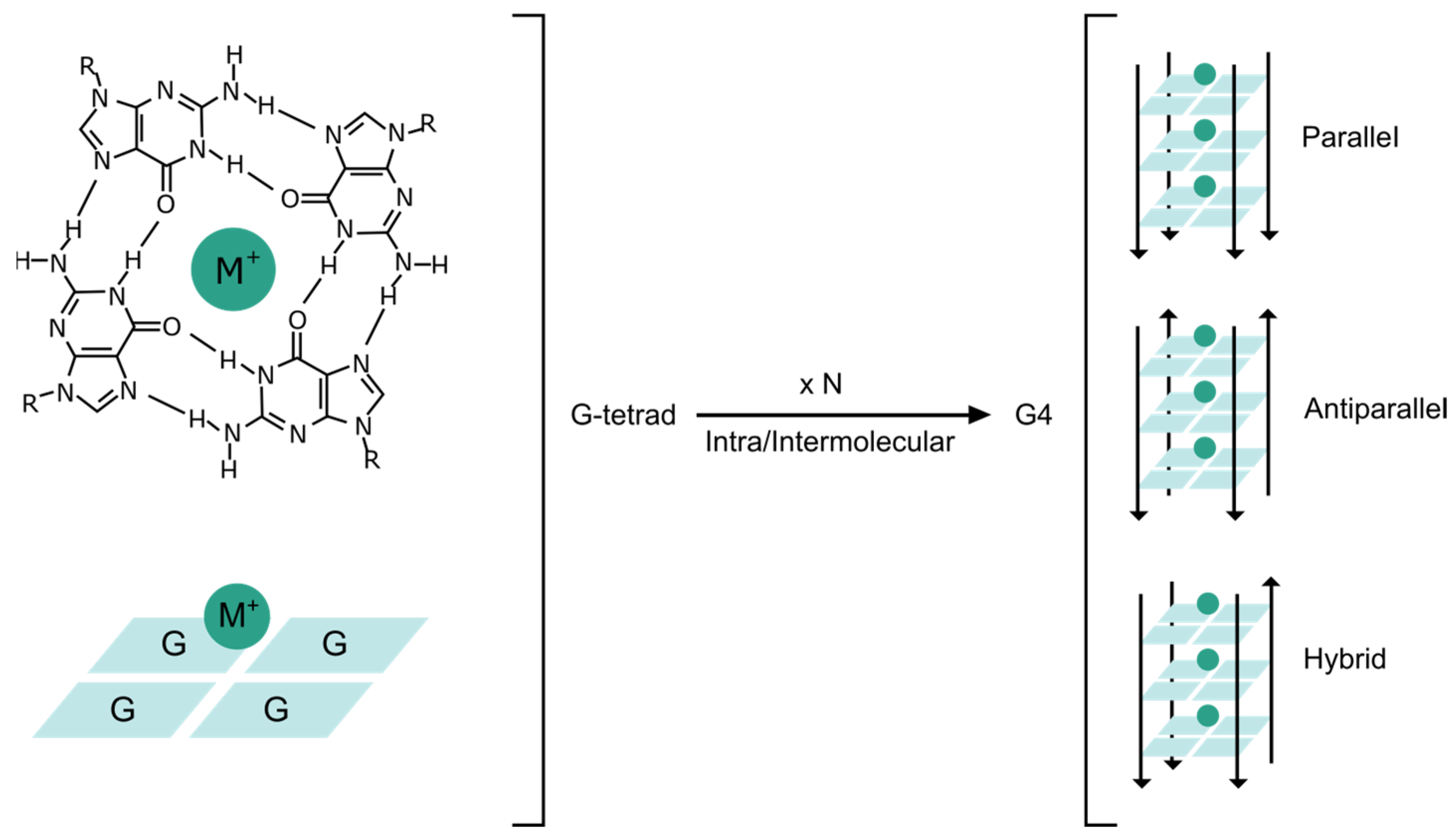
:1. Introduction
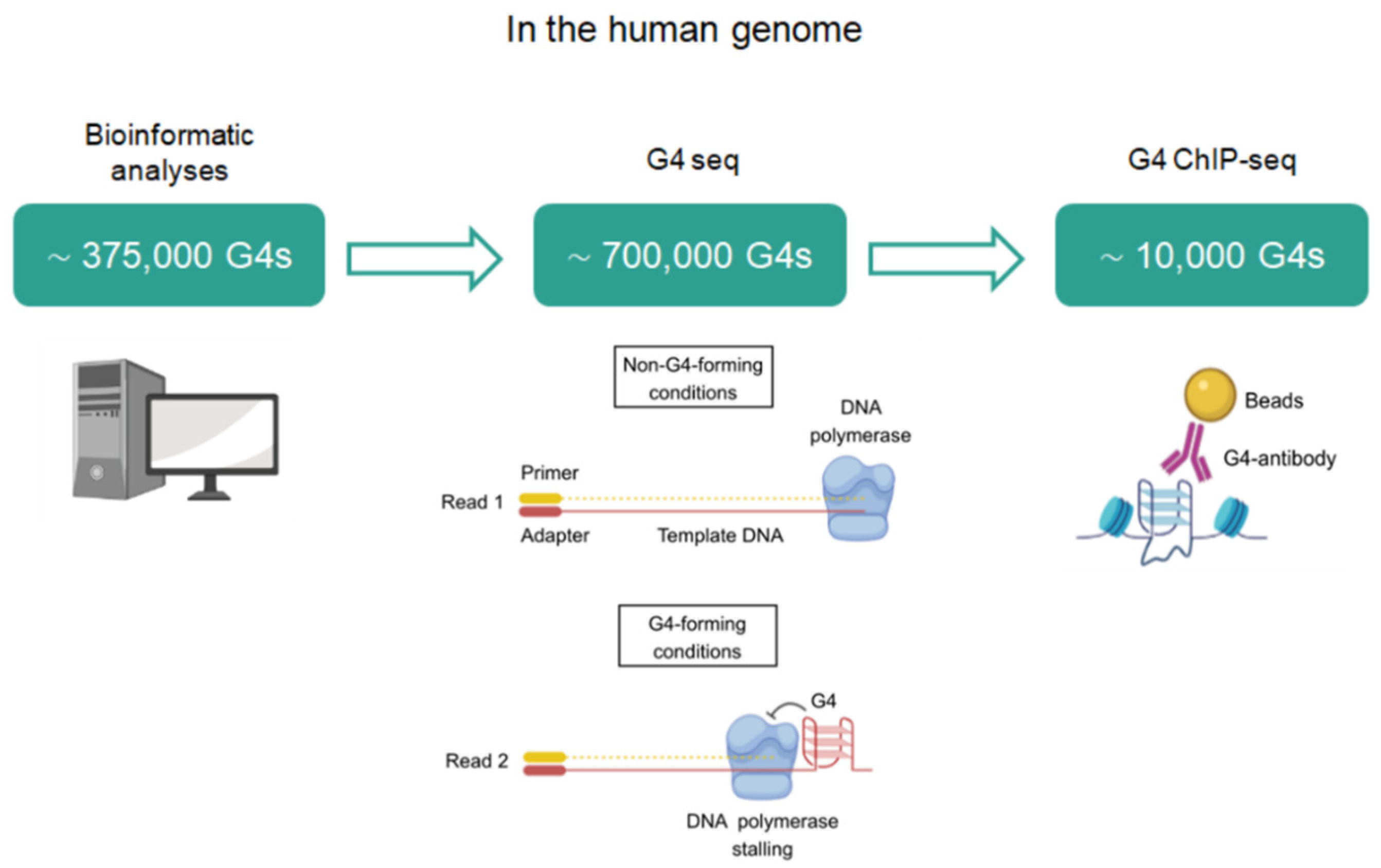
2. Identification of G4BPs
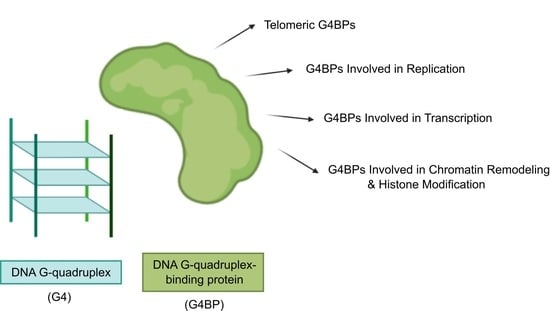
3. G4BPs
3.1. Telomeric G4BPs
3.2. G4BPs Involved in Replication
3.3. G4BPs Involved in Transcription
3.4. G4BPs Involved in Chromatin Remodeling and Histone Modification
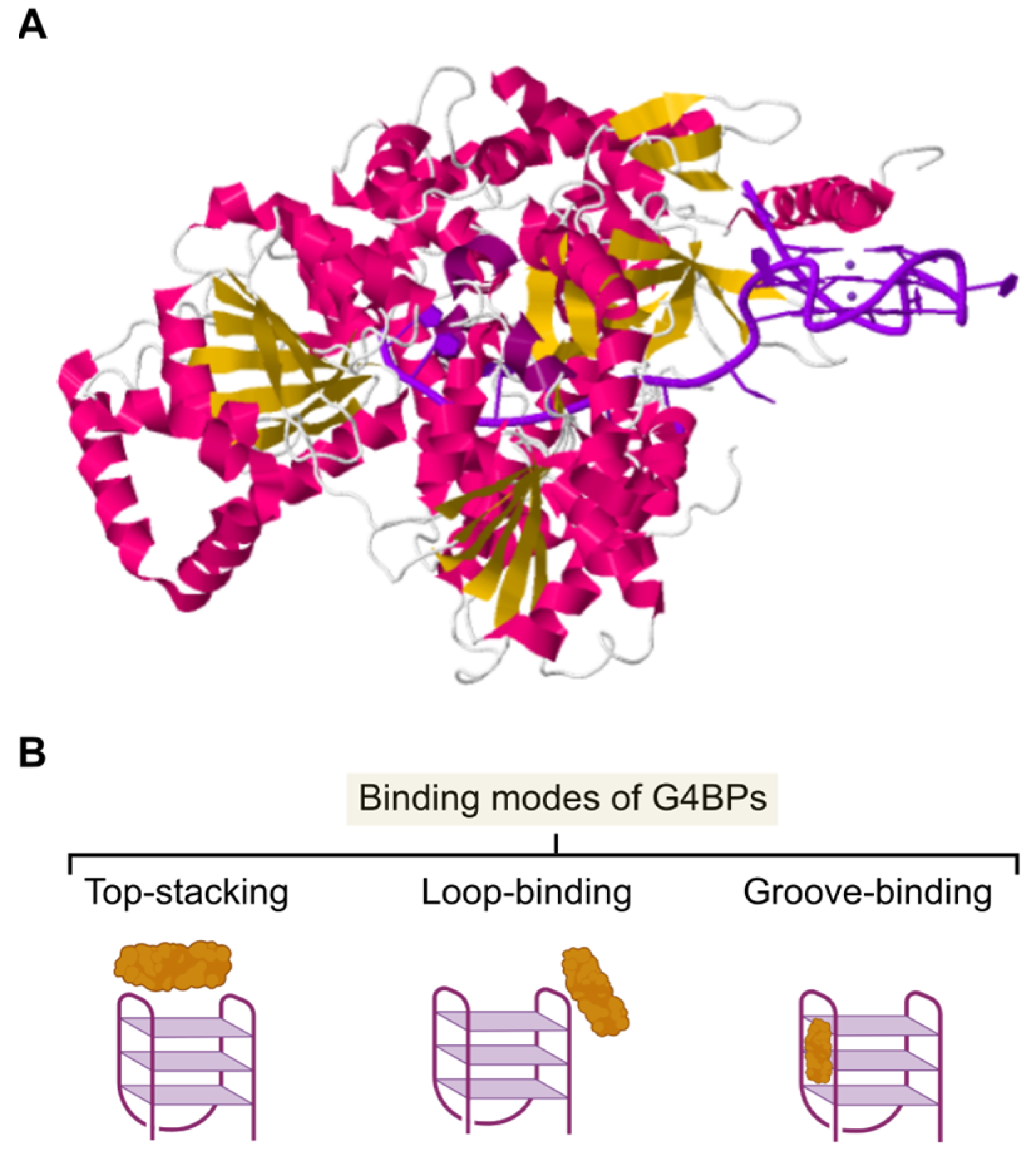
4. Structural Properties of G4BPs
5. Concluding Remarks
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bang, I. Untersuchungenüber die Guanylsäure. Biochem. Z 1910, 26, 293–311. [Google Scholar]
- Gellert, M.; Lipsett, M.N.; Davies, D.R. Helix formation by guanylic acid. Proc. Natl. Acad. Sci. USA 1962, 48, 2013–2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neidle, S.; Balasubramanian, S. Quadruplex Nucleic Acids; RSC Publishing: London, UK, 2006. [Google Scholar] [CrossRef]
- Sen, D.; Gilbert, W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature 1988, 334, 364–366. [Google Scholar] [CrossRef] [PubMed]
- Todd, A.K.; Johnston, M.; Neidle, S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005, 33, 2901–2907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marsico, G.; Chambers, V.S.; Sahakyan, A.B.; McCauley, P.; Boutell, J.M.; Di Antonio, M.; Balasubramanian, S. Whole genome experimental maps of DNA G-quadruplexes in multiple species. Nucleic Acids Res. 2019, 47, 3862–3874. [Google Scholar] [CrossRef] [Green Version]
- Shen, W.; Gorelick, R.J.; Bambara, R.A. HIV-1 nucleocapsid protein increases strand transfer recombination by promoting dimeric G-quartet formation. J. Biol. Chem. 2011, 286, 29838–29847. [Google Scholar] [CrossRef] [Green Version]
- Cahoon, L.A.; Seifert, H.S. An alternative DNA structure is necessary for pilin antigenic variation in Neisseria gonorrhoeae. Science 2009, 325, 764–767. [Google Scholar] [CrossRef] [Green Version]
- Smargiasso, N.; Gabelica, V.; Damblon, C.; Rosu, F.; De Pauw, E.; Teulade-Fichou, M.P.; Rowe, J.A.; Claessens, A. Putative DNA G-quadruplex formation within the promoters of Plasmodium falciparum var genes. BMC Genom. 2009, 10, 362. [Google Scholar] [CrossRef] [Green Version]
- Huppert, J.; Balasubramanian, S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2006, 35, 406–413. [Google Scholar] [CrossRef] [Green Version]
- Haensel-Hertsch, R.; Beraldi, D.; Lensing, S.V.; Marsico, G.; Zyner, K.; Parry, A.; Di Antonio, M.; Pike, J.; Kimura, H.; Narita, M.; et al. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016, 48, 1267–1272. [Google Scholar] [CrossRef] [Green Version]
- Spiegel, J.; Adhikari, S.; Balasubramanian, S. The Structure and Function of DNA G-Quadruplexes. Trends Chem. 2019, 2, 123–136. [Google Scholar] [CrossRef] [Green Version]
