Abstract
Background: Antigen tests for SARS-CoV-2 testing are rapid and inexpensive but usually have lower sensitivity than RT-qPCR and are only validated for nasopharyngeal/throat swabs; the latter are considered the gold standard in terms of material collection but are not tolerated by patients with frequent sampling. The present study, therefore, investigates the extent to which SARS-CoV-2 antigen testing is comparable to RT-qPCR from an easily obtained gargle solution compared to nasopharyngeal swabs. Methods: The performance of a high-quality POC fluorescence immune antigen test in single nasal swab samples and gargle samples compared to RT-qPCR was investigated (total n = 620 samples (gargle samples = 309, and nasal swabs = 311)). Findings: In our setting, the detection of SARS-CoV2 with an antigen test was reliable up to a Ct value of 30 for single nasal swab samples and was reduced to Ct:20 for single gargle samples. The overall antigen-test sensitivity is 83.92% (swab samples) and 75.72% (gargle samples). Interpretation: Antigen tests showed reliable results up to a detection limit of Ct: 30 with only nasal swab samples but not gargle samples. If the use of gargle samples is preferred due to their advantages, such as painless testing, easy handling, and the lack of a need to involve trained personnel for sample taking, reliable results can only be achieved with RT-qPCR.
1. Introduction
At the beginning of the COVID-19 pandemic, only material from nasopharyngeal swabs was recommended for virus detection by the WHO [1]. Later, gargle samples were also shown to be a suitable source for testing; using gargle samples brings different benefits: (a) Its collection is neither painful nor unpleasant and is easy to perform, thus increasing test acceptance [2]; (b) it is safer for healthcare personnel since gargling can be performed without contact by the test persons themselves; (c) when pooling samples to increase test capacity and save resources, gargle samples offer an easier and safer option, since every individual can throw their sample into the pooling container, which also reduces the chance of contamination and mixing samples [3,4,5]. A disadvantage of gargle samples may be that the test sensitivity is lower due to dilution effects caused by the gargle amount [6,7,8].
Although RT-qPCR is the gold standard for detecting SARS-CoV-2 [9], it requires special equipment, skilled laboratory personnel with a background in molecular biology, and at least 3 h of processing time, which are not feasible in all settings, thus limiting its application. In contrast, antigen tests, which are mainly based on fluorescence immunoassays and the detection of a specific nucleocapsid protein derived from SARS-CoV-2 [10], are faster, cheaper, easier to perform, and robust in almost any situation. Numerous antigen and RT-qPCR tests are now on the market, and the most common tests require nasopharyngeal swabs [11].
Here, we evaluate if one could combine the convenience of gargling with the simple testing procedure of antigen tests. Thus, we compared the sensitivity and specificity of a fluorescence-based antigen-test (STANDARD™ F COVID-19 Ag FIA kit (SD BIOSENSOR Inc., Suwon-si, Korea) with those of RT-qPCR, first using nasopharyngeal swabs and, in a second step, gargle samples. Finally, we tested how the gargle samples performed with a selection of other antigen tests and with RT-qPCR pool testing.
2. Methods
2.1. Study Cohort and Sampling
In total, 309 gargle samples and 311 nasal swabs were collected for routine testing in hospitals of the Order of St. John in Regensburg and Straubing, Germany, from October 2019 until April 2020, when the original variant and the Alpha Variant of SARS-CoV-2 were predominant in Germany. Two nasal swabs were collected concomitantly by medical personnel in the emergency room. One nasal swab was transferred immediately to the extraction buffer from a STANDARD™ F COVID-19 Ag FIA kit (SD BIOSENSOR Inc., Suwon, Korea) to be tested with an antigen test, and a second nasal swab from the same patient was transported to the laboratory for RT-PCR testing on the same day for quality control.
Gargle samples were provided by patients and medical students by gargling for approximately 30 s with 10 mL of sterile water (Ampuwa). The recovered gargle fluid of approximately 10 mL was then transferred to a 250 mL container, and antigen against SARS-CoV-2 was analyzed in each gargle sample immediately after sampling. The remaining sample fluid was kept for quality control and RT-qPCR analysis. All samples analyzed here were leftovers from routine testing and were anonymized before the analysis in this study was performed.
