Simple Formulae, Deep Learning and Elaborate Modelling for the COVID-19 Pandemic
Abstract
:1. Introduction
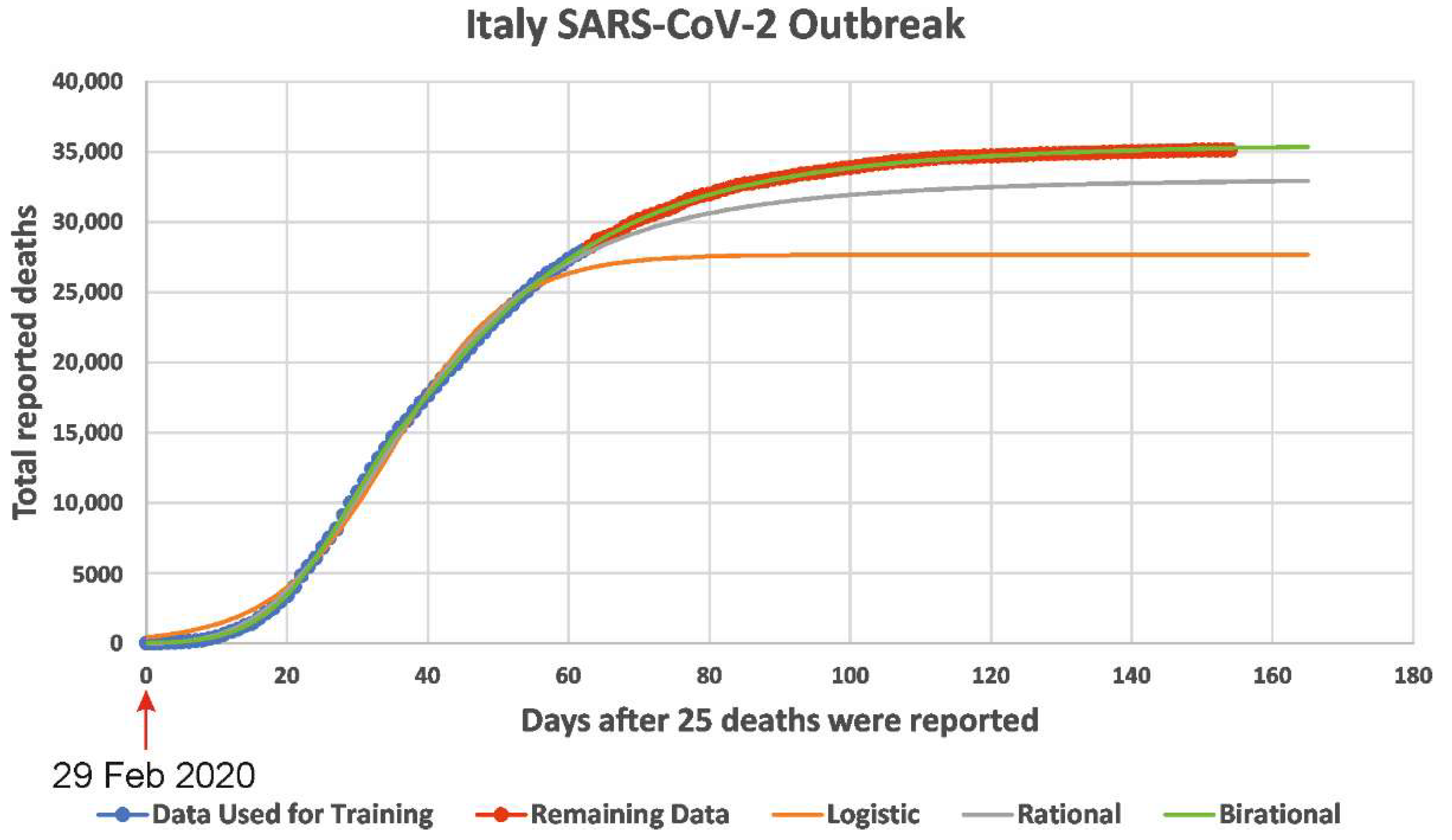
2. The Success of the Novel Formulae as Opposed to the Logistic Formula
3. Deep Learning
4. A Typical Susceptible-Exposed-Infected-Recovered (SEIR) Type Model
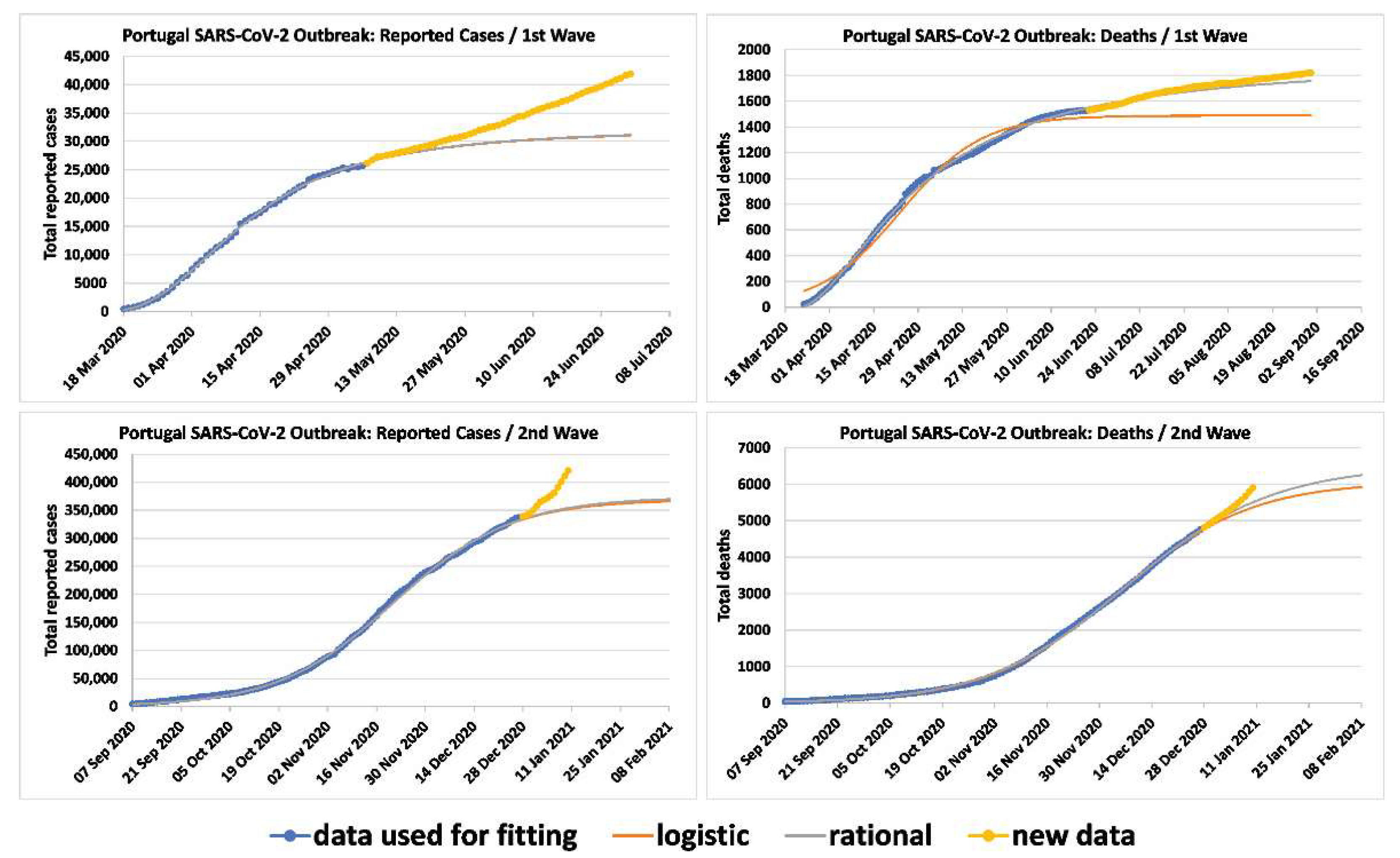
5. From the First to the Second Wave
5.1. Predictions for the Number of Reported Infected Cases and the Number of the Deceased for the Second Wave in Portugal
5.2. Higher Infectivity and Lower Virulence
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Holmdahl, I.; Buckee, C. Wrong but useful-What Covid-19 epidemiologic models can and cannot tell us. N. Engl. J. Med. 2020, 383, 303–305. [Google Scholar] [CrossRef] [PubMed]
- Fokas, A.S.; Dikaios, N.; Kastis, G.A. Mathematical models and deep learning for predicting the number of individuals reported to be infected with SARS-CoV-2. J. R. Soc. Interface 2020, 17, 20200494. [Google Scholar] [CrossRef] [PubMed]