- Burge, S.; Parkinson, G.N.; Hazel, P.; Todd, A.K.; Neidle, S. Quadruplex DNA: Sequence, topology and structure. Nucleic Acids Res. 2006, 34, 5402–5415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grün, J.T.; Schwalbe, H. Folding dynamics of polymorphic G-quadruplex structures. Biopolymers 2022, 113, e23477. [Google Scholar] [CrossRef] [PubMed]
- Lane, A.N.; Chaires, J.B.; Gray, R.D.; Trent, J.O. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008, 36, 5482–5515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huppert, J.L.; Balasubramanian, S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005, 33, 2908–2916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chambers, V.S.; Marsico, G.; Boutell, J.M.; Di Antonio, M.; Smith, G.P.; Balasubramanian, S. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015, 33, 877–881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hänsel-Hertsch, R.; Spiegel, J.; Marsico, G.; Tannahill, D.; Balasubramanian, S. Genome-wide mapping of endogenous G-quadruplex DNA structures by chromatin immunoprecipitation and high-throughput sequencing. Nat. Protoc. 2018, 13, 551–564. [Google Scholar] [CrossRef] [PubMed]
- Varshney, D.; Spiegel, J.; Zyner, K.; Tannahill, D.; Balasubramanian, S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 2020, 21, 459–474. [Google Scholar] [CrossRef]
- Mendoza, O.; Bourdoncle, A.; Bouí, J.-B.; Brosh, R.M.; Mergny, J.-L. G-quadruplexes and helicases. Nucleic Acids Res. 2016, 44, 1989–2006. [Google Scholar] [CrossRef] [Green Version]
- Linke, R.; Limmer, M.; Juranek, S.A.; Heine, A.; Paeschke, K. The relevance of g-quadruplexes for dna repair. Int. J. Mol. Sci. 2021, 22, 12599. [Google Scholar] [CrossRef]
- Robinson, J.; Raguseo, F.; Nuccio, S.P.; Liano, D.; Di Antonio, M. DNA G-quadruplex structures: More than simple roadblocks to transcription? Nucleic Acids Res. 2021, 49, 8419–8431. [Google Scholar] [CrossRef] [PubMed]
- Meier-Stephenson, V. G4-quadruplex-binding proteins: Review and insights into selectivity. Biophys. Rev. 2022, 14, 635–654. [Google Scholar] [CrossRef] [PubMed]
- Shu, H.; Zhang, R.; Xiao, K.; Yang, J.; Sun, X. G-Quadruplex-Binding Proteins: Promising Targets for Drug Design. Biomolecules 2022, 12, 648. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, K.; Okamoto, K.; Okabe, S.; Fujii, R.; Ueda, K.; Ohashi, K.; Seimiya, H. G-quadruplex-forming nucleic acids interact with splicing factor 3B subunit 2 and suppress innate immune gene expression. Genes Cells 2021, 26, 65–82. [Google Scholar] [CrossRef]
