Abstract
Despite extensive worldwide vaccination, the current COVID-19 pandemic caused by SARS-CoV-2 continues. The Omicron variant is a recently emerged variant of concern and is now overtaking the Delta variant. To characterize the potential antigenicity of the Omicron variant, we examined the distributions of SARS-CoV-2 nonself mutations (in reference to the human proteome) as five amino acid stretches of short constituent sequences (SCSs) in the Omicron and Delta proteomes. The number of nonself SCSs did not differ much throughout the Omicron, Delta, and reference sequence (RefSeq) proteomes but markedly increased in the receptor binding domain (RBD) of the Omicron spike protein compared to those of the Delta and RefSeq proteins. In contrast, the number of nonself SCSs decreased in non-RBD regions in the Omicron spike protein, compensating for the increase in the RBD. Several nonself SCSs were tandemly present in the RBD of the Omicron spike protein, likely as a result of selection for higher binding affinity to the ACE2 receptor (and, hence, higher infectivity and transmissibility) at the expense of increased antigenicity. Taken together, the present results suggest that the Omicron variant has evolved to have higher antigenicity and less virulence in humans despite increased infectivity and transmissibility.
1. Introduction
Despite worldwide efforts for vaccination, the COVID-19 pandemic still persists as of December 2021, two years after its pathogenic emergence caused by a novel coronavirus, SARS-CoV-2 [1,2,3,4]. Recently, a new variant emerged from South Africa, which was reported on 25 November 2021 [5] and was designated the Omicron variant (B.1.1.529), one of the variants of concern announced by the World Health Organization (WHO) on 26 November 2021 [6]. A risk assessment of the Omicron variant was urgently released on 2 December 2021 [7]. Currently, the Omicron variant is spreading worldwide, including in Denmark [8] and the United States of America [9,10], displacing a previous variant of concern, the Delta variant (B.1.617.2) [6,11]. Prompt characterization of the Omicron variant is of high importance for public health.
Early and preliminary data indicated that the Omicron variant is less virulent but more transmissible than previous variants [12,13,14,15]. Possible functional changes associated with multiple mutations unique to the Omicron genome have been detected and evaluated by several computational studies [16,17,18,19,20,21,22,23,24,25,26,27,28]. Many mutations have been localized in the receptor binding domain (RBD) of the spike (S) protein, possibly contributing to higher affinity to the ACE2 (angiotensin-converting enzyme 2) receptor and lower affinity to pre-existing infection-induced or vaccine-induced antibodies [15,18,19,20,21,22,23,24,25]. An increase in hydrophobic amino acid residues [23] or in electrostatic interactions [26,27] introduced by mutations has been linked to potentially higher infectivity and transmissibility. Computational analyses, including a structural analysis [15], an analysis based on artificial intelligence (AI) trained with numerous experimental data [28], and an analysis with amino acid interaction (AAI) networks [16], have revealed the potential resistance of the Omicron variant against pre-existing antibodies. One of the studies also indicated that infectivity of the Omicron variant is likely to be 2.8-fold higher than that of the Delta variant [28]. Furthermore, lower replication ability has been linked to less severity of the symptoms associated with Omicron [29,30].
According to a report from the United States [10], the average weekly age-standardized incidence of COVID-19 cases in the Delta predominance period was 460.1 and 90.9 in unvaccinated and fully vaccinated persons, respectively, and that in the Omicron emergence period was 725.6 and 230.9, respectively. Clearly, COVID-19 cases increased. In contrast, the average weekly age-standardized incidence of COVID-19-associated deaths in the Delta predominance period was 11.4 and 0.7 in unvaccinated and vaccinated persons, respectively, and the incidence in the Omicron emergence period was 9.7 and 0.5, respectively. Clearly, the incidence of COVID-19-associated deaths decreased. Similarly, an early study from South Africa reported lower clinical severity of the Omicron variant (23.4%) than the previous Delta variant (62.5%) [31,32]. High transmission of the Omicron variant has been demonstrated in a designated quarantine hotel, which challenges the zero COVID policy in Hong Kong [33].
A large-scale SARS-CoV-2 genome analysis suggested that Omicron mutations have been selected for vaccine resistance [34]. In fact, in vitro functional analyses of the Omicron variant have suggested significant escape from pre-existing infection-induced or vaccine-induced antibodies [35,36,37,38]. Thus, vaccines developed before the emergence of the Omicron variant may be less effective, but pre-existing T cell immunity may still be effective [39,40,41], and booster shots appear to reactivate the immune system by increasing neutralizing antibodies [10,41]. The evasion of immunity induced by the previous vaccination or infection by the Omicron variant may be explained by structural changes in the Omicron spike proteins due to accumulated mutations [42,43,44,45]. Structural analyses also revealed that the binding affinity of the Omicron spike protein to ACE2 receptor has been enhanced or maintained despite multiple mutations [42,43,44,45].
These studies have already addressed some of the concerns associated with the Omicron variant, but alternative and complementary methods to further characterize new variants will also be helpful. Here, we employed a novel method simply based on amino acid sequences of SARS-CoV-2 and human proteomes to evaluate the antigenicity of the Omicron variant. This method is based on self/nonself identification of SARS-CoV-2 amino acid sequences with respect to human amino acid sequences.
The human immune system recognizes foreign proteins based on short amino acid sequences presented as peptides by MHC (major histocompatibility complex) molecules [46,47,48,49]. The basis of self/nonself discrimination is antigen presentation of peptide-MHC complexes by antigen presenting cells (APCs) and their recognition via a diverse repertoire of T cell receptors (TCRs) by T cells [49,50]. Traditionally, it has been postulated that T cells with high-affinity TCRs to the presented self-peptides are eliminated during the period of T cell education in the thymus [50,51]. This postulate can be translated into the following research idea. In the present study, a stretch of amino acid sequence, when it exists as a part of a protein, is called a short constituent sequence (SCS; pronounced as [es/si/es] or [ʃɔks] like shocks but may also be pronounced as [sɔks] like socks or [sæks] like sax). Operationally speaking, the human immune system stores memories of all possible SCSs from the human proteome, which are here called self SCSs. Every peptide presented by MHC molecules is collated with a dataset of self SCSs. When a sequence of a given peptide presented by MHC molecules is found in the dataset, it is recognized as “self”, a part of a human body. In contrast, when a sequence of a peptide is not found in the dataset, it is recognized as “nonself”, a foreign object to be eliminated by the immune system. These SCSs in a protein are called nonself SCSs, and they are antigenic by definition, although their antigenicity in vitro and in vivo should be determined experimentally. When the quantity of a given peptide sequence increases rapidly to emergency levels when a microbe or virus infects a human body, the immune system likely produces antibodies against it even if it is found in a dataset. However, self SCSs may still be less antigenic than nonself SCSs.
