Abstract
São Paulo state has been the epicenter of the Coronavirus Disease 2019 (COVID-19) in Brazil, ranking first by state with over six million reported cases. In February 2021, the P.4 lineage was reported in 21 cities across the state by public health authorities due to the L452R mutation. Here, by analyzing 17,304 genome sequences of SARS-CoV-2 sampled between February and August of 2021 in 476 distinct cities in São Paulo, we assess the transmission dynamics of the P.4 lineage and other SARS-CoV-2 variants that were, at the time of the study, co-circulating in the state. Additionally, clinical parameters from the city of Araras, São Paulo (N = 251) were considered to estimate the potential risk and mortality rate associated with the P.4 lineage since its higher prevalence was observed in that city. Our data suggest a low frequency (0.55%) of the P.4 lineage across the state, with the gamma variant being the dominant form in all regions (90%) at that time. Furthermore, no evidence of increased transmissibility and disease severity related to the P.4 lineage was observed. The displacement through the time of different lineages in São Paulo highlights how challenging genomic surveillance appears to track the emergence of new SARS-CoV-2 lineages, which could better guide the implementation of control measures.
1. Introduction
Several genetic variants of the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) have emerged in the second year of the Coronavirus Disease 2019 (COVID-19) pandemic and are currently circulating around the world [1,2,3]. In most cases, those emerging variants have been associated with increased transmissibility, increased affinity for the human ACE2 receptor, and more severe disease [3,4,5]. Genomic surveillance studies have allowed the monitoring of circulating strains in different parts of the world, guiding a better implementation of control measures [6].
SARS-CoV-2 is a positive-sense single-stranded RNA virus with among the largest and most complex RNA viral genomes known, ranging from 26 to 32 kb [7]. Like any RNA virus, its mutation rate is generally higher than that of DNA viruses [8], which enhances the ability of these viruses to adapt to new conditions, including emerging into new hosts and circumventing vaccine-induced immunity [9,10]. Coronaviruses, however, experience fewer mutations than most RNA viruses because their RNA-dependent RNA polymerase (RDRP) has proofreading activity. Therefore, numerous SARS-CoV-2 variants can be explained in part by the low fidelity of their viral RNA polymerase [8,11,12].
Five main genetic variants emerged and sustained a trajectory of increasing in frequency: the Alpha (B.1.1.7), Beta (B.1.351), Delta (B.1.617.2), Gamma (P.1), and Omicron (B.1.1.529). These are considered “Variants of Concern” (VOC) because of increased transmissibility, hospitalizations, and deaths, which has alarmed national and international organizations.
Brazil’s first COVID-19 case was reported on 26 February 2020 in São Paulo state in a traveler returning from Italy. As of 13 October 2022, more than 6 million cases and more than 170,000 deaths have been reported in the state, which makes it the epicenter of the COVID-19 epidemic in Brazil. The Gamma VOC and the Zeta (P.2) VOI (Variant of Interest) have been established as the most prevalent lineages circulating across all Brazilian territories, with the higher prevalence of Gamma in São Paulo state up to February 2021 [13]. In February 2021, a new sub-lineage of the ancestral B.1.1.28 harboring the L452R spike mutation, named P.4, was first identified in Porto Ferreira city, São Paulo, Brazil [14].
Since its first detection, an increase in frequency was observed in more than 21 cities of the state [14]. In previous studies, this mutation has been associated with heightened adaptiveness of the virus, decreased sensibility to neutralizing antibodies, escape from cellular immunity, and increased infectivity [15,16,17,18]. In this respect, the Network for Pandemic Alert of Emerging SARS-CoV-2 Variants based in the state of São Paulo and led by the Butantan Institute analyzed n = 17,304 SARS-CoV-2 genome sequences to assess the transmission dynamics of the variants that were co-circulating in the state. In this report, we provide a retrospective genomic and clinical overview regarding the SARS-CoV-2 P.4 lineage with the intent to evaluate its potential impact.
