Genetic Diversity and Spatiotemporal Distribution of SARS-CoV-2 Alpha Variant in India
Abstract
1. Introduction
2. Materials and Methods
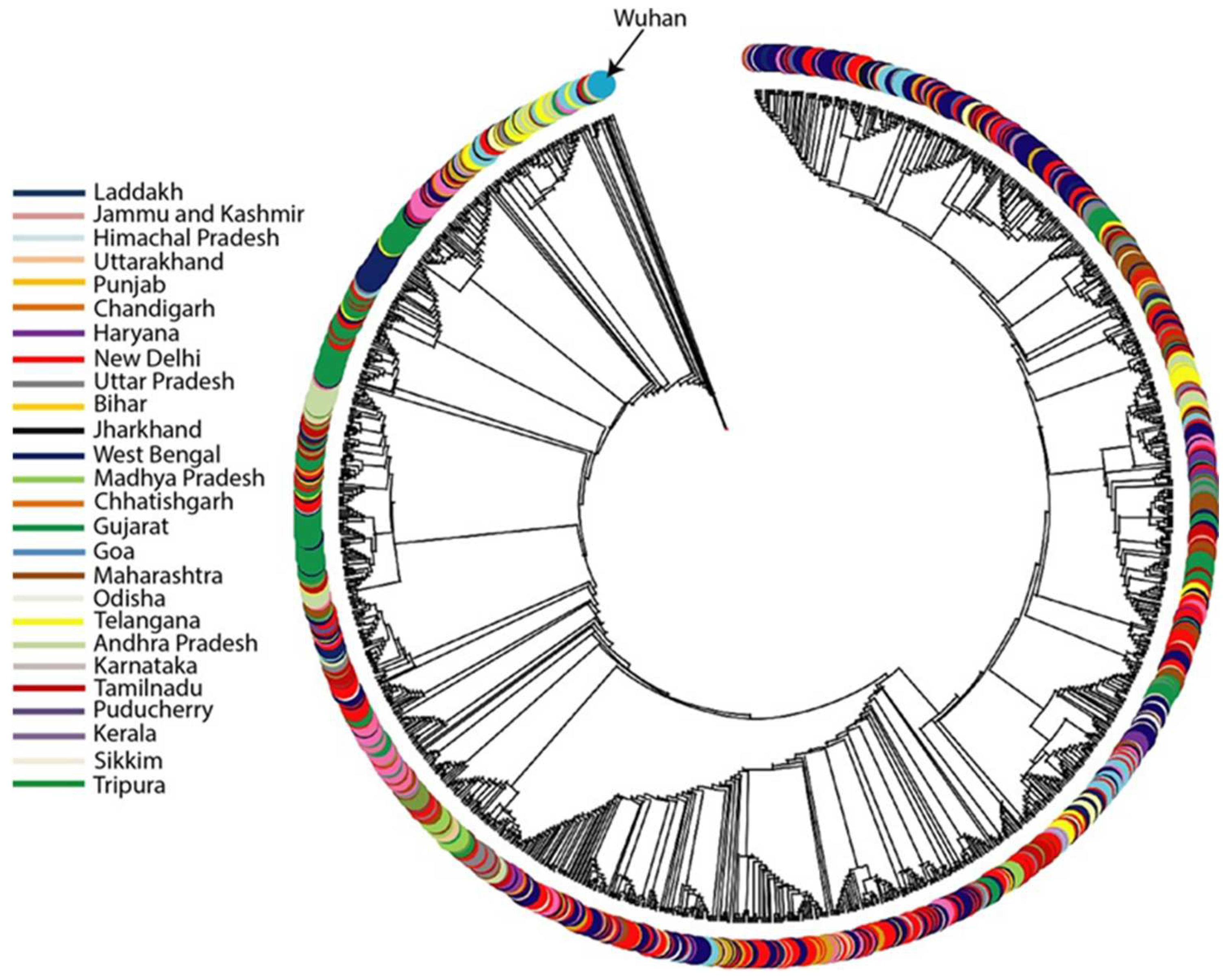
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- MOHFW-GoI. Available online: http://www.ub.edu/dnasp/ (accessed on 3 December 2021).