These theoretical considerations are focused on linear epitopes and cannot readily account for conformational epitopes, but combinatorial sets of linear epitopes may later be taken into account. At present, with such a potential limitation, we reasoned that the relative abundance of self and nonself SCSs in a virus can be used as an indicator to evaluate the antigenicity and thus the virulence of that virus because a nonself SCS is more antigenic than a self SCS for the host immune system to avoid autoimmunity. When the number of nonself SCSs increases in a viral proteome during evolution, this means that the virus is more discoverable by the host immune system because nonself SCSs can be easily recognized as foreign objects. As a result, viral virulence may decrease.
Although protein analysis methods based on SCSs have been developed since 2005 [52,53,54,55,56,57,58], we introduced the self/nonself concept in SCS-based analysis for the first time in a previous study [59]. In that study, we showed that when using five amino acid stretches as SCS units, most SCSs in the SARS-CoV-2 proteome are self SCSs in reference to the human proteome [59]. In other words, nonself SCSs are scattered in a sea of self SCSs. We also discovered nonself SCS clusters in the spike protein that may serve as excellent candidate epitopes for vaccines that efficiently induce immunity without inducing autoimmunity [59]. Importantly, this finding has been supported by experimental determination of SARS-CoV-2 epitopes [60,61,62,63,64,65,66]; the antigenicity of the nonself SCS clusters has been validated experimentally [59]. In the present study, we attempted to develop a simple method for predicting antigenicity based on self/nonself SCSs to understand the relationship between the Omicron variant and the human immune system.
2. Materials and Methods
The human reference proteome and the SARS-CoV-2 proteome sequences were obtained from NCBI (the National Center for Biotechnology Information, Bethesda, MD, USA) as described in Otaki et al., (2021) [59]. In addition to the SARS-CoV-2 proteome reference sequence (ASM985889v3), the Delta and Omicron variant proteomes were obtained similarly (accessed on 30 November 2021 and 15 December 2021). As of 30 November 2021, there were three proteomes of the Omicron variants available at NCBI: OL698718, OL672836, and OL677199. One of them (OL698718) contained numerous X (unknown) amino acids and thus was excluded from the analysis. Because two of them (OL672836 and OL677199) had completely identical sequences, as confirmed with SIM [67] (https://web.expasy.org/sim/sim_notes.html; accessed on 27 December 2021) at Expasy (Swiss Bioinformatics Resource Portal, operated by the SIB Swiss Institute of Bioinformatics, Lausanne, Switzerland), we focused on OL672836. The Delta variant used here was OL822485, one of the most recent Delta variant proteomes available at the time of data extraction.
SCSs (containing five amino acids) were extracted from the SARS-CoV-2 proteome by sliding one amino acid residue at a time from the N-terminus to the C-terminus. All SARS-CoV-2 SCSs were categorized into either self (existent in the human reference proteome) or nonself (nonexistent in the human reference proteome) SCSs as described in Otaki et al., (2021) [59]. We assigned each SCS in the SARS-CoV-2 proteome a 0 (self; invisible from the host immune system) or 1 (nonself; visible from the host immune system) at the first position of its amino acid in a protein sequence [59]. The numbers of nonself SCSs were counted and assigned in sequence maps manually based on the self (0) or nonself (1) assignments calculated by our program and exported into Microsoft Excel (Supplementary Files S1 and S2). For analyses, ORF7b was excluded because some proteome files did not show this protein as being translated. ORF1a was also excluded because its sequence was completely redundant with that of ORF1ab.
3. Results
We first characterized the numbers of nonself SCSs in the proteomes of the Delta and Omicron variants in comparison with that in the RefSeq (reference sequence) proteome. The numbers of nonself SCSs in the proteomes of the Delta and Omicron variants did not differ much from those in the RefSeq proteome (Table 1). The number of nonself SCSs in the Delta variant proteome increased by just one, and the number of nonself SCSs in the Omicron variant proteome decreased by just one, although these data did not indicate that there were no self/nonself status changes in these variants. The increase or decrease in the number of nonself SCSs in each open reading frame (ORF) or proteome, ΔN, in reference to RefSeq was not remarkable, mostly either 0 or ±1.

Table 1.
Number of nonself SCSs in the SARS-CoV-2 reference sequence (RefSeq) and in the Delta and Omicron variants.
We next focused on the spike protein. Nonself SCSs were not concentrated in the receptor binding domain (RBD) of the spike protein of the Delta variant (Figure 1). In contrast, many nonself SCSs were localized in the RBD of the spike protein of the Omicron variant (Figure 2). Focusing on the RBD, the Delta variant had two nonself SCSs created by mutations (self-to-nonself status changes), and no nonself SCSs found in RefSeq disappeared (nonself-to-self status changes) (Figure 1 and Figure 3). Thus, the net increase in nonself SCSs was +2. In contrast, the Omicron variant had seven nonself SCSs (due to the following seven mutations, G339D, S375F, S477N, T478K, Q498R, N501Y, and Y505H, among 15 mutations), and three nonself SCSs found in RefSeq disappeared (Figure 2 and Figure 3). Thus, the net increase in nonself SCSs was +4.
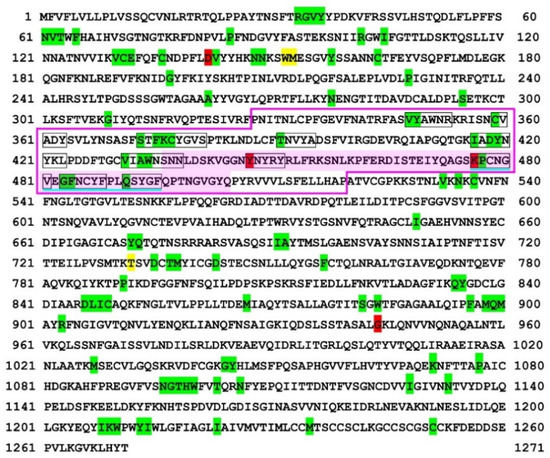
Figure 1.