2.2. SARS-CoV-2 Antigen Testing Procedures
Gargle and swab samples were tested for SARS-CoV-2 antigen right after sampling. Either the nasal swab or 150 µL of gargle sample was transferred into an extraction buffer tube provided with the STANDARD™ F COVID-19 Ag FIA kit (SD BIOSENSOR Inc., Suwon, Korea), followed by treatment according to the manufacturer’s instructions. Briefly, after closing the buffer tube with the provided nozzle cap, the tube was squeezed 10 times to mix the sample with the extraction buffer. Then, we applied 4 drops of the extracted specimen to the well of the respective test cassette. After 15 min of incubation at room temperature, the test cassette was loaded into the analyzer (SD BIOSENSOR), and the COI as a numerical representation of the measured fluorescence signal was calculated automatically by the analyzer. A COI ≥ 1.0 represents a positive result for SARS-CoV-2 nucleoproteins, according to the manufacturer.
To better evaluate our data from the antigen testing of the gargle, we checked our results against two other antigen tests—one with a similar (76%) and one with a lower sensitivity (36%) [10]: Eight positive PCR gargle samples with increasing Ct values were tested with the CLINITES Rapid COVID-19 Antigen Test (SIEMENS Healthineers., Houston, TX, USA)—a test with similar sensitivity—and with the NADAL COVID-19 Ag test (Ref.243103N-20, nal von minden., Moers, Germany). For both tests, the sample and buffer were mixed at a ratio of 1:1. For the CLINITES, the waiting time was 1 min, and then 4 drops were added, followed by 15 min of incubation time. For the NADAL kit, the waiting time was 2 min after mixing the sample and buffer, and then 2 drops were added into the sample well, followed by an incubation of 15 min.
2.3. RT-qPCR Testing Procedures
To detect SARS-CoV-2 genomic RNA in gargling samples, we performed RNA extraction as a first step by using BEXS Ready Viral DNA/RNA kits (Inno-train Diagnostik, Kronberg, Germany). We added a fixed amount (10 µL per sample) of a 70-base-pair fragment of Equine Arteritis Virus (EAV, TIB Molbiol, Berlin, Germany) as an extraction control to each sample. Then, we conducted one-step RT-qPCR with the LightCycler® Multiplex RNA Virus Master (target E gene) on a Light Cycler 480 II Instrument (Roche Diagnostics). A positive sample was confirmed with a second qualitative test system: the Xpert Xpress™ SARS-CoV-2 assay (cartridge system including an extraction step and amplification targeting the E- and N2-genes) on a GeneXpert instrument (Cepheid, Sunnyvale, CA, USA).
To investigate the detectability of individual positive gargle samples in a standardized gargle pool in our PCR setting, we added 1 mL of each positive sample with different Ct values 20 mL of a negative gargle pool of 20 participants. Then, RNA was extracted from both single and pool samples by using the MagnifiQ™ RNA buffer kit (A&A Biotechnology, Gdansk, Poland)) on an Auto-Pure96 Nucleic Acid Purification System (Hangzhou Allsheng Instruments, Shanghai, China) according to the manufacturer’s protocol. RT-PCR-based SARS-CoV-2 RNA detection was performed on a BIORAD Real-Time PCR System using the single-well dual target (ORF1b and N2 gene). Further information regarding the primer and probe sequences is available in Supplementary Table S1.
To show that the BIORAD and Light Cycler 480 II Instrument (Roche Diagnostics) have very similar sensitivities and specificities for detecting SARS-CoV-2, we ran identical samples on both systems (Supplementary Table S2).
3. Results
We performed antigen tests and RT-qPCR tests by using the LightCycler® Multiplex RNA Virus Master (target E gene—TibMolBiol) on 311 nasal swabs and 309 gargle samples (total n = 620). Out of these, 47 swab samples (Figure 1A) and 64 gargle samples (Figure 1B) were determined to be positive by RT-qPCR (duplicate testing in two test systems) with constant Ct values below 40.
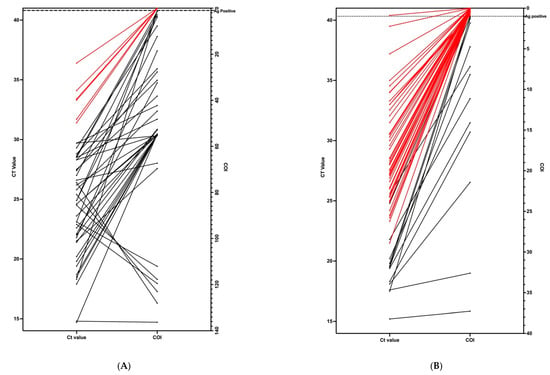
Figure 1.