2. Materials and Methods
2.1. SARS-CoV-2 Samples
The study included a total of 17,304 SARS-CoV-2 genome sequence data (Table S1) from the Network for Pandemic Alert of Emerging SARS-CoV-2 Variants sampled between 21 February and 7 August 2021. Total nucleic acid was extracted from nasopharyngeal and oropharyngeal swab samples using the Extract kit (Loccus, Cotia, São Paulo, Brasil) in an automated system (Extracta 96, Loccus, Cotia, São Paulo, Brasil) and submitted to real-time RT-PCR using different commercial assays (AllPlex 2019-nCoV Assay-Seegene; GeneFinderTM COVID-19 Plus RealAmp Kit; USA/CDC guidelines for N1 and N2 genes). SARS-CoV-2 positive samples with Cq levels lower than 30 were selected for genomic surveillance. This study was approved by the Ethics Committee of the School of Animal Science and Food Engineering, University of Sao Paulo (protocol number: 46827521.3.0000.5422), following Brazilian regulations and international ethical standards.
2.2. SARS-CoV-2 Gamma//VOC Detection by Real-Time PCR
During the period in which the samples were collected for this study, there was a high prevalence of circulation of VOC Gamma (P.1) in the São Paulo state. Therefore, we used the frequency of the NSP6 deletion among real-time RT-PCR-positive cases as a reliable indicator for frequency of VOC Gamma (P.1) to filter samples.
A total of 1246 samples were evaluated for Gamma/VOC by a real-time PCR screening test developed by FioCruz/ILMD [4]. The test used a forward primer (Gamma/VOCs-FNF 5′-GGGTGATGCGTATTATGACATGGTTGG) a reverse primer (Gamma/VOCs-FNR 5′-CTAGCACCATCATCATACACAGTTCTTGC) and a probe (Gamma/VOCs-FNP 5′(ZEN)-TGGTTGATACTAGTTTGAAGCTAAAA) to detect the deletion in the ORF1b (NSP6: S106del, G107del, F108del) found in the three VOCs (Gamma, Alpha, and Beta). Both primers were used at 300 nM, and the probe was used at 150 nM (final concentration) with AllPlex master Mix (Seegene Inc., Seoul, South Korea). Thus, a target sampling for SARS-CoV2 sequencing was performed using just negative samples for NSP6 PCR (N = 29) to identify new lineages. These samples are included in our 17,304 SARS-CoV-2 genome sequence data.
2.3. SARS-CoV-2 Sequencing
The libraries were constructed using Illumina COVIDSeqTM Test (Illumina, San Diego, CA, EUA), according to the manufacturer’s instructions. The cDNA was synthesized by reverse transcriptase with random hexamers. The virus genome was amplified using two pool primers in separate PCR reactions. The PCR-amplified product was processed for tagmentation and adapter ligation using IDT for Illumina Nextera UD Indexes Set A, B, C, and D (384 indexes). The enrichment and cleanup steps were carried out according to the manufacturer’s protocol. All samples were processed as batches in a 96-well plate; these 96 libraries were pooled together in a tube. Pooled samples were quantified using a Qubit dsDNA High Sensitivity assay kit on a Qubit fluorometer (Invitrogen Inc., Carlsbad, CA, USA), and the fragment sizes were analyzed in Agilent Fragment analyzer 5200 (Agilent Inc., Santa Clara, CA, USA). The pooled library was normalized to 4 nM concentration and denatured with 5 μL of 0.2 N NaOH. The 1.2 pM library was spiked with 1% PhiX control (PhiX Control v3) and was sequenced on an Illumina MiniSeq platform (Illumina) using a MiniSeq System Mid-Output Kit (300 cycles).
2.4. Data Processing and Identification of SARS-CoV-2 Lineages
FASTQ reads were generated by the Illumina pipeline at BaseSpace (https://basespace.illumina.com, accessed on 5 October 2021). The viral isolate sequences were aligned with the reference sequence for SARS-CoV-2 using the Illumina DRAGEN COVIDSeq Test pipeline (v3.5.3). Viral lineages were identified using the software Pangolin v.3.1.7 (http://pangolin.cog-uk.io/, accessed on 10 October 2021), and nucleotide and amino acid mutations were mapped using Nextclade v0.14.3 (https://clades.nextstrain.org/, accessed on 10 October 2021).