- Andrews, M.A.; Areekal, B.; Rajesh, K.R.; Krishnan, J.; Suryakala, R.; Krishnan, B.; Muraly, C.P.; Santhosh, P.V. First confirmed case of COVID-19 infection in India: A case report. Indian J. Med. Res. 2020, 151, 490–492. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.T.; Leung, K.; Bushman, M.; Kishore, N.; Niehus, R.; de Salazar, P.M.; Cowling, B.J.; Lipsitch, M.; Leung, G.M. Estimating clinical severity of COVID-19 from the transmission dynamics in Wuhan, China. Nat. Med. 2020, 26, 506–510. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Xiang, R.; Huo, S.; Zhou, Y.; Jiang, S.; Wang, Q.; Yu, F. Molecular mechanism of interaction between SARS-CoV-2 and host cells and interventional therapy. Sig. Transduct. Target Ther. 2021, 6, 233. Available online: https://www.nature.com/articles/s41392-021-00653-w (accessed on 4 December 2021). [CrossRef] [PubMed]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; COVID-19 Genomics UK (COG-UK) Consortium; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef] [PubMed]
- WHO. Tracking SARS-CoV-2 Variants. Available online: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ (accessed on 1 March 2022).
- Challen, R.; Brooks-Pollock, E.; Read, J.M.; Dyson, L.; Tsaneva-Atanasova, K.; Danon, L. Risk of mortality in patients infected with SARS-CoV-2 variant of concern 202012/1: Matched cohort study. BMJ 2021, 372, n579. [Google Scholar] [CrossRef] [PubMed]
- Campbell, F.; Archer, B.; Laurenson-Schafer, H.; Jinnai, Y.; Konings, F.; Batra, N.; Pavlin, B.; Vandemaele, K.; Van Kerkhove, M.D.; Jombart, T.; et al. Increased transmissibility and global spread of SARS-CoV-2 variants of concern. Eurosurveillance 2021, 26, 2100509. [Google Scholar] [CrossRef] [PubMed]
- Callaway, E. Delta coronavirus variant: Scientists brace for impact. Nature 2021, 595, 17–18. [Google Scholar] [CrossRef] [PubMed]
- Gobeil, K.J.; McDowell, S.; Mansouri, K.; Parks, R.; Stalls, V.; Kopp, M.F.; Manne, K.; Saunders, K.; Edwards, R.J.; Haynes, B.F.; et al. Effect of natural mutations of SARS-CoV-2 on spike structure, conformation, and antigenicity. Science 2021, 373, eabi6226. [Google Scholar] [CrossRef] [PubMed]
- Starr, T.N.; Greaney, A.J.; Hilton, S.K.; Ellis, D.; Crawford, K.H.D.; Dingens, A.S.; Navarro, M.J.; Bowen, J.E.; Tortorici, M.A.; Walls, A.C.; et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell 2020, 182, 1295–1310.e20. [Google Scholar] [CrossRef] [PubMed]
- Jaspe, R.C.; Loureiro, C.L.; Sulbaran, Y.; Moros, Z.C.; D’Angelo, P.; Rodríguez, L.; Zambrano, J.L.; Hidalgo, M.; Vizzi, E.; Alarcón, V.; et al. Introduction and rapid dissemination of SARS-CoV-2 Gamma Variant of Concern in Venezuela. ScienceDirect 2021, 96, 105147. [Google Scholar] [CrossRef] [PubMed]
- Thorne, V.P.G.; Bouhaddou, V.P.; Reuschl, V.P.-K.; Zuliani-Alvarez, V.P.; Polacco, V.P.; Pelin, V.P.; Batra, V.P.; Whelan, V.P.V.X.; Ummadi, V.P.; Rojc, V.P.; et al. Evolution of enhanced innate immune evasion by the SARS-CoV-2 B.1.1.7 UK variant. bioRxiv 2021. [Google Scholar] [CrossRef]
- Schmidt, F.; Weisblum, Y.; Rutkowska, M.; Poston, D.; DaSilva, J.; Zhang, F.; Bednarski, E.; Cho, A.; Schaefer-Babajew, D.J.; Gaebler, C.; et al. High genetic barrier to SARS-CoV-2 polyclonal neutralizing antibody escape. Nature 2021, 600, 512–516. [Google Scholar] [CrossRef] [PubMed]
- GISAID—Initiative. Available online: https://www.gisaid.org/ (accessed on 10 January 2022).
- MAFFT [Computer Software]. Multiple Alignment Program for Amino Acid or Nucleotide Sequences, Version 7. 2020. Available online: https://mafft.cbrc.jp/alignment/server/ (accessed on 15 January 2022).
- MEGAX [Computer Software], (10.2.4); Pennsylvania State University: State College, PA, USA. Available online: https://www.megasoftware.net/ (accessed on 15 January 2022).
- Google Sheets. 2009. Available online: https://www.google.com/sheets/about/ (accessed on 15 January 2022).
- Lorenz, M.; Kayser-Bril, N.; Aisch, G. Datawrapper. 2012. Available online: https://www.datawrapper.de/ (accessed on 5 February 2022).
- Julio Rozas, A.F.-M.; Guirao-Rico, D.; Librado, P. DnaSP [Computer Software], (6.12.03); Universitat de Barcelona: Barcelona, Spain, 2019. Available online: http://www.ub.edu/dnasp/ (accessed on 15 January 2022).