- Fokas, A.S.; Dikaios, N.; Kastis, G.A. Covid-19: Predictive mathematical formulae for the number of deaths during lockdown and possible scenarios for the post-lockdown period. Proc. R. Soc. A 2021, 477, 20200745. [Google Scholar] [CrossRef] [PubMed]
- Fokas, A.S.; Kastis, G.A. SARS-CoV-2: The Second Wave in Europe. J. Med. Internet Res. 2021, 23, e22431. [Google Scholar] [CrossRef] [PubMed]
- Fokas, A.S.; Cuevas-Maraver, J.; Kevrekidis, P.G. A quantitative framework for exploring exit strategies from the COVID-19 lockdown. Chaos Solitons Fractals 2020, 140, 110244. [Google Scholar] [CrossRef]
- Fokas, A.S.; Cuevas-Maraver, J.; Kevrekidis, P.G. Easing COVID-19 lockdown measures while protecting the older restricts the deaths to the level of the full lockdown. Sci. Rep. 2021, 11, 5839. [Google Scholar] [CrossRef] [PubMed]
- Fokas, A.S.; Oberai, A.A.; Yortsos, Y.C. A novel SEIR type model exhibiting algebraic behavior. in preparation.
- Luhar, M.; Oberai, A.A.; Fokas, A.S.; Yortsos, Y.C. Accounting for Super-Spreader Events and Algebraic Decay in SIR Models. Comput. Methods Appl. Mech. Eng. 2022. submitted. [Google Scholar]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef]
- Hochreiter, S.; Schmidhuber, J. Long Short-Term Memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef]
- Graves, A.; Schmidhuber, J. Framewise phoneme classification with bi-directional LSTM and other neural network architectures. Neural Netw. 2005, 18, 602–610. [Google Scholar] [CrossRef] [PubMed]
- Schuster, M.; Paliwal, K.K. Bidirectional recurrent neural networks. IEEE Trans. Signal Process. 1997, 45, 2673–2681. [Google Scholar] [CrossRef]
- Ferguson, N.M.; Laydon, D.; Nedjati-Gilani, G.; Imai, N.; Ainslie, K.; Baguelin, M.; Ghani, A.C. Impact of Non-Pharmaceutical Interventions (NPIs) to Reduce COVID-19 Mortality and Healthcare Demand. Imperial College, London, March 2020. Available online: https://www.imperial.ac.uk/mrc-global-infectious-disease-analysis/covid-19/ (accessed on 20 March 2020).