2.5. Evolutionary Analyses
The phylogenetic tree was estimated using FastTree v2.1.10 [19] under a GTR (General Time Reversible) substitution model. Testing for informativeness regarding dating estimates was performed in TempEst v1.5.3 using the best-fitting root function [20]. Ancestral area reconstruction was conducted under a Bayesian Binary MCMC model in RASP 4.0 [21]. The Bayesian Binary MCMC (BBM) method infers the probabilities of each area at each internal node by Markov Chain Monte Carlo sampling. Therefore, each pie chart depicts the area(s) with the largest probability of having been occupied by the ancestor represented by that node. Assessment of substitutions relative to the Wuhan-Hu-1 genome (GenBank accession: NC_045512.2) was performed in NextClade CLI 1.4.0 (https://clades.nextstrain.org/, accessed on 30 October 2021).
2.6. Clinical Data and Statistical Analysis
The clinical data of the 251 patients from the city of Araras, São Paulo state (SP), Brazil, were obtained between 31 March and 11 June 2021 using the epidemiological management system that evaluates the development of clinical signs, hospitalization, and death of patients. The analysis of variance (ANOVA) was performed using SAS software (9.4; SAS Institute Inc., Cary, NC, USA) to evaluate associations between non-P.4 and P.4 lineage and cohort characteristics (age, sex, disease stage, hospitalization, and death). A p-value < 0.05 was considered statistically significant.
3. Results
3.1. Detection of P.4 SARS-CoV-2 Lineage
A total of 17,304 SARS-CoV-2 genome sequences from 476 cities sampled between February to August 2021 were analyzed in this study to assess the circulation of multiple SARS-CoV-2 lineages, including the P.4 lineage, over time in the state of São Paulo. The genomes reported in this study were assigned to 31 lineages based on the dynamic nomenclature of Phylogenetic Assignment of Named Global Outbreak Lineages (PANGOLIN version 3.1.7), with the Gamma lineage predominating in 86% of the samples (Figure 1A). The first case of the P.4 lineage was observed in the 10th epidemiological week (7–13 March 2021) in São Paulo city. Among the 17,304 genomes sequenced, a total of 94 were identified as the P.4 lineage, which was distributed in 30 cities (Figure 1B), suggesting a low frequency of this lineage (0.55%).
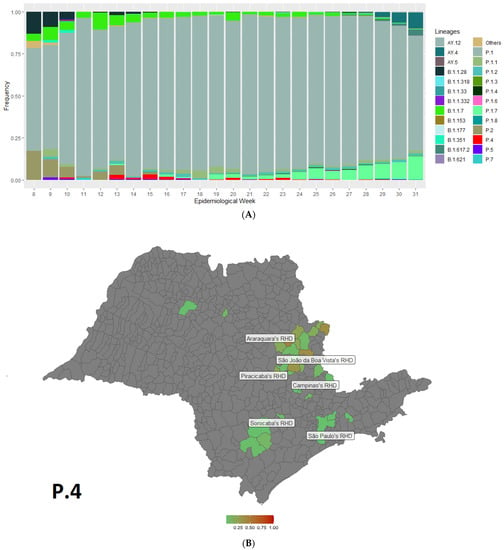
Figure 1.
(A) Temporal distributions of SARS-CoV-2 lineages in the São Paulo state, Brazil, between February and August 2021 estimated from the complete genome sequences; (B) Map of the São Paulo state showing the frequency of the P.4 lineage from genomes generated in this study in the different Regional Health Departments (RHD) of the state. RHD represents territorial divisions of a political-administrative nature in the São Paulo state. Araraquara’s RHD includes the city of Descalvado and Porto Ferreira; Campinas’ RHD: Hortolândia, Socorro, Sumaré, and Valinhos; São Paulo’s RHD: Guararema, Guarulhos, and Sao Paulo; Piracicaba’s RHD: Araras, Conchal, Cordeirópolis, Ipeuna, Leme, Pirassununga, and Rio Claro; São João da Boa Vista’s RHD: Aguaí, Caconde, Casa Branca, Itapira, Santa Cruz das Palmeiras, São José do Rio Pardo, Tambaú, and Tapiratiba; São José do Rio Preto’s RHD: José Bonifácio and Pindorama; and Sorocaba’s RHD: Capao Bonito, Ipero, Itapetininga, and São Miguel Arcanjo.