Correlation between Ct values from RT-qPCR (left side) and COI values from antigen tests (right side). Positive samples that were negative on antigen tests are shown in red. (A) (on the left) shows the results in 47 PCR-positive swab samples. (B) (right one) shows the results in 64 PCR-positive gargle samples.
The false-negative rate of the antigen test—when the antigen test was negative but the RT-qPCR test was positive—was high in gargle samples at 16.18% (50 out of 309), while it was only 1.92% (6 out of 311) in the swab samples. Regarding the false positive rate—when the antigen test was positive, but the RT-qPCR test was negative—it was higher in the swab samples at 11.57% (36 out of 311) than in gargle samples at 8.09% (25 out of 309). In general, the sensitivity of the antigen test was higher in the swab samples (83.92%) than the gargle samples (75.73%). The antigen test’s sensitivity for PCR-positive samples with Ct values of 15–20 was 100% in both swab (n = 8) and gargle (n = 10) samples. In the group of PCR-positive samples with Ct values of 20–25 and 25–30, the antigen test’s efficiency stayed at 100% for the swab samples (n = 17, n = 16), but dropped to 25% (Ct 20–25, n = 12) and 4.1% (Ct 25–30, n = 24) for the gargle samples. Our results show that the antigen test did not detect SARS-CoV-2 in samples with Ct values above 30 in either swab or gargle samples. We had n = 220 antigen-negative results from each of the swab and gargle samples, which we could confirm as negative samples with RT-qPCR.
The other two antigen tests (CLINITEST/NADAL) were tested as indicators for our selected SD BIOSENSOR FIA test; neither of the other two test kits was more appropriate (Table 1). Furthermore, we explored the possibility that a simple dilution effect introduced by using gargle samples may have reduced the sensitivity. Therefore, we calculated the effect of lowering the positive cutoff index in the SD BIOSENSOR test for gargle samples on the sensitivity and specificity of the test results, as shown in Table 2. That procedure did not significantly increase the sensitivity, while the specificity was massively decreased at the same time.

Table 1.
Comparison of different antigen test kits with positive gargle samples.

Table 2.
Comparison of the efficiency of detecting positive gargle samples and the rate of false positives for negative gargle samples by changing the COI.
Based on these test results, we determined that gargle samples cannot easily be used with currently available antigen tests that are licensed for the use with swab samples. To explore if gargle samples could be pooled for PCR testing to bring the costs of testing down to areas similar to those of antigen testing and to explore if pooled gargle samples in combination with PCR testing would have similar or higher detection limits compared to those of antigen tests, we tested if single positive gargle samples of Ct values between 32 and 38 could be detected by PCR testing in standardized gargle pools of 20 + 1 samples. Therefore, we added 1 mL of positive gargle fluid to 20 mL of pooled gargle fluid from negative individuals and performed RNA extraction followed by RT-qPCR, as described for single-sample PCR (Table 3). Indeed, even when pools of 21 were used, positive gargle samples with a set Ct value of up to 35 could be detected in the pools.

Table 3.
Comparison of different RT-qPCR Ct values of the single samples and in the pool of 21 individuals.
4. Discussion
Our data show that swab samples containing viral loads correlating to Ct values of up 30 (in our PCR testing setup) can reliably be detected with high-quality antigen testing. Using 10 mL of gargle samples with the same antigen test reduces the detection limit to Ct values of 20. When gargle samples were analyzed in pools of 21 by PCR, single positive samples with Ct values of up to 35 were reliably detected within the pools.
For our experiment, we used a well-established antigen test that is based on fluorescence detection and provides a numerical output for measurements. That the antigen test by Biosensor Inc. gives reliable results was shown previously in several reports: In a study published in the spring of 2021 on 359 nasopharyngeal swab samples from Italy, the antigen test showed good sensitivity for samples with Ct values lower than 25 [11]. In a further study, the STANDARD Q COVID-19 antigen test showed the sensitivity of 74.4% in 289 swab samples, in which 31 out of 39 positives were detected with the antigen test, and all positive samples had RT-qPCR Ct values lower than 27 [12].