- Ramaswamy, H.; Oberai, A.A.; Luhar, M.; Yortsos, Y.C. Understanding the R0 of pademics. In Proceedings of the 42nd Ibero-Latin-American Congress on Computational Methods in Engineering (XLII CILAMCE) and the 3rd Pan American Congress on Computational Mechanics (III PANACM), Rio de Janeiro, Brazil, 9–12 November 2021; p. 9449. [Google Scholar]
- Lauring, A.S.; Andino, R. Quasispecies theory and the behavior of RNA viruses. PLoS Pathog. 2010, 6, e1001005. [Google Scholar] [CrossRef] [PubMed]
- Ghedin, E.; Sengamalay, N.A.; Shumway, M.; Zaborsky, J.; Feldblyum, T.; Subbu, V.; Spiro, D.J.; Sitz, J.; Koo, H.; Bolotov, P.; et al. Large-scale sequencing of human influenza reveals the dynamic nature of viral genome evolution. Nature 2005, 437, 1162–1166. [Google Scholar] [CrossRef]
- Badua, C.; Baldo, K.A.T.; Medina, P.M.B. Genomic and proteomic mutation landscapes of SARS-CoV-2. J. Med. Virol. 2021, 93, 1702–1721. [Google Scholar] [CrossRef]
- Artesi, M.; Bontems, S.; Gobbels, P.; Franckh, M.; Maes, P.; Boreux, R.; Meex, C.; Melin, P.; Hayette, M.P.; Bours, V.; et al. A Recurrent Mutation at Position 26340 of SARS-CoV-2 Is Associated with Failure of the E Gene Quantitative Reverse Transcription-PCR Utilized in a Commercial Dual-Target Diagnostic Assay. J. Clin. Microbiol. 2020, 58, e01598-20. [Google Scholar] [CrossRef]
- Al Khatib, H.A.; Benslimane, F.M.; Elbashir, I.E.; Coyle, P.V.; Al Maslamani, M.A.; Al-Khal, A.; Al Thani, A.A.; Yassine, H.M. Within-Host Diversity of SARS-CoV-2 in COVID-19 Patients With Variable Disease Severities. Front. Cell Infect. Microbiol. 2020, 10, 575613. [Google Scholar] [CrossRef]
- Pandit, B.; Bhattacharjee, S.; Bhattacharjee, B. Association of clade-G SARS-CoV-2 viruses and age with increased mortality rates across 57 countries and India. Infect. Genet. Evol. 2021, 90, 104734. [Google Scholar] [CrossRef]
- Andreano, E.; Piccini, G.; Licastro, D.; Casalino, L.; Johnson, N.V.; Paciello, I.; Monego, S.D.; Pantano, E.; Manganaro, N.; Manenti, A.; et al. SARS-CoV-2 escape from a highly neutralizing COVID-19 convalescent plasma. Proc. Natl. Acad. Sci. USA 2021, 118, e2103154118. [Google Scholar] [CrossRef]
- Amanat, F.; Strohmeier, S.; Rathnasinghe, R.; Schotsaert, M.; Coughlan, L.; García-Sastre, A.; Krammer, F. Introduction of two prolines and removal of the polybasic cleavage site lead to higher efficacy of a recombinant spike-based SARS-CoV-2 vaccine in the mouse model. mbio 2021, 12, e02648-20. [Google Scholar] [CrossRef]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827.e19. [Google Scholar] [CrossRef] [PubMed]
- Mansbach, R.A.; Chakraborty, S.; Nguyen, K.; Montefiori, D.C.; Korber, B.; Gnanakaran, S. The SARS-CoV-2 Spike Variant D614G Favors an Open Conformational State. Sci. Adv. 2021, 7, eabf3671. [Google Scholar] [CrossRef] [PubMed]
- Long, S.W.; Olsen, R.J.; Christensen, P.A.; Bernard, D.W.; Davis, J.J.; Shukla, M.; Nguyen, M.; Saavedra, M.O.; Yerramilli, P.; Pruitt, L.; et al. Molecular Architecture of Early Dissemination and Massive Second Wave of the SARS-CoV-2 Virus in a Major Metropolitan Area. mBio 2020, 11, e02707-20. [Google Scholar] [CrossRef] [PubMed]
- Volz, E.; Hill, V.; McCrone, J.T.; Price, A.; Jorgensen, D.; O’Toole, Á.; Southgate, J.; Johnson, R.; Jackson, B.; Nascimento, F.F.; et al. Evaluating the Effects of SARS-CoV-2 Spike Mutation D614G on Transmissibility and Pathogenicity. Cell 2021, 184, 64–75. [Google Scholar] [CrossRef]
- COVID-19 Situation Update for the EU/EEA, as of Week 7 2021, Updated 25 February 2021. European Center for Disease Control. Available online: https://www.ecdc.europa.eu/en/cases-2019-ncov-eueea (accessed on 1 March 2021).
- COVID-19 Situation Worldwide, as of Week 7 2021, Updated 25 February 2021. European Center for Disease Control. Available online: https://www.ecdc.europa.eu/en/geographical-distribution-2019-ncov-cases (accessed on 1 March 2021).