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef] [PubMed]
- El-Shabasy, R.M.; Nayel, M.A.; Taher, M.M.; Abdelmonem, R.; Shoueir, K.R.; Kenawy, E.R. Three wave changes, new variant strains, and vaccination effect against COVID-19 pandemic. Int. J. Biol. Macromol. 2022, 204, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Samarasekera, U. India grapples with second wave of COVID-19. Lancet Microbe 2021, 2, e238. [Google Scholar] [CrossRef] [PubMed]
- Sarah Walker, O.G.; Pritchard, E.; Jones, J.; House, T. Tracking the Emergence of SARS-CoV-2 Alpha Variant in the United Kingdom. N. Engl. J. Med. 2021, 385, 2582–2585. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Carballa, A.; Pardo-Seco, J.; Bello, X.; Martinón-Torres, F.; Salas, A. Superspreading in the emergence of COVID-19 variants. Trends Genet. 2021, 37, 1069–1080. [Google Scholar] [CrossRef] [PubMed]
- Anti-Farm Law Protest Sites Becoming Super Spreader, COVID Cases Underreported in Rural PUNJAB: Report. Available online: https://www.opindia.com/2021/05/covid-surge-in-rural-punjab-villagers-returning-from-delhi-border-protest-positive-death/ (accessed on 4 February 2023).
- Jodhka, S.S. Why are the farmers of Punjab protesting? J. Peasant Stud. 2021, 48, 1356–1370. [Google Scholar] [CrossRef]

| States and Union Territories | Total Number of Samples | Haplotype Diversity |
|---|---|---|
| Chandigarh | 92 | 0.997 |
| Haryana | 46 | 0.997 |
| Gujarat | 194 | 0.995 |
| New Delhi | 681 | 0.995 |
| Telangana | 74 | 0.993 |
| Andhra Pradesh | 56 | 0.992 |
| Maharashtra | 120 | 0.989 |
| Tamil Nadu | 70 | 0.986 |
| Jammu Kashmir | 189 | 0.985 |
| Uttar Pradesh | 2 | 0.983 |
| Uttarakhand | 66 | 0.982 |
| Odisha | 29 | 0.978 |
| Rajasthan | 3 | 0.976 |
| Puducherry | 35 | 0.974 |
| Madhya Pradesh | 237 | 0.971 |
| West Bengal | 171 | 0.971 |
| Chhattisgarh | 92 | 0.961 |
| Jharkhand | 22 | 0.956 |
| Himachal Pradesh | 48 | 0.948 |
| Assam | 2 | 0.933 |
| Tripura | 5 | 0.915 |
| Karnataka | 131 | 0.895 |
| Sikkim | 8 | 0.863 |
| Punjab | 692 | 0.861 |
| Goa | 5 | 0.8 |
| Meghalaya | 15 | 0.798 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parasar, J.; Pandey, R.K.; Patel, Y.; Singh, P.P.; Srivastava, A.; Mishra, R.K.; Kumar, B.; Rai, N.; Mishra, V.N.; Shrivastava, P.; et al. Genetic Diversity and Spatiotemporal Distribution of SARS-CoV-2 Alpha Variant in India. COVID 2023, 3, 472-479. https://doi.org/10.3390/covid3040035
Parasar J, Pandey RK, Patel Y, Singh PP, Srivastava A, Mishra RK, Kumar B, Rai N, Mishra VN, Shrivastava P, et al. Genetic Diversity and Spatiotemporal Distribution of SARS-CoV-2 Alpha Variant in India. COVID. 2023; 3(4):472-479. https://doi.org/10.3390/covid3040035
Chicago/Turabian StyleParasar, Jahnavi, Rudra Kumar Pandey, Yashvant Patel, Prajjval Pratap Singh, Anshika Srivastava, Rahul Kumar Mishra, Bhupendra Kumar, Niraj Rai, Vijaya Nath Mishra, Pankaj Shrivastava, and et al. 2023. "Genetic Diversity and Spatiotemporal Distribution of SARS-CoV-2 Alpha Variant in India" COVID 3, no. 4: 472-479. https://doi.org/10.3390/covid3040035
APA StyleParasar, J., Pandey, R. K., Patel, Y., Singh, P. P., Srivastava, A., Mishra, R. K., Kumar, B., Rai, N., Mishra, V. N., Shrivastava, P., Kishor, P. B. K., Suravajhala, P., Tamang, R., Pathak, A. K., & Chaubey, G. (2023). Genetic Diversity and Spatiotemporal Distribution of SARS-CoV-2 Alpha Variant in India. COVID, 3(4), 472-479. https://doi.org/10.3390/covid3040035