Self/nonself mapping of the SARS-CoV-2 spike protein of the Delta variant. The first amino acids of nonself SCSs are indicated by green (present in RefSeq) or red (new in this variant) shading. The first amino acids of new self SCSs in this variant (not present in RefSeq) are indicated by yellow shading. Other SCSs with no such indications are self SCSs. The receptor binding domain (RBD) [60] is boxed in pink lines. The receptor binding motif (RBM) [61,68] is shaded in pink. The nonself SCSs in the RBM are boxed in black lines. A potentially important nonself SCS region for vaccine development in the RBD is underlined in blue. For self/nonself mapping of RefSeq, see Otaki et al., (2021) [59].
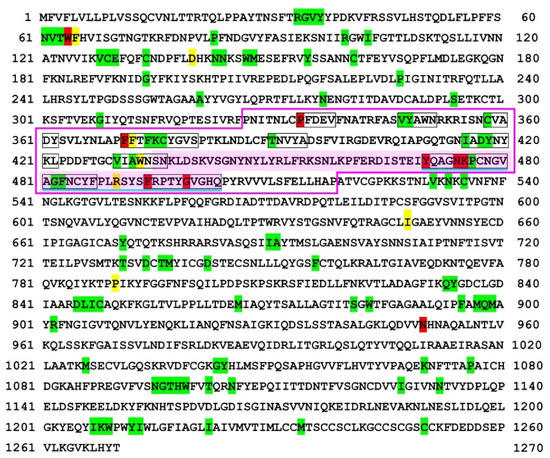
Figure 2.
Self/nonself mapping of the SARS-CoV-2 spike protein of the Omicron variant. For colors and underlines, see legend in Figure 1. The nonself SCS at 63, TWFHV, is produced from TWFHA in RefSeq but without a status change and, hence, is shaded in green despite its amino acid change.
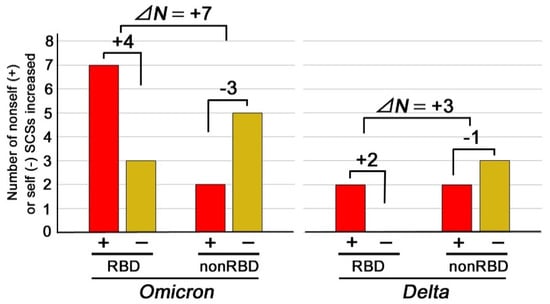
Figure 3.
The number of nonself (+) and self (−) SCSs increased in the RBD or non-RBD regions in the Omicron and Delta variants. Newly emerged nonself SCSs (self-to-nonself status changes) are shown in red bars, whereas newly emerged self SCSs (nonself-to-self status changes) are shown in yellow bars. Note the difference between RBD and non-RBD and the difference between the Omicron and Delta variants. ΔN in this figure indicates the differences in the net changes between the RBD and non-RBD regions of the Omicron or Delta variant.
Interestingly, this tendency of an increase in the number of nonself SCSs in the RBD was not observed in the non-RBD regions. Instead, the net changes in the number of nonself SCSs in the non-RBD region of the Omicron and Delta variants were −3 and −1, respectively (Figure 3). The differences in the net changes between the RBD and non-RBD regions of the Omicron and Delta variants, ΔN, were +7 and +3, respectively (Figure 3). In both variants, an increase in the number of nonself SCSs in the RBD was compensated for by a decrease in the number of nonself SCSs (an increase in the number of self SCSs) in the non-RBD regions (Figure 3), resulting in a net spike change of just +1 (Table 1, Figure 3).
Notably, in the receptor binding motif (RBM) of the Omicron variant, three novel nonself SCSs (YQAGN, NKPCN, and KPCNG) were localized immediately at the N-terminal side of the potential vaccine epitope identified in a previous study [59], extending the epitope region toward the N-terminal side (Figure 2). Furthermore, two novel nonself SCSs (FRPTY and GVGHQ) were localized immediately at the C-terminal side of the potential epitope, extending the epitope region toward the C-terminal side up to the end of the RBM (Figure 2). On the other hand, one nonself SCS (QSYGF) found in RefSeq and the Delta variant disappeared (Figure 1 and Figure 2). Together, in a 34 amino acid stretch of the C-terminal end of the RBM, eight nonself SCSs (YQAGN, NKPCN, KPCNG, PCNGV, GFNCY, FNCYF, FRPTY, and GVGHQ) were concentrated tandemly.
4. Discussion
In the present study, we reasoned that the number of nonself SCSs in a proteome or in a protein can impact its antigenicity. We further reasoned that antigenicity is probably reflected directly in the virulence of the virus. Not only the number but also the locations of nonself SCSs in a protein (e.g., in a binding domain, at an active site, or in proximity to one another) would matter to determine the antigenicity of nonself SCSs. Based on the above logic, we endeavoured to urgently characterize the Omicron variant of SARS-CoV-2. Although immunological validity should be experimentally evaluated in further studies in vitro and in vivo, this simple in silico study proposes a novel method of predicting viral virulence based on the amino acid sequences of viral proteins. Accordingly, this study did not produce much data (in comparison to other bioinformatics and genomics studies) but did illustrate a new approach, which is often dismissed but very important in current biology research [69].
Our first results on the Omicron variant were obtained immediately after the emergence of the variant and were released as of 3 January 2022 [70]. At that time, only preliminary data were presented about this variant in the literature database. As of February 2022, many studies on the Omicron variant have been published, as described in the Introduction section. These studies confirmed diminished virulence of the Omicron variant, which is reported previously [10,11,12,13] and largely consistent with the current study, although the underlying reasons for this lower virulence appear to be different between the present and previous reports [24,25,26,27,28,29,30]. Several studies have pointed out slow replication and cellular infection processes for less virulence [29,30]. Our study may be unique in that it focused on the antigenicity of amino acid sequences in proteins.