- Lee, C.-Y.; McNerney, C.; Myong, S. G-Quadruplex and Protein Binding by Single-Molecule FRET Microscopy. Methods Mol. Biol. 2019, 2035, 309–322. [Google Scholar] [PubMed]
- Engelen, E.; Brandsma, J.H.; Moen, M.J.; Signorile, L.; Dekkers, D.H.; Demmers, J.; Kockx, C.E.; Ozgür, Z.; van IJcken, W.F.; van den Berg, D.L.; et al. Proteins that bind regulatory regions identified by histone modification chromatin immunoprecipitations and mass spectrometry. Nat. Commun. 2015, 6, 7155. [Google Scholar] [CrossRef] [Green Version]
- Huang, Z.-L.; Dai, J.; Luo, W.-H.; Wang, X.-G.; Tan, J.-H.; Chen, S.-B.; Huang, Z.-S. Identification of G-Quadruplex-Binding Protein from the Exploration of RGG Motif/G-Quadruplex Interactions. J. Am. Chem. Soc. 2018, 140, 17945–17955. [Google Scholar] [CrossRef]
- Zhang, X.; Spiegel, J.; Cuesta, S.M.; Adhikari, S.; Balasubramanian, S. Chemical profiling of DNA G- quadruplex-interacting proteins in live cells. Nat. Chem. 2021, 13, 626–633. [Google Scholar] [CrossRef]
- Su, H.; Xu, J.; Chen, Y.; Wang, Q.; Lu, Z.; Chen, Y.; Chen, K.; Han, S.; Fang, Z.; Wang, P.; et al. Photoactive G-Quadruplex Ligand Identifies Multiple G-Quadruplex-Related Proteins with Extensive Sequence Tolerance in the Cellular Environment. J. Am. Chem. Soc. 2021, 143, 1917–1923. [Google Scholar] [CrossRef]
- Mishra, S.K.; Tawani, A.; Mishra, A.; Kumar, A. G4IPDB: A database for G-quadruplex structure forming nucleic acid interacting proteins. Sci. Rep. 2016, 6, 38144. [Google Scholar] [CrossRef] [Green Version]
- Sun, Z.-Y.; Wang, X.-N.; Cheng, S.-Q.; Su, X.-X.; Ou, T.-M. Developing Novel G-Quadruplex Ligands: From Interaction with Nucleic Acids to Interfering with Nucleic Acid-Protein Interaction. Molecules 2019, 24, 396. [Google Scholar] [CrossRef] [Green Version]
- Henderson, E.; Hardin, C.C.; Walk, S.K.; Tinoco, I., Jr.; Blackburn, E.H. Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine·guanine base pairs. Cell 1987, 51, 899–908. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.; Bandaria, J.N.; Qureshi, M.H.; Yildiz, A.; Balci, H. G-quadruplex formation in telomeres enhances POT1/TPP1 protection against RPA binding. Proc. Natl. Acad. Sci. USA 2014, 111, 2990–2995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaires, J.B.; Gray, R.D.; Dean, W.L.; Monsen, R.; DeLeeuw, L.W.; Stribinskis, V.; O Trent, J. Human POT1 unfolds G-quadruplexes by conformational selection. Nucleic Acids Res. 2020, 48, 31. [Google Scholar] [CrossRef] [Green Version]
