In Silico Screening of Prospective MHC Class I and II Restricted T-Cell Based Epitopes of the Spike Protein of SARS-CoV-2 for Designing of a Peptide Vaccine for COVID-19
Abstract
1. Introduction
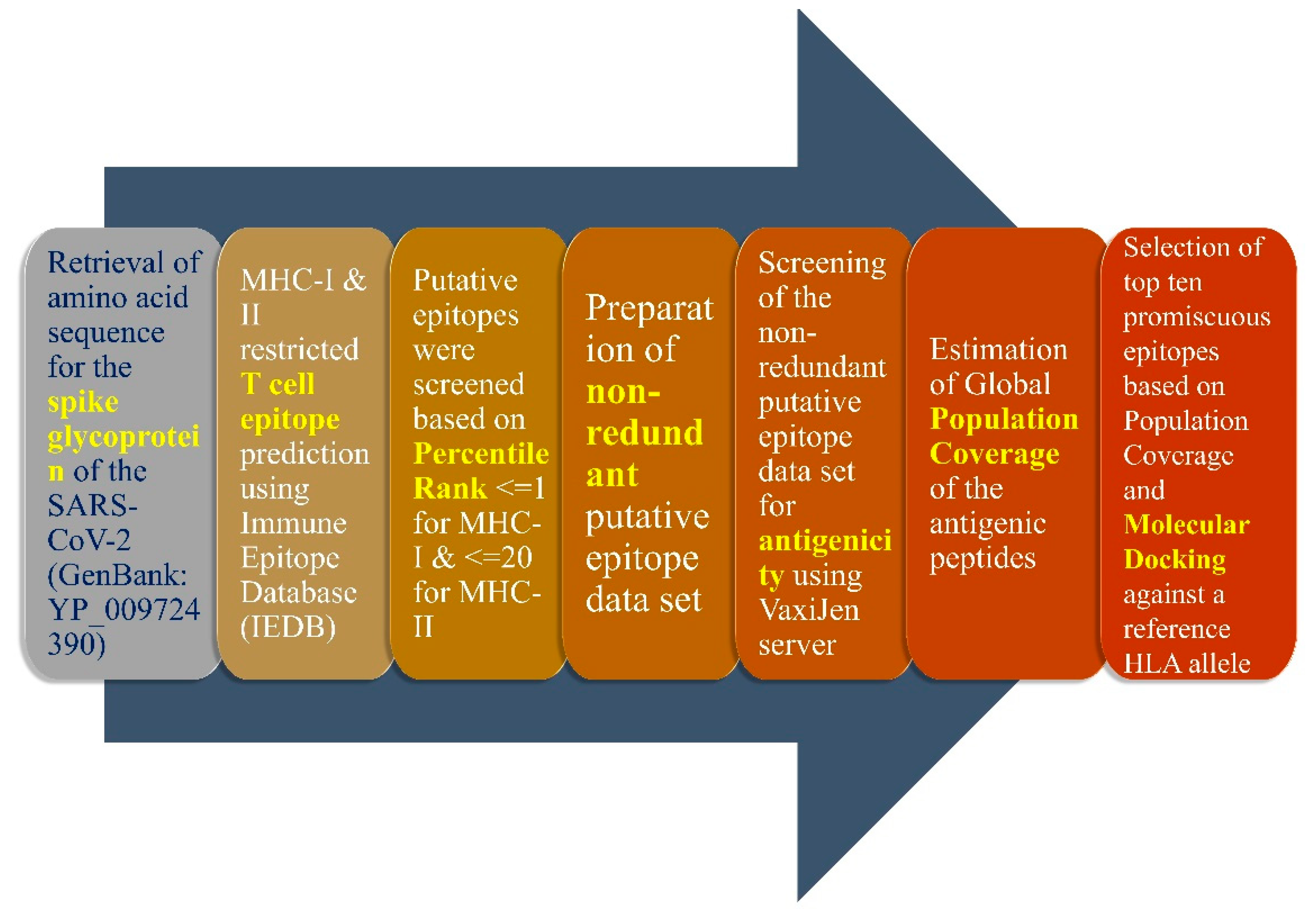
2. Materials and Methods
2.1. Screening of MHC Class-I and Class-II Epitopes
2.2. Prediction of Suitable Epitopes on the Basis of Antigenicity
2.3. B Cell-Based Epitope Prediction
3. Results
3.1. Selection of Class I and Class II MHC Restricted Epitopes Based on Population Coverage and Antigenicity
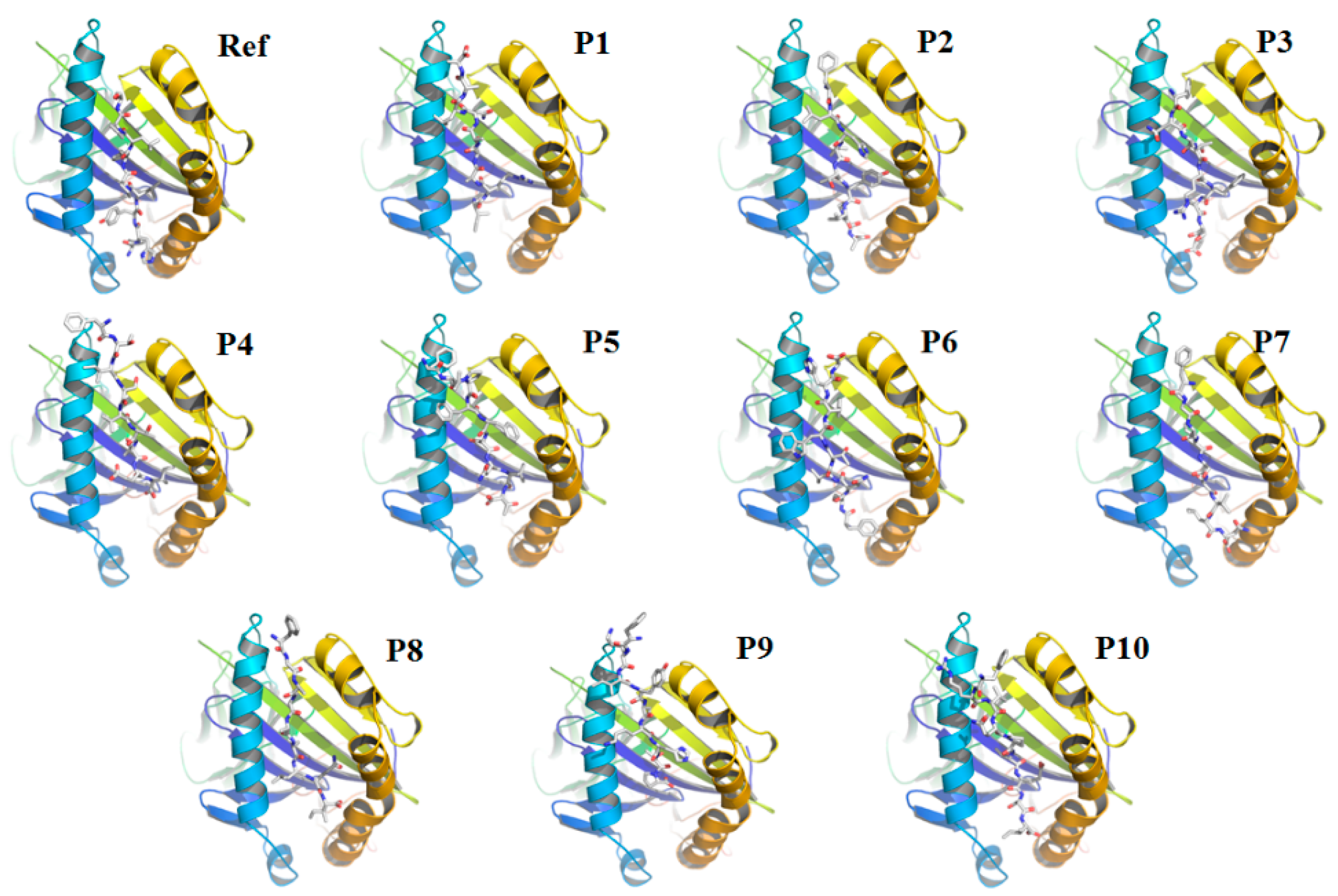
3.2. Re-Docking of the Peptides Confirmed the Specificity of the Selected Peptides
3.3. T Cell Epitope Prediction with Restriction to Class I and Class II MHC Molecules
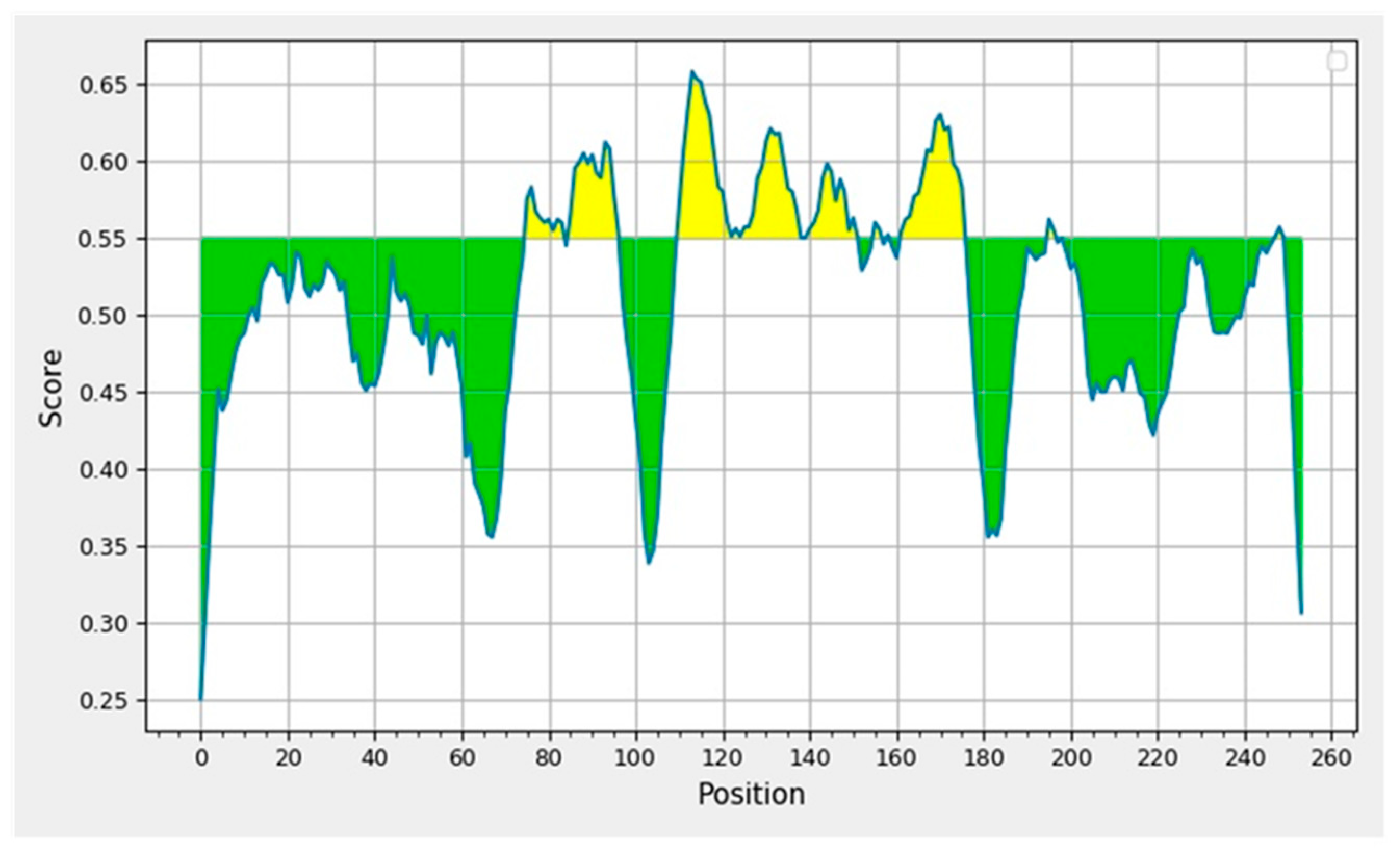
3.4. Prediction and Determination of Linear and Discontinuous Epitopes of B Cells
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Polyiam, K.; Phoolcharoen, W.; Butkhot, N.; Srisaowakarn, C.; Thitithanyanont, A.; Auewarakul, P.; Hoonsuwan, T.; Ruengjitchatchawalya, M.; Mekvichitsaeng, P.; Roshorm, Y.M. Immunodominant linear B cell epitopes in the spike and membrane proteins of SARS-CoV-2 identified by immunoinformatics prediction and immunoassay. Sci. Rep. 2021, 11, 20383. [Google Scholar] [CrossRef] [PubMed]
- Guo, E.; Guo, H. CD8 T cell epitope generation toward the continually mutating SARS-CoV-2 spike protein in genetically diverse human population: Implications for disease control and prevention. PLoS ONE 2020, 15, e0239566. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Yang, H.; Ji, W.; Wu, W.; Chen, S.; Zhang, W.; Duan, G. Virology, epidemiology, pathogenesis, and control of COVID-19. Viruses 2020, 12, 372. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.H.; Nitsche, A.; Müller, M.A.; Drosten, C.; Pöhlmann, S.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef]