Theoretically, a virus evolves under selection pressure for higher infectivity and transmissibility, and in this evolutionary process, the amino acid residues of the binding site for its receptor are mutated for higher affinity, as in the case of the Omicron variant [23,24,25,26,27,28]. This type of evolution is here called “offensive” evolution. Offensive evolution may attenuate when any functional disadvantage unavoidably associated with excessive mutations for higher affinity overwhelms the mutational functional advantage. Offensive evolution may also attenuate when nonself SCSs accumulate for higher antigenicity.
Alternatively, a virus theoretically evolves for higher sequence mimicry to the host SCS repertoire defined by the host proteome [59], and in this evolutionary process, nonself SCSs decrease in number for lower antigenicity. This type of evolution is here called “defensive” evolution. Defensive mimicry for host SCSs by increasing self SCSs is advantageous for viruses because they cannot be readily detected by the host immune system. Excessive mimicry evolution may also be attenuated due to an unavoidable functional compromise.
Both offensive and defensive evolutionary scenarios are selectively advantageous for a virus. Given a sufficient period of time, an equilibrium state between offensive and defensive evolution may be established through a functional compromise of a virus. However, in reality, a virus may evolve within a limited period (i.e., replications). Either offensive or defensive evolution may settle at local maxima of survival, depending on various environmental and host conditions. The Omicron variant has clearly taken an offensive evolutionary route, judging from the high-affinity mutations [23,24,25,26,27,28], and might have settled tentatively at a local maximum.
It seems that for the Omicron variant, the selection pressure for higher binding affinity to ACE2 (i.e., higher infectivity and transmissibility) is so large that an increase in the number of nonself SCSs in the RBD was unavoidably allowed despite its immunological disadvantage for the virus. In other words, an increase in nonself SCSs in the RBD likely means a compromise of the virus, which probably indicates low virulence. In support of this view, nonself mutations in the RBD appear to contribute directly to higher affinity to ACE2 [28].
Based on the idea that SCS frequencies in proteomes are related to the phylogenetic and parasitic status of organisms [71,72], we speculated previously that the SARS-CoV-2 proteome will eventually evolve to accumulate self SCSs to defray molecular and cellular attack from the human immune system [59]. However, this defensive evolution based on the sequence mimicry hypothesis was not detected clearly in the Omicron and Delta variants. An increase in self SCS was detected only in the non-RBD regions in the Omicron and Delta variants.
We discovered that the Omicron variant has accumulated self-to-nonself status change mutations at the RBD of the spike protein. The Omicron mutant had seven new nonself SCSs in the RBD, in contrast to just two additions in the Delta variant. The net increase in nonself SCSs within the RBD was +4 in the Omicron variant, in contrast to +2 in the Delta variant. This increase is likely immunologically significant, considering that the RBD is readily accessible to other proteins, such as ACE2, antibodies, and T cell receptors [24,25,39,40,41,42,43,44,45]. Interestingly, there were just two newly added nonself SCSs in the non-RBD regions in both the Omicron and Delta variants, but in the Omicron variant, more new self SCSs were introduced in the non-RBD regions than in the non-RBD regions in the Delta variant and the RBD of the Omicron variant. This increase in self SCSs in the non-RBD regions in the Omicron variant may compensate for the unavoidable increase in nonself SCSs (hence, the unavoidable increase in antigenicity) in the RBD to make the net nonself SCS increase as small as possible and, thus, to make an overall degree of the antigenicity increase as small as possible. Indeed, the differences in nonself SCS changes between the RBD and non-RBD regions, ΔN, were +7 and +3 in the Omicron and Delta variants, respectively.
In addition to the above discussion on the number of nonself SCSs, the location of nonself SCSs within the RBM in the RBD warrants further discussion. We discovered a candidate stretch of amino acids that contained nonself SCSs within the RBM of the RefSeq spike protein, a potential epitope for vaccine development to avoid vaccine-induced autoimmunity [37]. Interestingly, in the Omicron variant but not in the Delta variant, three additional nonself SCSs (YQAGN, NKPCN, and KPCNG) are present at the N-terminal side of the potential epitope region, and furthermore, two additional nonself SCSs (FRPTY and GVGHQ) are present at the C-terminal side, although one nonself SCS in RefSeq is not present. Together, the candidate epitope region in RefSeq has been extended to both the N-terminal and C-terminal sides. Because this region in the RBM is important for ACE2 binding, the expansion of this nonself SCS epitope region here was probably unavoidable to increase the binding affinity for ACE2 at the expense of an increase in antigenicity in the RBM. This result supports the previous finding that this region is likely a good (probably the best) epitope candidate for vaccine development [59].
At first glance, the present results may not seem to be consistent with recent studies on the Omicron variant, which suggested possible immune escape based on mutations for higher affinity to ACE2 and for lower affinity to pre-existing neutralizing antibodies [23,24,25,26,27,28,34,35,36,37,38]. These studies evaluated whether the pre-existing immunological memory induced by previous vaccines or infection continues to be effective against the Omicron variant. In contrast, in the present study, the antigenicity of the Omicron variant itself was evaluated. The present study suggests that the Omicron variant has reduced virulence because of its relatively high antigenicity in the RBD and may not cause severe symptoms. However, due to its high infectivity and transmissibility, the Omicron variant should still be regarded with alarm.
Antigenicity defined by nonself SCSs is just a single factor to explain virulence. A virus with low virulence might preferentially affect tissues/organs such as the digestive tract and testes, leading to nonlife-threatening but long-term effects, rather than tissues/organs such as the lungs, the effects of which could be life-threatening. For example, infection may cause infertility if testicular cells expressing ACE2 are preferentially infected. Moreover, due to high infectivity and transmissibility, even if virulence is low, the absolute number of COVID-19 cases may not decrease in the Omicron surge in comparison with the Delta surge. Therefore, the Omicron variant should be considered highly threatening in terms of public health.
The frequency of random mutations is directly proportional to the number of replications (i.e., the number of infected people), but selection pressure shapes the direction of viral evolution. The higher binding affinity to ACE2 and higher antigenicity together caused by the accumulated mutations in the Omicron variant suggest that a hampered transmission state despite a large number of replications was a driving force for the evolution of the Omicron variant. Ironically, such an unnatural state was created by worldwide vaccination, which might have helped the emergence of the Omicron variant, as suggested by a recent study [34].