- Qureshi, M.H.; Ray, S.; Sewell, A.L.; Basu, S.; Balci, H. Replication Protein A Unfolds G-Quadruplex Structures with Varying Degrees of Efficiency. J. Phys. Chem. 2012, 8, 5588–5594. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharjee, A.; Wang, Y.; Diao, J.; Price, C.M. Dynamic DNA binding, junction recognition and G4 melting activity underlie the telomeric and genome-wide roles of human CST. Nucleic Acids Res. 2017, 45, 12311–12324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biffi, G.; Tannahill, D.; Balasubramanian, S. An intramolecular G-quadruplex structure is required for binding of telomeric repeat-containing RNA to the telomeric protein TRF2. J. Am. Chem. Soc. 2012, 134, 11974–11976. [Google Scholar] [CrossRef]
- Purohit, G.; Mukherjee, A.K.; Sharma, S.; Chowdhury, S. Extratelomeric Binding of the Telomere Binding Protein TRF2 at the PCGF3 Promoter Is G-Quadruplex Motif-Dependent. Biochemistry 2018, 57, 2317–2324. [Google Scholar] [CrossRef]
- Wu, W.; Rokutanda, N.; Takeuchi, J.; Lai, Y.; Maruyama, R.; Togashi, Y.; Nishikawa, H.; Arai, N.; Miyoshi, Y.; Suzuki, N.; et al. HERC2 Facilitates BLM and WRN Helicase Complex Interaction with RPA to Suppress G-Quadruplex DNA. Cancer Res. 2018, 78, 6371–6385. [Google Scholar] [CrossRef] [Green Version]
- Lin, W.; Sampathi, S.; Dai, H.; Liu, C.; Zhou, M.; Hu, J.; Huang, Q.; Campbell, J.; Shin-Ya, K.; Zheng, L.; et al. Mammalian DNA2 helicase/nuclease cleaves G-quadruplex DNA and is required for telomere integrity. EMBO J. 2013, 32, 1425–1439. [Google Scholar] [CrossRef] [Green Version]
- Vannier, J.-B.; Sandhu, S.; Petalcorin, M.I.; Wu, X.; Nabi, Z.; Ding, H.; Boulton, S.J. RTEL1 is a replisome-associated helicase that promotes telomere and genome-wide replication. Science 2013, 342, 239–242. [Google Scholar] [CrossRef]
- Takahama, K.; Takada, A.; Tada, S.; Shimizu, M.; Sayama, K.; Kurokawa, R.; Oyoshi, T. Regulation of telomere length by G-quadruplex telomere DNA- and TERRA-binding protein TLS/FUS. Chem. Biol. 2013, 20, 341–350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krüger, A.C.; Raarup, M.K.; Nielsen, M.; Kristensen, M.; Besenbacher, F.; Kjems, J.; Birkedal, V. Interaction of hnRNP A1 with telomere DNA G-quadruplex structures studied at the single molecule level. Eur. Biophys. J. 2010, 39, 1343–1350. [Google Scholar] [CrossRef] [PubMed]
- Hudson, J.S.; Ding, L.; Le, V.; Lewis, E.; Graves, D. Recognition and Binding of Human Telomeric G-Quadruplex DNA by Unfolding Protein 1. Biochemistry 2014, 53, 3347–3356. [Google Scholar] [CrossRef] [PubMed]