Because of the high prevalence of Gamma VOC in the São Paulo state, it was difficult to track the P.4 variant in a randomized genomic surveillance study. Therefore, we genotyped all SARS-CoV-2-positive samples (N = 1036) received between 23 and 29 May 2021 (21st epidemiology week) by the real-time PCR assay to detect the deletion at ORF1b (NSP6: S106del, G107del, F108del), which is a genetic signature of the VOCs Gamma (P.1), Alpha (B.1.1.7), and Beta (B.1.351), to eliminate these samples from our sequencing and better estimate the trajectory of P.4 in this region. Twenty-nine of the SARS-CoV-2-positive samples failed for deletion, supporting our sequencing results that indicate the high prevalence of the Gamma (P.1) lineage. Thus, we sequenced those failed samples and observed that 1.93% (20/1036) were classified as the P.4 lineage, of which 45% (9/20) were from the city of Araras, SP, Brazil.
3.2. Phylogenetic Analysis of P.4 Genomes
Phylogenetic analysis was performed for a dataset of 94 P.4 genomes identified in this study (Table S2). All P.4 genomes appeared in a monophyletic group, whose most recent common ancestor was dated to April 2021 from the city of Palmares Paulista (Figure 2). Within the P.4 lineage, we found that 100% of the sequences presented the following nonsynonymous mutations: ORF1ab: A3143V, P971L, P314L, and Y822C; N: G204R and R203K; and S: D614G, I720V, and V1176F. Only two samples were found without the L452R mutation in the Spike protein (VOI) (Table S3).
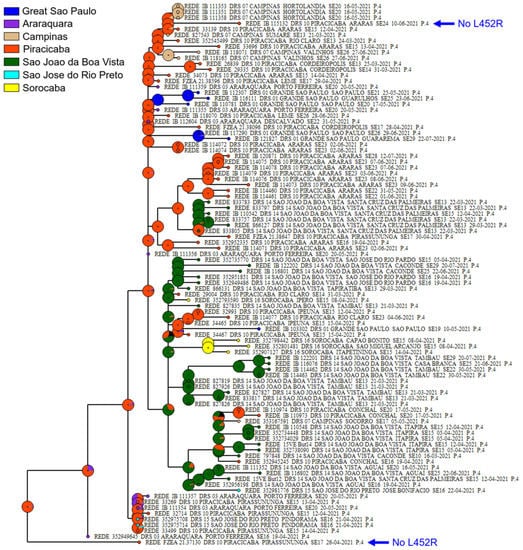
Figure 2.
Phylogenetic tree of P.4 lineage based on a dataset with 17,304 genome sequences. Sequences from this study that do not have an L452R mutation are highlighted with a blue arrow. Each pie chart depicts the area(s) with the largest probability of having been occupied by the ancestor represented by that node.
The TempEst analysis focused on the P.4 subtree returned an R2 close to zero, indicating that there is no sufficient signal in the P.4 genomes to allow divergence dating, which is possibly due to collection times being relatively shallow regarding evolutionary time. The biogeographic analysis of ancestral area estimation suggested the Piracicaba’s RHD as the likely origin of this lineage (Figure 2).
3.3. Targeted Detection in the City of Araras-SP and Clinical Outcome of P4 in Patients
Considering that most sequences from lineage P.4 were from Araras-SP, we genotyped all SARS-CoV-2-positive samples (N = 183) received between 6 and 12 June 2021 (23rd epidemiological week) from Araras-SP. Eleven samples failed for the NSP6 deletion (6%) and were therefore submitted for SARS-CoV-2 genome sequencing. From those, eight samples (4.37%) were confirmed to be P.4 lineage (Table S4).
Subsequently, clinical data from a total of 251 patients with COVID-19 from the city of Araras-SP, obtained in the period between 31 March and 11 June 2021, were analyzed to estimate whether the lineage P.4 reflects severe cases, hospitalizations, and increased mortality. Of these, 23 patients were assigned for P.4 lineage and 228 for non-P.4 lineage by genome sequence. Overall, it was possible to determine the low incidence of hospitalizations (1/23–4%) and no deaths associated with P.4 (Table 1). There were no significant differences in sex and mortality rate between groups.

Table 1.
Cohort characteristics by SARS-CoV-2 non-P.4 and P.4 lineages.