5. Conclusions
Through an application of the SCS concept to the human-SARS-CoV-2 system together with immunological self/nonself considerations, in the present study, a novel method was used to characterize the Omicron and Delta variants. We found that nonself SCSs increased in the RBD of the Omicron spike protein, which probably results in faster identification and elimination by the immune system because nonself SCSs are more antigenic than self SCSs. Considering that the Omicron spike protein binds to ACE2 with higher affinity [23,24,25,26,27,28], it appears that the Omicron variant increased its infectivity and transmissibility at the expense of higher antigenicity and lower virulence. Thus, the symptoms of individuals infected with the Omicron variant may be less severe than those of individuals infected with the Delta variant. We also confirmed that a specific stretch in the RBM is a good candidate epitope for vaccines. Since SARS-CoV-2 evolution might have been driven by selection pressure imposed by worldwide vaccination, vaccination-focused strategies against the Omicron variant may further enhance a current mode of evolution. Alternatives to vaccination-focused strategies [73,74,75,76,77,78,79] may also be useful. Continuous caution regarding the Omicron variant is necessary. We further envision that the present study may open a way to clarify a mystery of self/nonself discrimination in the human immune system.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/covid2030029/s1. Supplementary File S1: Omicron OL672836 self/nonself assignment (Excel file) and Supplementary File S2: Delta OL822485 self/nonself assignment (Excel file).
Author Contributions
Conceptualization, J.M.O.; methodology, W.N. and M.N.; software, W.N. and M.N.; validation, J.M.O.; formal analysis, J.M.O. and W.N.; investigation, J.M.O., W.N. and M.N.; resources, W.N. and M.N.; data curation, J.M.O., W.N. and M.N.; writing—original draft preparation, J.M.O.; writing—review and editing, J.M.O.; visualization, J.M.O.; supervision, J.M.O. and M.N.; project administration, J.M.O.; funding acquisition, J.M.O. and M.N. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by basic research funds to J.M.O. and M.N. from the University of the Ryukyus. The APC was funded by a basic research fund from the Faculty of Science, University of the Ryukyus.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data that support the conclusions of the study are included in this paper and the related Supplementary Information. The source codes for human SCS analysis and for SARS-CoV-2 SCS analysis are freely available at https://adslab-uryukyu.github.io/scs-sars-cov-2/ (accessed on 4 October 2021).
Acknowledgments
The authors would like to acknowledge Tatsuya Ishibashi for confirming self/nonself assignments using an independent program of his own and Hideo Yamasaki, Wataru Taira, and other laboratory members of the BCPH Unit of Molecular Physiology for discussion.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Tan, Y.; Ling, Y.; Lu, G.; Liu, F.; Yi, Z.; Jia, X.; Wu, M.; Shi, B.; Xu, S.; et al. Viral and host factors related to the clinical outcome of COVID-19. Nature 2020, 583, 437–440. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Liu, Z.; Chen, Z.; Huang, X.; Xu, M.; He, T.; Zhang, Z. The establishment of reference sequence for SARS-CoV-2 and variation analysis. J. Med. Virol. 2020, 92, 667–674. [Google Scholar] [CrossRef]
- South African Institute for Communicable Diseases (NICD) Division of the National Health Laboratory Service. New COVID-19 Variant Detected in South Africa. Available online: https://www.nicd.ac.za/new-covid-19-variant-detected-in-south-africa/ (accessed on 20 December 2021).
- World Health Organization. Tracking SARS-CoV-2 Variants. Updated on 13 December 2021. Available online: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ (accessed on 20 December 2021).
- European Centre for Disease Prevention and Control (ECDC). Threat Assessment Brief: Implications of the Further Emergence and Spread of the SARS CoV 2 B.1.1.529 Variant of Concern (Omicron) for the EU/EEA First Update. Available online: https://www.ecdc.europa.eu/en/publications-data/covid-19-threat-assessment-spread-omicron-first-update (accessed on 20 December 2021).
- Espenhain, L.; Funk, T.; Overvad, M.; Edslev, S.M.; Fonager, J.; Ingham, A.C.; Rasmussen, M.; Madsen, S.L.; Espersen, C.H.; Sieber, R.N.; et al. Epidemiological characterisation of the first 785 SARS-CoV-2 Omicron variant cases in Denmark, December 2021. Eurosurveillance 2021, 26, 2101146. [Google Scholar] [CrossRef]
- CDC COVID-19 Response Team. SARS-CoV-2 B.1.1.529 (Omicron) Variant—United States, December 1–8, 2021. MMWR Morb. Mortal. Wkly. Rep. 2021, 70, 1731–1734. [Google Scholar] [CrossRef]
- Johnson, A.G.; Amin, A.B.; Ali, A.R.; Hoots, B.; Cadwell, B.L.; Arora, S.; Avoundjian, T.; Awofeso, A.O.; Barnes, J.; Bayoumi, N.S.; et al. COVID-19 Incidence and Death Rates Among Unvaccinated and Fully Vaccinated Adults with and Without Booster Doses During Periods of Delta and Omicron Variant Emergence—25 U.S. Jurisdictions, April 4–December 25, 2021. MMWR Morb. Mortal. Wkly. Rep. 2022, 71, 132–138. [Google Scholar] [CrossRef]