- Paudel, B.P.; Moye, A.L.; Assi, H.A.; El-Khoury, R.; Cohen, S.B.; Holien, J.K.; Birrento, M.L.; Samosorn, S.; Intharapichai, K.; Tomlinson, C.G.; et al. A mechanism for the extension and unfolding of parallel telomeric g-quadruplexes by human telomerase at single-molecule resolution. Elife 2020, 9, e56428. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, A.V.; Kubareva, E.A.; Monakhova, M.V.; Zvereva, M.I.; Dolinnaya, N.G. Impact of g-quadruplexes on the regulation of genome integrity, dna damage and repair. Biomolecules 2021, 11, 1284. [Google Scholar] [CrossRef]
- Sauer, M.; Paeschke, K. G-quadruplex unwinding helicases and their function in vivo. Biochem. Soc. Trans. 2017, 45, 1173–1182. [Google Scholar] [CrossRef] [Green Version]
- Castillo Bosch, P.; Segura-Bayona, S.; Koole, W.; Heteren, J.T.; Dewar, J.M.; Tijsterman, M.; Knipscheer, P. FANCJ promotes DNA synthesis through G-quadruplex structures. EMBO J. 2014, 33, 2521–2533. [Google Scholar] [CrossRef] [Green Version]
- Guo, M.; Hundseth, K.; Ding, H.; Vidhyasagar, V.; Inoue, A.; Nguyen, C.H.; Zain, R.; Lee, J.S.; Wu, Y. A distinct triplex DNA unwinding activity of ChlR1 helicase. J. Biol. Chem. 2015, 290, 5174–5189. [Google Scholar] [CrossRef] [Green Version]
- Paeschke, K.; Bochman, M.L.; Daniela Garcia, P.; Cejka, P.; Friedman, K.L.; Kowalczykowski, S.C.; Zakian, V.A. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature 2013, 497, 458–464. [Google Scholar] [CrossRef] [Green Version]
- Byrd, A.K.; Bell, M.R.; Raney, K.D. Pif1 helicase unfolding of G-quadruplex DNA is highly dependent on sequence and reaction conditions. J. Biol. Chem. 2018, 293, 17792–17802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brázda, V.; Hároníková, L.; Liao, J.C.C.; Fridrichová, H.; Jagelská, E. Strong preference of BRCA1 protein to topologically constrained non-B DNA structures. BMC Mol. Biol. 2016, 17, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balasubramanian, S.; Hurley, L.H.; Neidle, S. Targeting G-quadruplexes in gene promoters: A novel anticancer strategy? Nat. Rev. Drug Discov. 2011, 10, 261–275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spiegel, J.; Cuesta, S.M.; Adhikari, S.; Hänsel-Hertsch, R.; Tannahill, D.; Balasubramanian, S. G-quadruplexes are transcription factor binding hubs in human chromatin. Genome Biol. 2021, 22, 117. [Google Scholar] [CrossRef]
- Raiber, E.A.; Kranaster, R.; Lam, E.; Nikan, M.; Balasubramanian, S. A non-canonical DNA structure is a binding motif for the transcription factor SP1 in vitro. Nucleic Acids Res. 2012, 40, 1499–1508. [Google Scholar] [CrossRef] [Green Version]