4. Discussion
Coronaviruses pose a major threat to human health, and SARS-CoV-2 has caused more deaths than SARS or MERS [22]. It has been reported that SARS-CoV-2 accumulates an average of one or two mutations per month [15]. This study described for the first time the retrospective temporal monitoring window of the P.4 lineage in São Paulo state, southeast Brazil. We analyzed 17,304 genome sequences from 476 cities of the São Paulo state (Tables S1 and S5) over seven months and observed the first case at the beginning of March; since then, the relative frequency of this lineage decreased. Cases of the P.4 lineage have already been detected in the south region of Brazil since October 2020 [16].
The cases of P.4 attracted attention because, in addition to increasing the numbers of cases from one week to another following a random sampling strategy for genomic surveillance, the variant carries the spike L452R mutation (VOI), which has been reported as one of the mutations associated with adaptiveness of the virus [15] and reduced antibody neutralization [17,23]. Moreover, it was found that L452R mutation increases the free energy of the RBD-ACE2 binding complex, resulting in stronger virus–cell attachment and, consequently, rising infectivity [14,17]. However, in our study, the presence of this mutation did not lead to an increased virulence or infectivity compared to Gamma (P.1). The L452R mutation is also associated with other known variants such as the B.1.427/B.1.429 variant identified in California, USA [17]; the A.27 variant in Germany [24]; and, most recently, the B.1.617.2 (Delta) variant identified in India [5].
A combined targeted RT-qPCR screening (N = 1246) for a deletion of NSP6 and a target sequencing approach using negative samples for this deletion was performed to evaluate epidemiological hotspots in the region. We aimed to select samples for sequencing to monitor the population distribution and frequency of the P.4 lineage in our region. We observed a higher prevalence in the city of Araras-SP (45%), although the P.4 variant frequency remained low (1.93%). A low frequency (0.68%) of the P.4 variant in Mexico between March 2020 and February 2021 has also been reported [25]. According to these authors, the P.4 lineage is the result of an early P.2 entry in Mexico followed by local evolution. Sant’Anna et al. also reported an increase in the P.2 lineage almost concurrently with P.4.1 in South Brazil [16]. The circulation of P.4 was then also reported in 35% of the samples analyzed in June 2021 in the city of Porto Ferreira-São Paulo, a city located 60 Km from Araras-SP [14].
The replacement of a lineage or the ability to contain it within a country or region depends on its survival benefits [26]. Some variants emerge and quickly become the reference lineage, while others increase the number of cases, collapse, and then disappear [12]. Although the L452R mutation indicates strong positive selection, this was not enough to maintain the P.4 lineage, which had reduced cases from the 17th epidemiological week onwards. Therefore, the combination of this with other mutations and epistatic effects may have impacted virus transmissibility, thus limiting the spread of P.4 to other cities in the region. On the other hand, recently released data suggest that the displacement of different VOCs such Gamma with Delta and most recently Delta with Omicron was observed in São Paulo state, demonstrating a selective advantage of those strains over all the others [3,27,28,29,30,31,32].
To our knowledge, this is the first study that reports a retrospective genomic and clinical overview of the P.4 lineage in Brazil. We did not find a higher risk of severe illness, hospitalizations, transmissibility, or overall mortality associated with this lineage, but our results highlight the importance of intense genomic monitoring in order to follow the real-time evolution of SARS-CoV-2 lineages through time [30].
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/covid2120127/s1, Table S1: Sequences from this study, Table S2: Dataset of the 94 genomes identified as P.4 lineage, Table S3: Frequency of the nonsynonymous mutations in the 94 genomes of P.4 lineage, Table S4: SARS-CoV-2-positive samples from the city of Araras genotyped to deletion at ORF1b (NSP6: S106del, G107del, and F108del), which is a genetic signature of the VOCs (P.1, B.1.1.7, and B.1.351), Table S5: List of laboratories that generated and shared the SARS-CoV-2 genome sequence data.