- Pallesen, J.; Wang, N.; Corbett, K.S.; Wrapp, D.; Kirchdoerfer, R.N.; Turner, H.L.; Cottrell, C.A.; Becker, M.M.; Wang, L.; Shi, W.; et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. USA 2017, 114, E7348–E7357. [Google Scholar] [CrossRef] [PubMed]
- Bertoletti, A.; Tan, A.T.; Le Bert, N. The T-cell response to SARS-CoV-2: Kinetic and quantitative aspects and the case for their protective role. Oxf. Open Immunol. 2021, 2, iqab006. [Google Scholar] [CrossRef]
- Sadat, S.M.; Aghadadeghi, M.R.; Yousefi, M.; Khodaei, A.; Larijani, M.S.; Bahramali, G. Bioinformatics analysis of SARS-CoV-2 to approach an effective vaccine candidate against COVID-19. Mol. Biotechnol. 2021, 63, 389–409. [Google Scholar] [CrossRef]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; COVID-19 Genomics UK (COG-UK) Consortium; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef]
- Wieczorek, M.; Abualrous, E.T.; Sticht, J.; Álvaro-Benito, M.; Stolzenberg, S.; Noé, F.; Freund, C. Major histocompatibility complex (MHC) class I and MHC class II proteins: Conformational plasticity in antigen presentation. Front. Immunol. 2017, 8, 292. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, C.; Sharma, A.R.; Bhattacharya, M.; Sharma, G.; Lee, S.-S. Immunoinformatics approach for the identification and characterization of T cell and B cell epitopes towards the peptide-based vaccine against SARS-CoV-2. Arch. Med. Res. 2021, 52, 362–370. [Google Scholar] [CrossRef] [PubMed]