- Yurkovetskiy, L.; Wang, X.; Pascal, K.E.; Tomkins-Tinch, C.; Nyalile, T.; Wang, Y.; Baum, A.; Diehl, W.E.; Dauphin, A.; Carbone, C.; et al. Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell 2020, 183, 739–751.e8. [Google Scholar] [CrossRef]
- Li, Q.; Wu, J.; Nie, J.; Zhang, L.; Hao, H.; Liu, S.; Zhao, C.; Zhang, Q.; Liu, H.; Nie, L.; et al. The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell 2020, 182, 1284–1294.e9. [Google Scholar] [CrossRef]
- Plante, J.A.; Liu, Y.; Liu, J.; Xia, H.; Johnson, B.A.; Lokugamage, K.G.; Zhang, X.; Muruato, A.E.; Zou, J.; Fontes-Garfias, C.R.; et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 2021, 592, 116–121. [Google Scholar] [CrossRef]
- Hou, Y.J.; Chiba, S.; Halfmann, P.; Here, C.; Kuroda, M.; Dinnon, K.H., III; Leist, S.R.; Schäfer, A.; Nakajima, N.; Takahashi, K.; et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science 2020, 370, 1464–1468. [Google Scholar] [CrossRef]
- Zhou, B.; Thao, T.T.N.; Hoffmann, D.; Taddeo, A.; Ebert, N.; Labroussaa, F.; Pohlmann, A.; King, J.; Steiner, S.; Kelly, J.N.; et al. SARS-CoV-2 spike D614G change enhances replication and transmission. Nature 2021, 592, 122–127. [Google Scholar] [CrossRef]
- Daniloski, Z.; Jordan, T.X.; Ilmain, J.K.; Guo, X.; Bhabha, G.; tenOever, B.R.; Sanjana, N.E. The D614G mutation in SARS-CoV-2 Spike increases transduction of multiple human cell types. eLife 2021, 10, e65365. [Google Scholar] [CrossRef]
- van Dorp, L.; Richard, D.; Tan, C.C.S.; Shaw, L.P.; Acman, M.; Balloux, F. No evidence for increased transmissibility from recurrent mutations in SARS-CoV-2. Nat. Commun. 2020, 11, 5986. [Google Scholar] [CrossRef] [PubMed]
- Callaway, E. The coronavirus is mutating-does it matter? Nature 2020, 585, 174–177. [Google Scholar] [CrossRef] [PubMed]
- Claro, I.M.; da Silva Sales, F.C.; Ramundo, M.S.; Candido, D.S.; Silva, C.A.M.; de Jesus, J.G.; Manuli, E.R.; de Oliveira, C.M.; Scarpelli, L.; Campana, G.; et al. Local Transmission of SARS-CoV-2 Lineage B.1.1.7, Brazil, December 2020. Emerg. Infect. Dis. 2021, 27, 970–972. [Google Scholar] [CrossRef] [PubMed]
- Dao, M.H.; Nguyen, H.T.; Nguyen, T.V.; Nguyen, A.H.; Luong, Q.C.; Vu, N.H.P.; Pham, H.T.T.; Nguyen, T.N.T.; Thach, D.H.; Nguyen, L.V.; et al. New SARS-CoV-2 Variant of Concern Imported from the United Kingdom to Vietnam, December 2020. J. Med. Virol. 2021, 93, 2628–2630. [Google Scholar] [CrossRef] [PubMed]
- Umair, M.; Ikram, A.; Salman, M.; Alam, M.M.; Badar, N.; Rehman, Z.; Tamim, S.; Khurshid, A.; Ahad, A.; Ahmad, H.; et al. Importation of SARS-CoV-2 Variant B.1.1.7 in Pakistan. J. Med. Virol. 2021, 93, 2623–2625. [Google Scholar] [CrossRef] [PubMed]
- Yadav, P.D.; Nyayanit, D.A.; Sahay, R.R.; Sarkale, P.; Pethani, J.; Patil, S.; Baradkar, S.; Potdar, V.; Patil, D.Y. Isolation and characterization of the new SARS-CoV-2 variant in travellers from the United Kingdom to India: VUI-202012/01 of the B.1.1.7 lineage. J. Travel Med. 2021, 28, taab009. [Google Scholar] [CrossRef]