Author Contributions
Conceptualized and designed the experiments: H.F.; Performed the analysis: M.D.P., J.C.C.L. and E.C.d.M.O.; Analyzed the data: M.D.P., E.C.d.M.O., J.C.C.L., J.S.L.P., G.R. and L.G.C.; Writing—original draft preparation: M.D.P.; Writing—review and edition: M.D.P., J.C.C.L., M.G., S.K., M.C.E., L.L.C. and H.F.; Molecular screening and produced SARS-CoV-2 genomic data: M.D.P., J.C.C.L., E.C.d.M.O., J.S.L.P., G.R., L.G.C., V.L.V., M.G., L.C.J.A., L.P.O.d.L., A.J.M., C.R.d.S.B., E.C.M., J.d.S.T.B., D.B.M., R.A.B., R.d.L.R.C.C., P.D.S.C.M., S.N.S., R.d.S.B., E.S.R., E.V.S., J.S.B., D.G.L.d.L.R., J.P.K., B.S., P.A.A., F.A.d.S.d.C., C.Á.B., L.S., B.d.C.M., R.M.T.G., J.A.S.-N., M.L.N., L.L.C., R.T.C., R.M.N., D.T.C., S.K., M.C.E., S.C.S. and H.F.; Clinical/epidemiological data: J.F.P.P. All authors have read and agreed to the published version of the manuscript.
Funding
This study was financed by Butantan Institute on behalf of the Rede de Alerta das Variantes da COVID-19, São Paulo Research Foundation (FAPESP) (Grant Numbers: 2020/10127-1; 2020/05367-3; and 2013/08135-2), the Central Public Health Laboratories, Blood Center of Ribeirao Preto, and it was supported by the Brazilian Ministry of Health, the Pan American Health Organization PAHO/WHO (APO21-00010098), and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (Grants Number: 401119/2020-3). J.C.C.L. is supported by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES)-Finance Code 001. M.G. is funded by PON “Ricerca e Innovazione” 2014-2020. M.G. is supported in part by the CRP—ICGEB Research Grant 2020 project CRP/BRA20-03, Contract CRP/20/03. M.L.N. is funded by NIH CREID Network/CreateNeo (1U01AI151807-01). J.A.S.-N. is funded by FAPESP (Grant Number: 2020/06136-5).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on an online repository. The name of the repository and accession number can be found in the supplementary material.
Acknowledgments
We thank all the authors who have kindly deposited and shared genome data on GISAID (Table S5). The authors acknowledge the technical support of Luciana de Araujo Pimenta, Luiz Aurelio de Campos Crispin, Gabriela Mauric Frossard Ribeiro, Glaucia Maria Rodrigues Borges, and Mariane Evaristo e Josiane Serrano Borges from the National Network for Pandemic Alert of SARS-CoV-2. We also acknowledge the contribution of all employees of General Coordination of Public Health Laboratories and professionals of Public Health Laboratories of Brazil and Network for Pandemic Alert of Emerging SARS-CoV-2 Variants for their contribution to the sequencing effort and their commitment and work during the the COVID-19 pandemic.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Koyama, T.; Platt, D.; Parida, L. Variant analysis of SARS-CoV-2 genomes. Bull. World Health Organ. 2020, 98, 495–504. [Google Scholar] [CrossRef]
- Iacobucci, G. COVID-19: New UK variant may be linked to increased death rate, early data indicate. BMJ 2021, 372, n230. [Google Scholar] [CrossRef] [PubMed]
- Tegally, H.; Wilkinson, E.; Giovanetti, M.; Iranzadeh, A.; Fonseca, V.; Giandhari, J.; Doolabh, D.; Pillay, S.; San, E.J.; Msomi, N.; et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature 2021, 592, 438–443. [Google Scholar] [CrossRef] [PubMed]
- Naveca, F.G.; Nascimento, V.; de Souza, V.C.; de Lima Corado, A.; Nascimento, F.; Silva, G.; Costa, Á.; Duarte, D.; Pessoa, K.; Mejía, M.; et al. COVID-19 in Amazonas, Brazil, was driven by the persistence of endemic lineages and P.1 emergence. Nat. Med. 2021, 27, 1230–1238. [Google Scholar] [CrossRef] [PubMed]