- de Sousa, E.; Ligeiro, D.; Lérias, J.R.; Zhang, C.; Agrati, C.; Osman, M.; El-Kafrawy, S.A.; Azhar, E.I.; Ippolito, G.; Wang, F.; et al. Mortality in COVID-19 disease patients: Correlating the association of major histocompatibility complex (MHC) with severe acute respiratory syndrome 2 (SARS-CoV-2) variants. Int. J. Infect. Dis. 2020, 98, 454–459. [Google Scholar] [CrossRef] [PubMed]
- Yashvardhini, N.; Kumar, A.; Jha, D.K. Immunoinformatics Identification of B-and T-Cell Epitopes in the RNA-Dependent RNA Polymerase of SARS-CoV-2. Can. J. Infect. Dis. Med. Microbiol. 2021, 2021, 6627141. [Google Scholar] [CrossRef] [PubMed]
- Weiskopf, D.; Angelo, M.A.; de Azeredo, E.L.; Sidney, J.; Greenbaum, J.A.; Fernando, A.N.; Broadwater, A.; Kolla, R.V.; de Silva, A.D.; de Silva, A.M.; et al. Comprehensive analysis of dengue virus-specific responses supports an HLA-linked protective role for CD8+ T cells. Proc. Natl. Acad. Sci. USA 2013, 110, E2046–E2053. [Google Scholar] [CrossRef] [PubMed]
- Greenbaum, J.; Sidney, J.; Chung, J.; Brander, C.; Peters, B.; Sette, A. Functional classification of class II human leukocyte antigen (HLA) molecules reveals seven different supertypes and a surprising degree of repertoire sharing across supertypes. Immunogenetics 2011, 63, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Doytchinova, I.A.; Flower, D.R. VaxiJen: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. 2007, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Jin, B.; Li, H.; Huang, S.-Y. HPEPDOCK: A web server for blind peptide–protein docking based on a hierarchical algorithm. Nucleic Acids Res. 2018, 46, W443–W450. [Google Scholar] [CrossRef]
- Peters, B.; Nielsen, M.; Sette, A. T cell epitope predictions. Annu. Rev. Immunol. 2020, 38, 123–145. [Google Scholar] [CrossRef] [PubMed]