Using gargle samples instead of swabs reduces the detection limit of the antigen test when applied reliably in swab samples by 210 fold. Dilution alone does not seem to be a reasonable explanation for this dramatic change, even though nucleoprotein antigens of SARS-CoV-2 are expected to be more concentrated in a nasal swab than in 10 mL of gargle fluid. If dilution would be the main factor, lowering the COI for determining a positive result could be a possible strategy for adjusting the detection limit. However, lowering the detection limit does not lead to a reasonable increase in sensitivity without an unreasonable decrease in specificity, as shown in Table 2. Therefore, we conclude that the chemistry of antigen tests would have to be specifically designed to work with gargle samples. This is not a specific feature of the test that we used here, but a common feature of antigen tests on the market that are designed to be used with nasal swabs, as we showed in our experiments. When different specimens were compared for antigen testing in another study, no positive gargle samples (n = 7, RT-qPCR Ct value: 26.3–36) were detected by the antigen test, which confirms our data showing that gargle samples with Ct values higher than 20 are not detectable with standard antigen tests. In that study, antigen tests showed a 44.4% sensitivity overall using a nasopharyngeal swab (8 out of 18 samples), and all positive samples found with the antigen test had Ct values lower than 25 [13]. Another study performed in Switzerland among hospitalized patients showed that the sensitivity of the SD Biosensor test for samples from asymptomatic COVID-19 patients was 28–33%, while it dropped to 25% in patients with 5 days of COVID-19 symptoms [14].
Taken together, our study seems to be the largest comparison of gargle versus nasal swab samples analyzed concomitantly with antigen tests and Rt-q PCR.
Simply combining the gargle procedure with current antigen tests failed. Still, it would be desirable to use gargle samples for the reasons mentioned above: It is a painless, safe, and well-accepted procedure that is especially suitable for children and repeated testing. The advantage of combining it with antigen testing would have been the simplicity of antigen testing in the field, the independence from laboratory logistics, and, not to be neglected, the attractive price of antigen tests compared to RT-qPCR. Thus, we investigated if gargle samples could be combined with pool PCR testing as an alternative.
Indeed, our results show that, by using 21 gargle samples in a pool, we are not only able to detect positive samples reliably and with costs comparable to those of antigen testing but can also do so with a significantly increased detection limit. We calculated the costs of our pool PCR to be less than EUR 1 per participating sample, taking material, personnel costs, and logistics into account (data available upon request from the authors). In our setting presented here and in a study on school testing that was recently published [3], we showed that the detection limit for pool testing is a Ct value of 35, increasing detection by a factor of 25 compared to antigen testing. In a pandemic situation, faster detection of positive individuals may be crucial; thus, using gargle pools in combination with RT-qPCR may be the key to successfully breaking infection chains without increasing costs. In situations where laboratory accessibility is limited, the combination of nasal swabs and a high-quality antigen test may be a feasible option.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/covid2060056/s1, Table S1: ORF1b and N2 gene primers and probes sequences information, Table S2: Comparison of sensitivity and specificity of two RT-qPCR settings (ROCHE, and BIORAD).
Author Contributions
Study design: M.K., P.K. and A.A.; Data collection: P.K., N.B., E.C. and T.W.; Laboratory analysis and data interpretation: P.K., N.B., E.C., T.W. and S.N.; Data Analysis: P.K., N.B. and E.C.; Manuscript writing: M.K., P.K. and N.B. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Bavarian State Ministry of Science and Arts (Grant STACADO).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the regional Ethics Committee of the University of Regensburg.
Informed Consent Statement
Patient consent was waived since SARS diagnostic was carried out as part of routine diagnostics in the context of medical care of patients during pandemic.
Conflicts of Interest
All authors declare that they have no competing financial or personal interests.