- Alpert, T.; Brito, A.F.; Lasek-Nesselquist, E.; Rothman, J.; Valesano, A.L.; MacKay, M.J.; Petrone, M.E.; Breban, M.I.; Watkins, A.E.; Vogels, C.B.F.; et al. Early introductions and community transmission of SARS-CoV-2 variant B.1.1.7 in the United States. Cell 2021, 184, 2595–2604.e13. [Google Scholar] [CrossRef]
- Firestone, M.J.; Lorentz, A.J.; Wang, X.; Como-Sabetti, K.; Vetter, S.; Smith, K.; Holzbauer, S.; Meyer, S.; Ehresmann, K.; Danila, R.; et al. First Identified Cases of SARS-CoV-2 Variant B.1.1.7 in Minnesota-December 2020–January 2021. MMWR Morb. Mortal. Wkly. Rep. 2021, 70, 278–279. [Google Scholar] [CrossRef]
- Galloway, S.E.; Paul, P.; MacCannell, D.R.; Johansson, M.A.; Brooks, J.T.; MacNeil, A.; Slayton, R.B.; Tong, S.; Silk, B.J.; Armstrong, G.L.; et al. Emergence of SARS-CoV-2 B.1.1.7 Lineage-United States, December 29, 2020–January 12, 2021. MMWR Morb. Mortal. Wkly. Rep. 2021, 70, 95–99. [Google Scholar] [CrossRef]
- Leung, K.; Shum, M.H.; Leung, G.M.; Lam, T.T.; Wu, J.T. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Eurosurveillance 2021, 26, 2002106. [Google Scholar] [CrossRef]
- Pfefferle, S.; Gunther, T.; Kobbe, R.; Czech-Sioli, M.; Nörz, D.; Santer, R.; Oh, J.; Kluge, S.; Oestereich, L.; Peldschus, K.; et al. SARS Coronavirus-2 variant tracing within the first Coronavirus Disease 19 clusters in northern Germany. Clin. Microbiol. Infect. 2021, 27, 130.e5–130.e8. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control. Risk Assessment: SARS-CoV-2-Increased Circulation of Variants of Concern and Vaccine Rollout in the EU/EEA, 14th Update. Available online: https://www.ecdc.europa.eu/en/publications-data/covid-19-risk-assessment-variants-vaccine-fourteenth-update-february-2021 (accessed on 1 March 2021).
- Mwenda, M.; Saasa, N.; Sinyang, N.; Busby, G.; Chipimo, P.J.; Hendry, J.; Kapona, O.; Yingst, S.; Hines, J.Z.; Minchella, P.; et al. Detection of B.1.351 SARS-CoV-2 Variant Strain-Zambia, December 2020. MMWR Morb. Mortal. Wkly. Rep. 2021, 70, 280–282. [Google Scholar] [CrossRef] [PubMed]
- Munoz, M.; Patino, L.H.; Ballesteros, N.; Paniz-Mondolfi, A.; Ramírez, J.D. Characterizing SARS-CoV-2 genome diversity circulating in South American countries: Signatures of potentially emergent lineages? Int. J. Infect. Dis. 2021, 105, 329–332. [Google Scholar] [CrossRef] [PubMed]
- Maggi, F.; Novazzi, F.; Genoni, A.; Baj, A.; Spezia, P.G.; Focosi, D.; Zago, C.; Colombo, A.; Cassani, G.; Pasciuta, R.; et al. Imported SARS-COV-2 Variant P.1 Detected in Traveler Returning from Brazil to Italy. Emerg. Infect. Dis. 2021, 27, 1249–1251. [Google Scholar] [CrossRef]
- Collier, D.A.; De Marco, A.; Ferreira, I.; Meng, B.; Datir, R.P.; Walls, A.C.; Kemp, S.A.; Bassi, J.; Pinto, D.; Silacci-Fregni, C.; et al. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature 2021, 593, 136–141. [Google Scholar] [CrossRef]
- Coronavirus (COVID-19) Infection Survey Technical Article: Waves and Lags of COVID-19 in England, June 2021. Available online: https://www.ons.gov.uk/peoplepopulationandcommunity/healthandsocialcare/conditionsanddiseases/articles/coronaviruscovid19infectionsurveytechnicalarticle/wavesandlagsofcovid19inenglandjune2021 (accessed on 15 August 2021).