- Viana, R.; Moyo, S.; Amoako, D.G.; Tegally, H.; Scheepers, C.; Althaus, C.L.; Anyaneji, U.J.; Bester, P.A.; Boni, M.F.; Chand, M.; et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature 2022. [Google Scholar] [CrossRef]
- Christie, B. Covid-19: Early studies give hope omicron is milder than other variants. BMJ 2021, 375, n3144. [Google Scholar] [CrossRef]
- Mahase, E. Covid-19: Hospital admission 50-70% less likely with omicron than delta, but transmission a major concern. BMJ 2021, 375, n3151. [Google Scholar] [CrossRef]
- Ingraham, N.E.; Ingbar, D.H. The omicron variant of SARS-CoV-2: Understanding the known and living with unknown. Clin. Transl. Med. 2021, 11, e685. [Google Scholar] [CrossRef]
- Kannan, S.R.; Spratt, A.N.; Sharma, K.; Chand, H.S.; Byrareddy, S.N.; Singh, K. Omicron SARS-CoV-2 variant: Unique features and their impact on pre-existing antibodies. J. Autoimmun. 2021, 126, 102779. [Google Scholar] [CrossRef]
- Miller, N.L.; Clark, T.; Raman, R.; Sasisekharan, R. Insights on the mutational landscape of the SARS-CoV-2 Omicron variant. bioRxiv 2021. [Google Scholar] [CrossRef]
- Kandeel, M.; Mohamed, M.E.M.; Abd El-Lateef, H.M.; Venugopala, K.N.; El-Beltagi, H.S. Omicron variant genome evolution and phylogenetics. J. Med. Virol. 2021, 94, 1627–1632. [Google Scholar] [CrossRef]
- Ren, S.-Y.; Wang, W.-B.; Gao, R.-D.; Zhou, A.-M. Omicron variant (B.1.1.529) of SARS-CoV-2: Mutation, infectivity, transmission, and vaccine resistance. World J. Clin. Cases 2022, 10, 1–11. [Google Scholar] [CrossRef]
- Kannan, S.; Shaik Syed Ali, P.; Sheeza, A. Omicron (B.1.1.529)—Variant of concern—Molecular profile and epidemiology: A mini review. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 8019–8022. [Google Scholar] [CrossRef]
- Araf, Y.; Akter, F.; Tang, Y.; Fatemi, R.; Parvez MS, A.; Zheng, C.; Hossain, M.G. Omicron variant of SARS-CoV-2: Genomics, transmissibility, and responses to current COVID-19 vaccines. J. Med. Virol. 2022. [Google Scholar] [CrossRef]
- Wang, L.; Cheng, G. Sequence analysis of the emerging SARS-CoV-2 variant Omicron in South Africa. J. Med. Virol. 2021, 94, 1728–1733. [Google Scholar] [CrossRef]
- Saxena, S.; Kumar, S.; Ansari, S.; Paweska, J.T.; Maurya, V.K.; Tripathi, A.K.; Abdel-Moneim, A. Characterization of the novel SARS-CoV-2 Omicron (B.1.1.529) Variant of Concern and its global perspective. J. Med. Virol. 2021, 94, 1738–1744. [Google Scholar] [CrossRef]
- Kumar, S.; Thambiraja, T.S.; Karuppanan, K.; Subramaniam, G. Omicron and Delta variant of SARS-CoV-2: A comparative computational study of spike protein. J. Med. Virol. 2021, 94, 1641–1649. [Google Scholar] [CrossRef]
- Shah, M.; Woo, H.G. Omicron: A heavily mutated SARS-CoV-2 variant exhibits stronger binding to ACE2 and potently escapes approved COVID-19 therapeutic antibodies. Front. Immunol. 2022, 12, 830527. [Google Scholar] [CrossRef] [PubMed]
- Omotuyi, O.; Olubiyi, O.; Nash, O.; Afolabi, E.; Oyinloye, B.; Fatumo, S.; Femi-Oyewo, M.; Bogoro, S. SARS-CoV-2 Omicron spike glycoprotein receptor binding domain exhibits super-binder ability with ACE2 but not convalescent monoclonal antibody. Comput. Biol. Med. 2022, 142, 105226. [Google Scholar] [CrossRef] [PubMed]
- Pawłowski, P. SARS-CoV-2 variant Omicron (B.1.1.529) is in a rising trend of mutations increasing the positive electric charge in crucial regions of the spike protein S. Acta Biochim. Pol. 2021, 69, 263–264. [Google Scholar] [CrossRef]
- Pascarella, S.; Ciccozzi, M.; Bianchi, M.; Benvenuto, D.; Cauda, R.; Cassone, A. The electrostatic potential of the Omicron variant spike is higher than in Delta and Delta-plus variants: A hint to higher transmissibility? J. Med. Virol. 2021, 94, 1277–1280. [Google Scholar] [CrossRef]
- Chen, J.; Wang, R.; Gilby, N.B.; Wei, G.-W. Omicron (B.1.1.529): Infectivity, vaccine breakthrough, and antibody resistance. arXiv 2021, arXiv:2112.01318v1. [Google Scholar] [CrossRef]
- Shuai, H.; Chan, J.F.-W.; Hu, B.; Chai, Y.; Yuen, T.T.-T.; Yin, F.; Huang, X.; Yoon, C.; Hu, J.-C.; Liu, H.; et al. Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature 2022. [Google Scholar] [CrossRef]
- Zhao, H.; Lu, L.; Peng, Z.; Chen, L.-L.; Meng, X.; Zhang, C.; lp, J.D.; Chan, W.-M.; Chu, A.W.-H.; Chan, K.-H.; et al. SARS-CoV-2 Omicron variant shows less efficient replication and fusion activity when compared with Delta variant in TMPRSS2-expressed cells. Emerg. Microbes Infect. 2022, 11, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Wolter, N.; Jassat, W.; Walaza, S.; Welch, R.; Moultrie, H.; Groome, M.; Amoako, D.G.; Everatt, J.; Bhiman, J.N.; Scheepers, C.; et al. Early assessment of the clinical severity of the SARS-CoV-2 omicron variant in South Africa: A data linkage study. Lancet 2022, 399, 437–446. [Google Scholar] [CrossRef]
- Nealon, J.; Cowling, B.J. Omicron severity: Milder but not mild. Lancet 2022, 399, 412–413. [Google Scholar] [CrossRef]
- Wong, S.-C.; Au, A.K.-W.; Chen, H.; Yuen, L.L.-H.; Li, X.; Lung, D.C.; Chu, A.W.-H.; Ip, J.D.; Chan, W.-M.; Tsoi, H.-W.; et al. Transmission of Omicron (B.1.1.529)—SARS-CoV-2 Variant of Concern in a designated quarantine hotel for travelers: A challenge of elimination strategy of COVID-19. Lancet Reg. Health West Pac. 2022, 18, 100360. [Google Scholar] [CrossRef]