- Arons, M.M.; Hatfield, K.M.; Reddy, S.C.; Kimball, A.; James, A.; Jacobs, J.R.; Taylor, J.; Spicer, K.; Bardossy, A.C.; Oakley, L.P.; et al. Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. N. Engl. J. Med. 2020, 382, 2081–2090. [Google Scholar] [CrossRef] [PubMed]
- Surveillances, V. The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19)—China, 2020. China CDC Wkly. 2020, 2, 113–122. [Google Scholar]
- Janice Oh, H.-L.; Ken-En Gan, S.; Bertoletti, A.; Tan, Y.-J. Understanding the T cell immune response in SARS coronavirus infection. Emerg. Microbes Infect. 2012, 1, 1–6. [Google Scholar] [CrossRef]
- Woldemeskel, B.A.; Garliss, C.C.; Blankson, J.N. SARS-CoV-2 mRNA vaccines induce broad CD4+ T cell responses that recognize SARS-CoV-2 variants and HCoV-NL63. J. Clin. Investig. 2021, 131, e149335. [Google Scholar] [CrossRef]
- Zhuang, Z.; Lai, X.; Sun, J.; Chen, Z.; Zhang, Z.; Dai, J.; Liu, D.; Li, Y.; Li, F.; Wang, Y.; et al. Mapping and role of T cell response in SARS-CoV-2–infected mice. J. Exp. Med. 2021, 218, e20202187. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; VanBlargan, L.A.; Bloyet, L.-M.; Rothlauf, P.W.; Chen, R.E.; Stumpf, S.; Zhao, H.; Errico, J.M.; Theel, E.S.; Liebeskind, M.J.; et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe 2021, 29, 477–488.e4. [Google Scholar] [CrossRef]
- Luo, M.; Liu, J.; Jiang, W.; Yue, S.; Liu, H.; Wei, S. IL-6 and CD8+ T cell counts combined are an early predictor of in-hospital mortality of patients with COVID-19. JCI Insight 2020, 5, e139024. [Google Scholar] [CrossRef] [PubMed]
- Shrotri, M.; van Schalkwyk, M.C.; Post, N.; Eddy, D.; Huntley, C.; Leeman, D.; Rigby, S.; Williams, S.V.; Bermingham, W.H.; Kellam, P.; et al. T cell response to SARS-CoV-2 infection in humans: A systematic review. PLoS ONE 2021, 16, e0245532. [Google Scholar] [CrossRef]