- Traini, M.C.; Caponi, C.; Ferrari, R.; De Socio, G.V. SARS-CoV-2 unreported cases in Italy: Analysis of serological survey and vaccination scenarios. Infect. Dis. Model. 2021, 6, 909–923. [Google Scholar] [CrossRef]
- How Many Cases of Covid-19 Are There? A Q&A. Available online: https://www.nuffieldtrust.org.uk/news-item/how-many-cases-of-covid-19-are-there-a-q-a (accessed on 15 August 2021).
- Campolieti, M. COVID-19 deaths in the USA: Benford’s law and under-reporting. J. Public Health 2021, fdab161. [Google Scholar] [CrossRef]
- Team, E.P.H.E.; Danis, K.; Fonteneau, L.; Danis, K.; Fonteneau, L.; Georges, S.; Daniau, C.; Bernard-Stoecklin, S.; Domegan, L.; O’Donnell, J.; et al. High impact of COVID-19 in long-term care facilities, suggestion for monitoring in the EU/EEA, May 2020. Eurosurveillance 2020, 25, 2000956. [Google Scholar]
- Horwitz, L.I.; Jones, S.A.; Cerfolio, R.J.; Francois, F.; Greco, J.; Rudy, B.; Petrilli, C.M. Trends in COVID-19 Risk-Adjusted Mortality Rates. J. Hosp. Med. 2021, 16, 90–92. [Google Scholar] [CrossRef]
- Gandhi, M.; Rutherford, G.W. Facial Masking for Covid-19-Potential for “Variolation” as We Await a Vaccine. N. Engl. J. Med. 2020, 383, e101. [Google Scholar] [CrossRef]

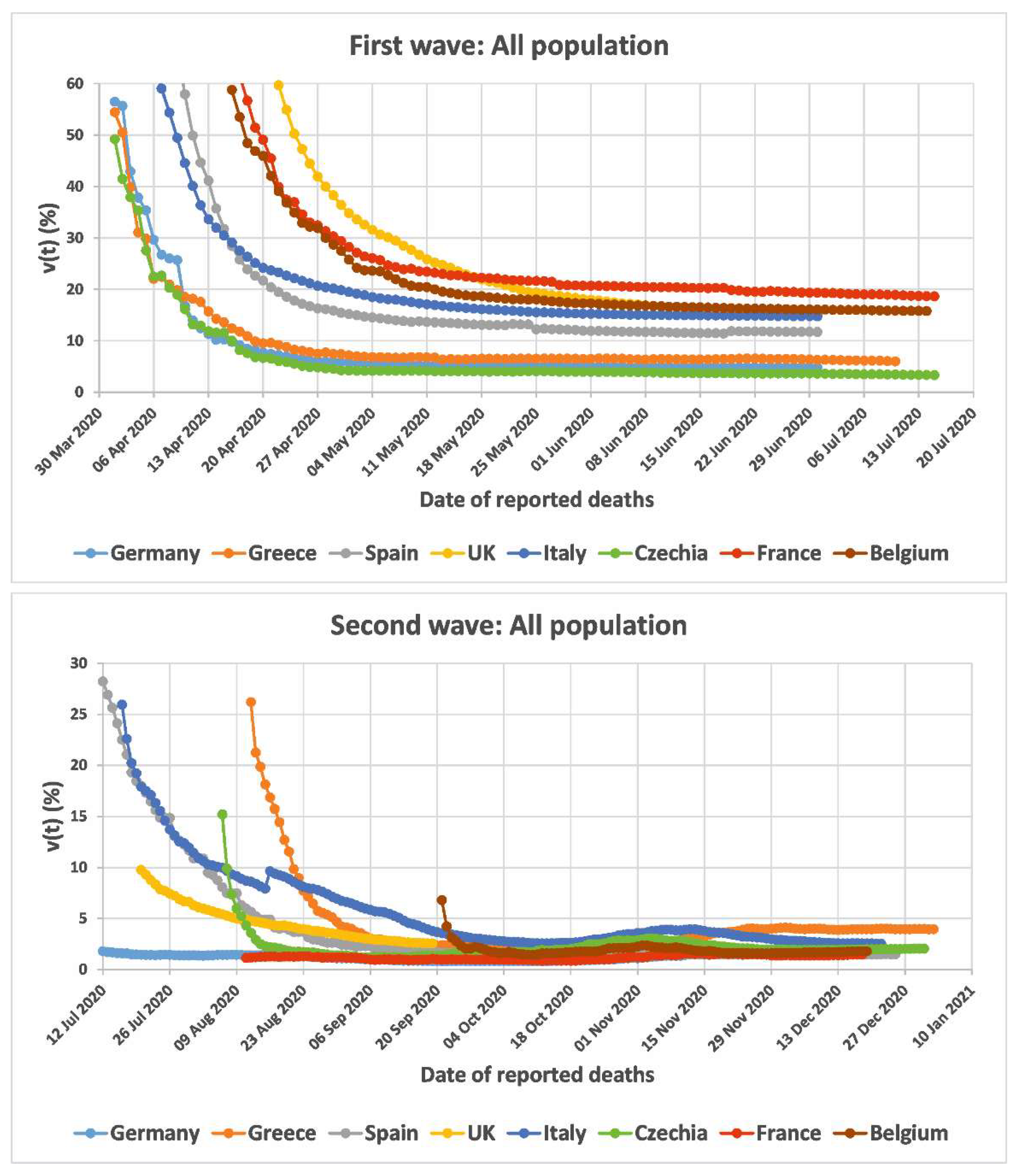
| All Population | 1st Wave | 2nd Wave | ||
|---|---|---|---|---|
| Date | v(t) (%) | Date | v(t) (%) | |
| Germany | 30/June/2020 | 4.86 | 26/November/2020 | 1.50 |