- Wang, R.; Chen, J.; Wei, G.-W. Mechanisms of SARS-CoV-2 evolution revealing vaccine-resistant mutations in Europe and America. J. Phys. Chem. Lett. 2021, 12, 11850–11857. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, Q.; Liang, Z.; Li, T.; Liu, S.; Cui, Q.; Nie, J.; Wu, Q.; Qu, X.; Huang, W.; et al. The significant immune escape of pseudotyped SARS-CoV-2 variant Omicron. Emerg. Microbes Infect. 2022, 11, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Cele, S.; Jackson, L.; Khoury, D.; Khan, K.; Moyo-Gwete, T.; Tegally, H.; San, J.E.; Cromer, D.; Scheepers, C.; Amoako, D.; et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature 2021, 602, 654–656. [Google Scholar] [CrossRef] [PubMed]
- Sievers, B.; Chakraborty, S.; Xue, Y.; Gelbart, T.; Gonzalez, J.C.; Cassidy, A.G.; Golan, Y.; Prahl, M.; Gaw, S.L.; Arunachalam, P.S.; et al. Antibodies elicited by SARS-CoV-2 infection or mRNA vaccines have reduced neutralizing activity against Beta and Omicron pseudoviruses. Sci. Transl. Med. 2022, 14, eabn7842. [Google Scholar] [CrossRef]
- VanBlargan, L.A.; Errico, J.M.; Halfmann, P.J.; Zost, S.J.; Crowe, J.E., Jr.; Purcell, L.A.; Kawaoka, Y.; Corti, D.; Fremont, D.H.; Diamond, M.S. An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibody. Nat. Med. 2022. [Google Scholar] [CrossRef]
- Redd, A.D.; Nardin, A.; Kared, H.; Bloch, E.M.; Abel, B.; Pekosz, A.; Laeyendecker, O.; Fehlings, M.; Quinn, T.C.; Tobian, A.R.R. Minimal cross-over between mutations associated with Omicron variant of SARS-CoV-2 and CD8+ T cell epitopes identified in COVID-19 convalescent individuals. bioRxiv 2021. [Google Scholar] [CrossRef]
- Ahmed, A.F.; Quadeer, A.A.; McKay, M.R. SARS-CoV-2 T cell responses elicited by COVID-19 vaccines or infection are expected to remain robust against Omicron. Viruses 2022, 14, 79. [Google Scholar] [CrossRef]
- Minka, S.O.; Minka, F.H. A tabulated summary of the evidence on humoral and cellular responses to the SARS-CoV-2 Omicron VOC, as well as vaccine efficacy against this variant. Immunol. Lett. 2022, 243, 38–43. [Google Scholar] [CrossRef]
- Han, P.; Li, L.; Liu, S.; Wang, Q.; Zhang, D.; Xu, Z.; Han, P.; Li, X.; Peng, Q.; Su, C.; et al. Receptor binding and complex structures of human ACE2 to spike RBD from omicron and delta SARS-CoV-2. Cell 2022, 185, 630–640.e10. [Google Scholar] [CrossRef]
- McCallum, M.; Czudnochowski, N.; Rosen, L.E.; Zepeda, S.K.; Bowen, J.E.; Walls, A.C.; Hauser, K.; Joshi, A.; Stewart, C.; Dillen, J.R.; et al. Structural basis of SARS-CoV-2 Omicron immune evasion and receptor engagement. Science 2022, 375, 864–868. [Google Scholar] [CrossRef]
- Mannar, D.; Saville, J.W.; Zhu, X.; Srivastava, S.S.; Berezuk, A.M.; Tuttle, K.S.; Marquez, A.C.; Sekirov, I.; Subramaniam, S. SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein-ACE2 complex. Science 2022, 375, 760–764. [Google Scholar] [CrossRef]
- Koley, T.; Kumar, M.; Goswami, A.; Ethayathulla, A.S.; Hariprasad, G. Structural modeling of Omicron spike protein and its complex with human ACE-2 receptor: Molecular basis for high transmissibility of the virus. Biochem. Biophys. Res. Commun. 2022, 592, 51–53. [Google Scholar] [CrossRef]
- Bjorkman, P.J.; Saper, M.A.; Samraoui, B.; Bennett, W.S.; Strominger, J.L.; Wiley, D.C. Structure of the human class I histocompatibility antigen, HLA-A2. Nature 1987, 329, 506–512. [Google Scholar] [CrossRef]
- Rossjohn, J.; Gras, S.; Miles, J.J.; Turner, S.J.; Godfrey, D.I.; McCluskey, J. T cell antigen receptor recognition of antigen-presenting molecules. Annu. Rev. Immunol. 2015, 33, 169–200. [Google Scholar] [CrossRef]
- Theodossis, A.; Guillonneau, C.; Welland, A.; Ely, L.K.; Clements, C.S.; Williamson, N.A.; Webb, A.I.; Wilce, J.A.; Mulder, R.J.; Dunstone, M.A.; et al. Constraints within major histocompatibility complex class I restricted peptides: Presentation and consequences for T-cell recognition. Proc. Natl. Acad. Sci. USA 2010, 107, 5534–5539. [Google Scholar] [CrossRef] [Green Version]
- Guermonprez, P.; Valladeau, J.; Zitvogel, L.; Thery, C.; Amigorena, S. Antigen presentation and T cell stimulation by dendritic cells. Ann. Rev. Immunol. 2002, 20, 621–667. [Google Scholar] [CrossRef]
- Bretscher, P.A. The historical postulate: Is it the basis, at the level of the system, for self-nonself discrimination? Scand. J. Immunol. 2021, 94, e13033. [Google Scholar] [CrossRef]
- Kieper, W.C.; Burghardt, J.T.; Surh, C.D. A role for TCR affinity in regulating naïve T cell homeostasis. J. Immunol. 2004, 172, 40–44. [Google Scholar] [CrossRef] [Green Version]
- Otaki, J.M.; Ienaka, S.; Gotoh, T.; Yamamoto, H. Availability of short amino acid sequences in proteins. Protein Sci. 2005, 14, 617–625. [Google Scholar] [CrossRef] [Green Version]
- Otaki, J.M.; Gotoh, T.; Yamamoto, H. Potential implications of availability of short amino acid sequences in proteins: An old and new approach to protein decoding and design. Biotechnol. Annu. Rev. 2008, 14, 109–141. [Google Scholar] [CrossRef]
- Otaki, J.M.; Tsutsumi, M.; Gotoh, T.; Yamamoto, H. Secondary structure characterization based on amino acid composition and availability in proteins. J. Chem. Inf. Model. 2010, 50, 690–700. [Google Scholar] [CrossRef]
- Tsutsumi, M.; Otaki, J.M. Parallel and antiparallel β-strands differ in amino acid composition and availability of short constituent sequences. J. Chem. Inf. Model. 2011, 51, 1457–1464. [Google Scholar] [CrossRef]