- Cogoi, S.; Paramasivam, M.; Membrino, A.; Yokoyama, K.K.; Xodo, L.E. The KRAS promoter responds to Myc-associated zinc finger and poly(ADP-ribose) polymerase 1 proteins, which recognize a critical quadruplex-forming GA-element. J. Biol. Chem. 2010, 285, 22003–22016. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Williams, P.; Ren, W.; Wang, M.Y.; Gao, Z.; Miao, W.; Huang, M.; Song, J.; Wang, Y. YY1 interacts with guanine quadruplexes to regulate DNA looping and gene expression. Nat. Chem. Biol. 2020, 17, 161–168. [Google Scholar] [CrossRef]
- González, V.; Guo, K.; Hurley, L.; Sun, D. Identification and characterization of nucleolin as a c-myc G-quadruplex-binding protein. J. Biol. Chem. 2009, 284, 23622–23635. [Google Scholar] [CrossRef] [Green Version]
- Lago, S.; Tosoni, E.; Nadai, M.; Palumbo, M.; Richter, S.N. The cellular protein nucleolin preferentially binds long-looped G-quadruplex nucleic acids. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1371–1381. [Google Scholar] [CrossRef]
- Dempsey, L.A.; Sun, H.; Hanakahi, L.A.; Maizels, N. G4 DNA binding by LR1 and its subunits, nucleolin and hnRNP D, a role for G-G pairing in immunoglobulin switch recombination. J. Biol. Chem. 1999, 274, 1066–1071. [Google Scholar] [CrossRef] [Green Version]
- Scognamiglio, P.L.; Di Natale, C.; Leone, M.; Poletto, M.; Vitagliano, L.; Tell, G.; Marasco, D. G-quadruplex DNA recognition by nucleophosmin: New insights from protein dissection. Biochim. Biophys. Acta Gen. Subj. 2014, 1840, 2050–2059. [Google Scholar] [CrossRef] [PubMed]
- Krishna Thakur, R.; Kumar, P.; Halder, K.; Verma, A.; Kar, A.; Parent, J.-L.; Basundra, R.; Kumar, A.; Chowdhury, S.; Ramachandran, G.N. Metastases suppressor NM23-H2 interaction with G-quadruplex DNA within c-MYC promoter nuclease hypersensitive element induces c-MYC expression. Nucleic Acids Res. 2009, 37, 172–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quante, T.; Otto, B.; Brázdová, M.; Kejnovská, I.; Deppert, W.; Tolstonog, G.V. Mutant p53 is a transcriptional co-factor that binds to G-rich regulatory regions of active genes and generates transcriptional plasticity. Cell Cycle 2012, 11, 3290–3303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petr, M.; Helma, R.; Polášková, A.; Krejčí, A.; Dvořáková, Z.; Kejnovská, I.; Navrátilová, L.; Adámik, M.; Vorlíčková, M.; Brázdová, M. Wild-type p53 binds to MYC promoter G-quadruplex. Biosci. Rep. 2016, 36, 397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xodo, L.; Paramasivam, M.; Membrino, A.; Cogoi, S. Protein hnRNPA1 binds to a critical G-rich element of KRAS and unwinds G-quadruplex structures: Implications in transcription. Nucleic Acids Symp. Ser. 2008, 52, 159–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishikawa, T.; Kuwano, Y.; Takahara, Y.; Nishida, K.; Rokutan, K. HnRNPA1 interacts with G-quadruplex in the TRA2B promoter and stimulates its transcription in human colon cancer cells. Sci. Rep. 2019, 9, 10276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gray, L.T.; Vallur, A.C.; Eddy, J.; Maizels, N. G quadruplexes are genomewide targets of transcriptional helicases XPB and XPD. Nat. Chem. Biol. 2014, 10, 313–318. [Google Scholar] [CrossRef]