References
- Wang, W.; Xu, Y.; Gao, R.; Lu, R.; Han, K.; Wu, G.; Tan, W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA 2020, 323, 1843–1844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kocagoz, T.; Can, O.; Uyar, N.Y.; Aksoy, E.; Polat, T.; Cankaya, D.; Karakus, B.; Mozioglu, E.; Kocagoz, S. Simple concentration method enables the use of gargle and mouthwash instead of nasopharyngeal swab sampling for the diagnosis of COVID-19 by PCR. Eur. J. Clin. Microbiol. 2021, 40, 2617–2622. [Google Scholar] [CrossRef] [PubMed]
- Kheiroddin, P.; Schöberl, P.; Althammer, M.; Cibali, E.; Würfel, T.; Wein, H.; Kulawik, B.; Buntrock-Döpke, H.; Weigl, E.; Gran, S.; et al. Results of WICOVIR Gargle Pool PCR Testing in German Schools Based on the First 100,000 Tests. Front. Pediatr. 2021, 9, 721518. [Google Scholar] [CrossRef] [PubMed]
- Malecki, M.; Lüsebrink, J.; Teves, S.; Wendel, A.F. Pharynx gargle samples are suitable for SARS-CoV-2 diagnostic use and save personal protective equipment and swabs. Infect. Control Hosp. Epidemiol. 2021, 42, 248–249. [Google Scholar] [CrossRef] [PubMed]
- Zander, J.; Scholtes, S.; Ottinger, M.; Kremer, M.; Kharazi, A.; Stadler, V.; Hauck, C.R. Self-Collected Gargle Lavage Allows Reliable Detection of SARS-CoV-2 in an Outpatient Setting. Microbiol. Spectr. 2021, 9, e0036121. [Google Scholar] [CrossRef] [PubMed]
- Gobeille Paré, S.; Bestman-Smith, J.; Fafard, J.; Doualla-Bell, F.; Jacob-Wagner, M.; Lavallée, C.; Charest, H.; Beauchemin, S.; Coutlée, F.; Dumaresq, J.; et al. Natural spring water gargle samples as an alternative to nasopharyngeal swabs for SARS-CoV-2 detection using a laboratory-developed test. J. Med. Virol. 2021, 94, 985–993. [Google Scholar] [CrossRef] [PubMed]
- Poukka, E.; Mäkelä, H.; Hagberg, L.; Vo, T.; Nohynek, H.; Ikonen, N.; Dub, T. Detection of SARS-CoV-2 Infection in Gargle, Spit, and Sputum Specimens. Microbiol. Spectr. 2021, 9, e0003521. [Google Scholar] [CrossRef] [PubMed]
- Biber, A.; Lev, D.; Mandelboim, M.; Lustig, Y.; Harmelin, G.; Shaham, A.; Schwartz, E. The role of mouthwash sampling in SARS-CoV-2 diagnosis. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 2199–2206. [Google Scholar] [CrossRef] [PubMed]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Drosten, C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scheiblauer, H.; Filomena, A.; Nitsche, A.; Puyskens, A.; Corman, V.M.; Drosten, C.; Nübling, C.M. Comparative sensitivity evaluation for 122 CE-marked rapid diagnostic tests for SARS-CoV-2 antigen, Germany, September 2020 to April 2021. Eurosurveillance 2021, 26, 2100441. [Google Scholar] [CrossRef] [PubMed]
- Liotti, F.M.; Menchinelli, G.; Lalle, E.; Palucci, I.; Marchetti, S.; Colavita, F.; Posteraro, B. Performance of a novel diagnostic assay for rapid SARS-CoV-2 antigen detection in nasopharynx samples. Clin. Microbiol. Infect. 2021, 27, 487–488. [Google Scholar] [CrossRef]
- Lindner, A.K.; Nikolai, O.; Kausch, F.; Wintel, M.; Hommes, F.; Gertler, M.; Krüger, L.J.; Gaeddert, M.; Tobian, F.; Lainati, F.; et al. Head-to-head comparison of SARS-CoV-2 antigen-detecting rapid test with self-collected nasal swab versus professional-collected nasopharyngeal swab. Eur. Respir. J. 2021, 57, 2003961. [Google Scholar] [CrossRef] [PubMed]
- Yamayoshi, S.; Sakai-Tagawa, Y.; Koga, M.; Akasaka, O.; Nakachi, I.; Koh, H.; Maeda, K.; Adachi, E.; Saito, M.; Nagai, H.; et al. Comparison of Rapid Antigen Tests for COVID-19. Viruses 2020, 12, E1420. [Google Scholar] [CrossRef] [PubMed]
- Coste, A.T.; Egli, A.; Greub, G. Self-testing for SARS-CoV-2: Importance of lay communication. Swiss Med. Wkly. 2021, 151, w20526. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).