- Motomura, K.; Fujita, T.; Tsutsumi, M.; Kikuzato, S.; Nakamura, M.; Otaki, J.M. Word decoding of protein amino acid sequences with availability analysis: A linguistic approach. PLoS ONE 2012, 7, e50039. [Google Scholar] [CrossRef]
- Motomura, K.; Nakamura, M.; Otaki, J.M. A frequency-based linguistic approach to protein decoding and design: Simple concepts, diverse applications, and the SCS Package. Comput. Struct. Biotechnol. J. 2013, 5, e201302010. [Google Scholar] [CrossRef] [Green Version]
- Endo, S.; Motomura, K.; Tsuhako, M.; Kakazu, Y.; Nakamura, M.; Otaki, J.M. Search for human-specific proteins based on availability scores of short constituent sequences: Identification of a WRWSH protein in human testis. In Computational Biology and Chemistry; Behzadi, P., Bernabò, N., Eds.; IntechOpen: London, UK, 2019; pp. 11–33. [Google Scholar] [CrossRef] [Green Version]
- Otaki, J.M.; Nakasone, W.; Nakamura, M. Self and nonself short constituent sequences of amino acids in the SARS-CoV-2 proteome for vaccine development. COVID 2021, 1, 555–574. [Google Scholar] [CrossRef]
- Zhang, B.Z.; Hu, Y.F.; Chen, L.L.; Yau, T.; Tong, Y.G.; Hu, J.C.; Cai, J.P.; Chan, K.-H.; Dou, Y.; Deng, J.; et al. Mining of epitopes on spike protein of SARS-CoV-2 from COVID-19 patients. Cell Res. 2020, 30, 702–704. [Google Scholar] [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef] [Green Version]
- Shi, R.; Shan, C.; Duan, X.; Chen, Z.; Liu, P.; Song, J.; Song, T.; Bi, X.; Han, C.; Wu, L.; et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 2020, 584, 120–124. [Google Scholar] [CrossRef]
- Barnes, C.O.; Jette, C.A.; Abernathy, M.E.; Dam, K.-M.A.; Esswein, S.R.; Gristick, H.B.; Malyutin, A.G.; Sharaf, N.G.; Huey-Tubman, K.E.; Lee, Y.E.; et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 2020, 588, 682–687. [Google Scholar] [CrossRef]
- Zost, S.J.; Gilchuk, P.; Case, J.B.; Binshtein, E.; Chen, R.E.; Nkolola, J.P.; Schäfer, A.; Reidy, J.X.; Trivette, A.; Nargi, R.S.; et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature 2020, 584, 443–449. [Google Scholar] [CrossRef]
- Yuan, M.; Wu, N.C.; Zhu, X.; Lee, C.-C.D.; So, R.T.Y.; Lv, H.; Mok, C.K.P.; Wilson, I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science 2020, 368, 630–633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shrock, E.; Fujimura, E.; Kula, T.; Timms, R.; Lee, I.-H.; Leng, Y.; Robinson, M.L.; Sie, B.M.; Li, M.Z.; Chen, Y.; et al. Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science 2020, 370, eabd4250. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Miller, W. A time-efficient, linear-space local similarity algorithm. Adv. Appl. Math. 1991, 12, 337–357. [Google Scholar] [CrossRef] [Green Version]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsiech, C.-L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [Green Version]
- Nurse, P. Biology must generate ideas as well as data. Nature 2021, 597, 305. [Google Scholar] [CrossRef]
- Otaki, J.M.; Nakasone, W.; Nakamura, M. Nonself mutations in the spike protein suggest an increase in the antigenicity and a decrease in the virulence of the Omicron variant of SARS-CoV-2. bioRxiv 2022. [Google Scholar] [CrossRef]
- Pe’er, I.; Felder, C.E.; Man, O.; Silman, I.; Sussman, J.L.; Beckmann, J.S. Proteomic signatures: Amino acid and oligopeptide compositions differentiates among phyla. Proteins 2004, 54, 20–40. [Google Scholar] [CrossRef]
- Zemková, M.; Zahradní, D.; Mokrejš, M.; Flegr, J. Parasitism as the main factor shaping peptide vocabularies in current organisms. Parasitology 2017, 144, 975–983. [Google Scholar] [CrossRef] [Green Version]
- Singh, T.U.; Parida, S.; Lingaraju, M.C.; Kesavan, M.; Kumar, D.; Singh, R.K. Drug repurposing approach to fight COVID-19. Pharmacol. Rep. 2020, 72, 1479–1508. [Google Scholar] [CrossRef]
- Chugh, H.; Awasthi, A.; Agarwal, Y.; Gaur, R.K.; Dhawan, G.; Chandra, R. A comprehensive review on potential therapeutics interventions for COVID-19. Eur. J. Pharmacol. 2021, 890, 173741. [Google Scholar] [CrossRef]
- Majumder, J.; Minko, T. Recent development on therapeutic and diagnostic approaches for COVID-19. AAPS J. 2021, 23, 14. [Google Scholar] [CrossRef]
- Gavriatopoulou, M.; Ntanasis-Stathopoulos, I.; Korompoki, E.; Fotiou, D.; Migkou, M.; Tzanninis, I.-G.; Psaltopoulou, T.; Kastritis, E.; Terpos, E.; Dimopoulos, M.A. Emerging treatment strategies for COVID-19 infection. Clin. Exp. Med. 2021, 21, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, H. Blood nitrate and nitrite modulating nitric oxide bioavailability: Potential therapeutic functions in COVID-19. Nitric Oxide 2020, 103, 29–30. [Google Scholar] [CrossRef]
- Yang, Y.; Islam, M.S.; Wang, J.; Li, Y.; Chen, X. Traditional Chinese Medicine in the treatment of patients infected with 2019-new coronavirus (SARS-CoV-2): A review and perspective. Int. J. Biol. Sci. 2020, 16, 1708–1717. [Google Scholar] [CrossRef]
- Liu, M.; Gao, Y.; Yuan, Y.; Yang, K.; Shi, S.; Zhang, J.; Tian, J. Efficacy and safety of Integrated Traditional Chinese and Western Medicine for corona virus disease 2019 (COVID-19): A systematic review and meta-analysis. Pharmacol. Res. 2020, 158, 104896. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).