- Callaway, E. Delta coronavirus variant: Scientists brace for impact. Nature 2021, 595, 17–18. [Google Scholar] [CrossRef]
- Lauring, A.S.; Hodcroft, E.B. Genetic Variants of SARS-CoV-2—What Do They Mean? JAMA 2021, 325, 529–531. [Google Scholar] [CrossRef]
- Snijder, E.J.; Bredenbeek, P.J.; Dobbe, J.C.; Thiel, V.; Ziebuhr, J.; Poon, L.L.M.; Guan, Y.; Rozanov, M.; Spaan, W.J.M.; Gorbalenya, A.E. Unique and Conserved Features of Genome and Proteome of SARS-coronavirus, an Early Split-off From the Coronavirus Group 2 Lineage. J. Mol. Biol. 2003, 331, 991–1004. [Google Scholar] [CrossRef]
- Robson, F.; Khan, K.S.; Le, T.K.; Paris, C.; Demirbag, S.; Barfuss, P.; Rocchi, P.; Ng, W.L. Coronavirus RNA Proofreading: Molecular Basis and Therapeutic Targeting. Mol. Cell 2020, 79, 710–727. [Google Scholar] [CrossRef]
- Duffy, S. Why are RNA virus mutation rates so damn high? PLoS Biol. 2018, 16, e3000003. [Google Scholar] [CrossRef]
- Elena, S.F.; Sanjuán, R. Adaptive value of high mutation rates of RNA viruses: Separating causes from consequences. J. Virol. 2005, 79, 11555–11558. [Google Scholar] [CrossRef]
- Peck, K.M.; Lauring, A.S. Complexities of Viral Mutation Rates. J. Virol. 2018, 92, 14. [Google Scholar] [CrossRef] [PubMed]
- Castonguay, N.; Zhang, W.; Langlois, M.-A. Meta-Analysis and Structural Dynamics of the Emergence of Genetic Variants of SARS-CoV-2. Front. Microbiol. 2021, 1, 1637. [Google Scholar] [CrossRef] [PubMed]
- Franceschi, V.B.; Ferrareze, P.A.G.; Zimerman, R.A.; Cybis, G.B.; Thompson, C.E. Mutation hotspots and spatiotemporal distribution of SARS-CoV-2 lineages in Brazil, February 2020-2021. Virus Res. 2021, 304, 198532. [Google Scholar] [CrossRef] [PubMed]
- Bittar, C.; Possebon, F.S.; Ullmann, L.S.; Geraldini, D.B.; da Costa, V.G.; de Almeida, L.G.P.; Paulo, P.R.; Nascimento-Júnior, N.M.; Cilli, E.M.; Artico Banho, C.; et al. The Emergence of the New P.4 Lineage of SARS-CoV-2 With Spike L452R Mutation in Brazil. Front. Public Health 2021, 9, 1465. [Google Scholar] [CrossRef] [PubMed]
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452. [Google Scholar] [CrossRef]
- Sant’Anna, F.H.; Varela, A.P.M.; Prichula, J.; Comerlato, J.; Comerlato, C.B.; Roglio, V.S.; Pereira, G.F.M.; Moreno, F.; Seixas, A.; Wendland, E.M. Emergence of the novel SARS-CoV-2 lineage VUI-NP13L and massive spread of P.2 in South Brazil. Emerg. Microbes Infect. 2021, 10, 1431–1440. [Google Scholar] [CrossRef]
- Tchesnokova, V.; Kulasekara, H.; Larson, L.; Bowers, V.; Rechkina, E.; Kisiela, D.; Sledneva, Y.; Choudhury, D.; Maslova, I.; Deng, K.; et al. Acquisition of the L452R mutation in the ACE2-binding interface of Spike protein triggers recent massive expansion of SARS-CoV-2 variants. J. Clin. Microbiol. 2021, 59, e00921-21. [Google Scholar] [CrossRef]
- Motozono, C.; Toyoda, M.; Zahradnik, J.; Saito, A.; Nasser, H.; Tan, T.S.; Ngare, I.; Kimura, I.; Uriu, K.; Kosugi, Y.; et al. SARS-CoV-2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host Microbe 2021, 29, 1124–1136.e11. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Rambaut, A.; Lam, T.; Max Carvalho, L.; Pybus, O. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol. 2016, 2, vew007. [Google Scholar] [CrossRef]