- Sun, H.; Yabuki, A.; Maizels, N. A human nuclease specific for G4 DNA. Proc. Natl. Acad. Sci. USA 2001, 98, 12444–12449. [Google Scholar] [CrossRef] [Green Version]
- Oyoshi, T.; Masuzawa, T. Modulation of histone modifications and G-quadruplex structures by G-quadruplex-binding proteins. Biochem. Biophys. Res. Commun. 2020, 531, 39–44. [Google Scholar] [CrossRef]
- Mao, S.Q.; Ghanbarian, A.T.; Spiegel, J.; Martínez Cuesta, S.; Beraldi, D.; Di Antonio, M.; Marsico, G.; Hänsel-Hertsch, R.; Tannahill, D.; Balasubramanian, S. DNA G-quadruplex structures mold the DNA methylome. Nat. Struct. Mol. Biol. 2018, 25, 951–957. [Google Scholar] [CrossRef]
- Cree, S.L.; Fredericks, R.; Miller, A.; Pearce, F.G.; Filichev, V.; Fee, C.; Kennedy, M.A. DNA G-quadruplexes show strong interaction with DNA methyltransferases in vitro. FEBS Lett. 2016, 590, 2870–2883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saha, D.; Singh, A.; Hussain, T.; Srivastava, V.; Sengupta, S.; Kar, A.; Dhapola, P.; Dhople, V.; Ummanni, R.; Chowdhury, S. Epigenetic suppression of human telomerase (hTERT) is mediated by the metastasis suppressor NME2 in a G-quadruplex-dependent fashion. J. Biol. Chem. 2017, 292, 15205–15215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pavlova, I.I.; Tsvetkov, V.B.; Isaakova, E.A.; Severov, V.V.; Khomyakova, E.A.; Lacis, I.A.; Lazarev, V.N.; Lagarkova, M.A.; Pozmogova, G.E.; Varizhuk, A.M. Transcription-facilitating histone chaperons interact with genomic and synthetic G4 structures. Int. J. Biol. Macromol. 2020, 160, 1144–1157. [Google Scholar] [CrossRef] [PubMed]
- Law, M.J.; Lower, K.M.; Voon, H.P.J.; Hughes, J.R.; Garrick, D.; Viprakasit, V.; Mitson, M.; De Gobbi, M.; Marra, M.; Morris, A.; et al. ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell 2010, 143, 367–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tikhonova, P.; Pavlova, I.; Isaakova, E.; Tsvetkov, V.; Bogomazova, A.; Vedekhina, T.; Luzhin, A.V.; Sultanov, R.; Severov, V.; Klimina, K.; et al. DNA G-quadruplexes contribute to CTCF recruitment. Int. J. Mol. Sci. 2021, 22, 7090. [Google Scholar] [CrossRef]
- Edwards, A.D.; Marecki, J.C.; Byrd, A.K.; Gao, J.; Raney, K.D. G-Quadruplex loops regulate PARP-1 enzymatic activation. Nucleic Acids Res. 2021, 49, 416–431. [Google Scholar] [CrossRef]