| Greece | 04/July/2020 | 6.18 | 02/January/2021 | 3.95 |
| Spain | 30/June/2020 | 11.72 | 25/December/2020 | 1.49 |
| UK | 30/June/2020 | 15.24 | 19/September/2020 | 2.55 |
| Italy | 30/June/2020 | 14.77 | 22/December/2020 | 2.54 |
| Czechia | 15/July/2020 | 3.33 | 31/December/2020 | 2.04 |
| France | 15/July/2020 | 18.62 | 18/December/2020 | 1.46 |
| Belgium | 14/July/2020 | 15.77 | 19/December/2020 | 1.78 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fokas, A.S.; Dikaios, N.; Tsiodras, S.; Kastis, G.A. Simple Formulae, Deep Learning and Elaborate Modelling for the COVID-19 Pandemic. Encyclopedia 2022, 2, 679-689. https://doi.org/10.3390/encyclopedia2020047
Fokas AS, Dikaios N, Tsiodras S, Kastis GA. Simple Formulae, Deep Learning and Elaborate Modelling for the COVID-19 Pandemic. Encyclopedia. 2022; 2(2):679-689. https://doi.org/10.3390/encyclopedia2020047
Chicago/Turabian StyleFokas, Athanassios S., Nikolaos Dikaios, Sotirios Tsiodras, and George A. Kastis. 2022. "Simple Formulae, Deep Learning and Elaborate Modelling for the COVID-19 Pandemic" Encyclopedia 2, no. 2: 679-689. https://doi.org/10.3390/encyclopedia2020047
APA StyleFokas, A. S., Dikaios, N., Tsiodras, S., & Kastis, G. A. (2022). Simple Formulae, Deep Learning and Elaborate Modelling for the COVID-19 Pandemic. Encyclopedia, 2(2), 679-689. https://doi.org/10.3390/encyclopedia2020047