- Yu, Y.; Harris, A.J.; Blair, C.; He, X. RASP (Reconstruct Ancestral State in Phylogenies): A tool for historical biogeography. Mol. Phylogenet. Evol. 2015, 87, 46–49. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Wu, T.; Liu, Q.; Yang, Z. The SARS-CoV-2 outbreak: What we know. Int. J. Infect. Dis. 2020, 94, 44–48. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Garcia-Knight, M.A.; Khalid, M.M.; Servellita, V.; Wang, C.; Morris, M.K.; Sotomayor-González, A.; Glasner, D.R.; Reyes, K.R.; Gliwa, A.S.; et al. Transmission, infectivity, and neutralization of a spike L452R SARS-CoV-2 variant. Cell 2021, 184, 3426. [Google Scholar] [CrossRef]
- Mor, O.; Mandelboim, M.; Fleishon, S.; Bucris, E.; Bar-Ilan, D.; Linial, M.; Nemet, I.; Kliker, L.; Lustig, Y.; Israel National Consortium for SARS-CoV-2 Sequencing; et al. The Rise and Fall of a Local SARS-CoV-2 Variant with the Spike Protein Mutation L452R. Vaccines 2021, 9, 937. [Google Scholar] [CrossRef] [PubMed]
- Barona-Gómez, F.; Delaye, L.; Díaz-Valenzuela, E.; Plisson, F.; Cruz-Pérez, A.; Díaz-Sánchez, M.; García-Sepúlveda, C.A.; Sanchez-Flores, A.; Pérez-Abreu, R.; Valencia-Valdespino, F.J.; et al. Phylogenomics and population genomics of SARS-CoV-2 in Mexico during the pre-vaccination stage reveals variants of interest B.1.1.28.4, B.1.1.222 or B.1.1.519 and the nucleocapsid mutation S194L associated with symptoms. Microb. Genom. 2021, 7, 000684. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Chong, K.C.; Wong, M.C.S.; Boon, S.S.; Huang, J.; Wang, M.H.; Ng, R.W.Y.; Lai, C.K.C.; Chan, P.K.S. A global analysis of replacement of genetic variants of SARS-CoV-2 in association with containment capacity and changes in disease severity. Clin. Microbiol. Infect. 2021, 27, 750–757. [Google Scholar] [CrossRef]
- Dejnirattisai, W.; Zhou, D.; Supasa, P.; Liu, C.; Mentzer, A.J.; Ginn, H.M.; Zhao, Y.; Duyvesteyn, H.M.E.; Tuekprakhon, A.; Nutalai, R.; et al. Antibody evasion by the P.1 strain of SARS-CoV-2. Cell 2021, 184, 2939–2954.e9. [Google Scholar] [CrossRef]
- Hoffmann, M.; Arora, P.; Groß, R.; Seidel, A.; Hörnich, B.F.; Hahn, A.S.; Krüger, N.; Graichen, L.; Hofmann-Winkler, H.; Kempf, A.; et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell 2021, 184, 2384–2393.e12. [Google Scholar] [CrossRef]
- Coutinho, R.M.; Marquitti, F.M.D.; Ferreira, L.S.; Borges, M.E.; da Silva, R.L.P.; Canton, O.; Portella, T.P.; Poloni, S.; Franco, C.; Plucinski, M.M.; et al. Model-based estimation of transmissibility and reinfection of SARS-CoV-2 P.1 variant. Commun. Med. 2021, 1, 48. [Google Scholar] [CrossRef]
- Faria, N.R.; Mellan, T.A.; Whittaker, C.; Claro, I.M.; da Silva Candido, D.; Mishra, S.; Crispim, M.A.E.; Sales, F.C.S.; Hawryluk, I.; McCrone, J.T.; et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science 2021, 372, abh2644. [Google Scholar] [CrossRef]
- Giovanetti, M.; Fonseca, V.; Wilkinson, E.; Tegally, H.; San, E.J.; Althaus, C.L.; Xavier, J.; Nanev Slavov, S.; Viala, V.L.; Ranieri Jerônimo Lima, A.; et al. Replacement of the Gamma by the Delta variant in Brazil: Impact of lineage displacement on the ongoing pandemic. Virus Evol. 2022, 8, veac024. [Google Scholar] [CrossRef] [PubMed]
- Alcantara, L.C.J.; Nogueira, E.; Shuab, G.; Tosta, S.; Fristch, H.; Pimentel, V.; Souza-Neto, J.A.; Coutinho, L.L.; Fukumasu, H.; Sampaio, S.C.; et al. SARS-CoV-2 epidemic in Brazil: How the displacement of variants has driven distinct epidemic waves. Virus Res. 2022, 315, 198785. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).