- Brázda, V.; Červeň, J.; Bartas, M.; Mikysková, N.; Coufal, J.; Pečinka, P. The Amino Acid Composition of Quadruplex Binding Proteins Reveals a Shared Motif and Predicts New Potential Quadruplex Interactors. Molecules 2018, 23, 2341. [Google Scholar] [CrossRef] [Green Version]
- Thandapani, P.; O’Connor, T.R.; Bailey, T.L.; Richard, S. Defining the RGG/RG Motif. Mol. Cell 2013, 50, 613–623. [Google Scholar] [CrossRef] [Green Version]
- González, V.; Hurley, L.H. The C-Terminus of Nucleolin Promotes the Formation of the c-MYC G-Quadruplex and Inhibits c-MYC Promoter Activity. Biochemistry 2010, 49, 9706–9714. [Google Scholar] [CrossRef] [Green Version]
- Sathyapriya, R.; Vishveshwara, S. Interaction of DNA with clusters of amino acids in proteins. Nucleic Acids Res. 2004, 32, 4109–4118. [Google Scholar] [CrossRef] [Green Version]
- Murzin, A.G. OB(oligonucleotide/oligosaccharide binding)-fold: Common structural and functional solution for non-homologous sequences. EMBO J. 1993, 12, 861–867. [Google Scholar] [CrossRef] [PubMed]
- Hwang, H.; Buncher, N.; Opresko, P.L.; Myong, S. POT1-TPP1 regulates telomeric overhang structural dynamics. Structure 2012, 20, 1872–1880. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Biological Function | Protein Name | Literature |
|---|---|---|
| Telomere lengthening | BLM | [40] |
| CST | [37] | |
| DNA2 | [41] | |
| hnRNPA1 | [44] | |
| hTERT | [46] | |
| Pif1 | [52] | |
| POT1 | [34] | |
| RPA | [36] | |
| RTEL1 | [42] | |
| TLS/FUS | [43] | |
| TRF2 | [38,39] | |
| UP1 | [45] | |
| WRN | [40] | |
| Replication | BLM | [48] |
| BRCA1 | [53] | |
| DDX11 | [50] | |
| FANCJ | [49] | |
| Pif1 | [51] | |
| WRN | [48] | |
| Transcription | GQN1 | [69] |
| hnRNPA1 | [66] | |
| MAZ | [57] | |
| NM23-H2 | [63] | |
| Nucleolin | [59] | |
| Nucleophosmin | [62] | |
| SP1 | [56] | |
| TP53 | [64,65] | |
| XPB | [68] | |
| XPD | [68] | |
| YY1 | [58] | |
| Chromatin remodeling and histone modification | ATRX | [75] |
| BRD3 | [74] | |
| CTCF | [76] | |
| DNMT1 | [71] | |
| REST-LSD1 | [73] | |
| SMARCA4 | [29] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sanchez-Martin, V. DNA G-Quadruplex-Binding Proteins: An Updated Overview. DNA 2023, 3, 1-12. https://doi.org/10.3390/dna3010001
Sanchez-Martin V. DNA G-Quadruplex-Binding Proteins: An Updated Overview. DNA. 2023; 3(1):1-12. https://doi.org/10.3390/dna3010001
Chicago/Turabian StyleSanchez-Martin, Victoria. 2023. "DNA G-Quadruplex-Binding Proteins: An Updated Overview" DNA 3, no. 1: 1-12. https://doi.org/10.3390/dna3010001
APA StyleSanchez-Martin, V. (2023). DNA G-Quadruplex-Binding Proteins: An Updated Overview. DNA, 3(1), 1-12. https://doi.org/10.3390/dna3010001