- Yadav, P.D.; Potdar, V.A.; Choudhary, M.L.; Nyayanit, D.A.; Agrawal, M.; Jadhav, S.M.; Majumdar, T.D.; Shete-Aich, A.; Basu, A.; Abraham, P.; et al. Full-genome sequences of the first two SARS-CoV-2 viruses from India. Indian J. Med. Res. 2020, 151, 200. [Google Scholar] [CrossRef] [PubMed]
- Hare, J.; Morrison, D.; Nielsen, M. Sampling SARS-CoV-2 proteomes for predicted CD8 T-cell epitopes as a tool for understanding immunogenic breadth and rational vaccine design. Front. Bioinform. 2021, 1, 1. [Google Scholar] [CrossRef] [PubMed]
- Almofti, Y.A.; Abd-elrahman, K.A.; Gassmallah, S.A.E.; Salih, M.A. Multi epitopes vaccine prediction against severe acute respiratory syndrome (SARS) coronavirus using immunoinformatics approaches. Am. J. Microbiol. Res. 2018, 6, 94–114. [Google Scholar] [CrossRef]
- Grifoni, A.; Sidney, J.; Vita, R.; Peters, B.; Crotty, S.; Weiskopf, D.; Sette, A. SARS-CoV-2 Human T cell Epitopes: Adaptive immune response against COVID-19. Cell Host Microbe 2021, 29, 1076–1092. [Google Scholar] [CrossRef] [PubMed]
- Heitmann, J.S.; Bilich, T.; Tandler, C.; Nelde, A.; Maringer, Y.; Marconato, M.; Reusch, J.; Jäger, S.; Denk, M.; Richter, M.; et al. A COVID-19 peptide vaccine for the induction of SARS-CoV-2 T cell immunity. Nature 2022, 601, 617–622. [Google Scholar] [CrossRef] [PubMed]
- Tartof, S.Y.; Slezak, J.M.; Fischer, H.; Hong, V.; Ackerson, B.K.; Ranasinghe, O.N.; Frankland, T.B.; Ogun, Q.A.; Zamparo, J.M.; Gray, S.; et al. Effectiveness of mRNA BNT162b2 COVID-19 vaccine up to 6 months in a large integrated health system in the USA: A retrospective cohort study. Lancet 2021, 398, 1407–1416. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, S.; Kode, V.; Bhojak, K.; Karunakaran, C.; Lee, K.; Manoharan, M.; Ramesh, A.; HV, S.; Srivastava, A.; Sathian, R.; et al. Immunodominant T-cell epitopes from the SARS-CoV-2 spike antigen reveal robust pre-existing T-cell immunity in unexposed individuals. Sci. Rep. 2021, 11, 13164. [Google Scholar] [CrossRef] [PubMed]
- Phan, I.Q.; Subramanian, S.; Kim, D.; Murphy, M.; Pettie, D.; Carter, L.; Anishchenko, I.; Barrett, L.K.; Craig, J.; Tillery, L.; et al. In silico detection of SARS-CoV-2 specific B-cell epitopes and validation in ELISA for serological diagnosis of COVID-19. Sci. Rep. 2021, 11, 4290. [Google Scholar] [CrossRef]
- Schwarz, T.; Heiss, K.; Mahendran, Y.; Casilag, F.; Kurth, F.; Sander, L.E.; Wendtner, C.M.; Hoechstetter, M.A.; Müller, M.A.; Sekul, R.; et al. SARS-CoV-2 proteome-wide analysis revealed significant epitope signatures in COVID-19 patients. Front. Immunol. 2021, 12, 629185. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.F.; Quadeer, A.A.; McKay, M.R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses 2020, 12, 254. [Google Scholar] [CrossRef] [PubMed]
- Chi, X.; Yan, R.; Zhang, J.; Zhang, G.; Zhang, Y.; Hao, M.; Zhang, Z.; Fan, P.; Dong, Y.; Yang, Y.; et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science 2020, 369, 650–655. [Google Scholar] [CrossRef]
- Poh, C.M.; Carissimo, G.; Wang, B.; Amrun, S.N.; Lee, C.Y.-P.; Chee, R.S.-L.; Fong, S.; Yeo, N.K.; Lee, W.; Torres-Ruesta, A.; et al. Two linear epitopes on the SARS-CoV-2 spike protein that elicit neutralising antibodies in COVID-19 patients. Nat. Commun. 2020, 11, 2806. [Google Scholar] [CrossRef]
- Zhang, B.; Hu, Y.; Chen, L.; Yau, T.; Tong, Y.; Hu, J.; Cai, J.; Chan, K.; Dou, Y.; Deng, J.; et al. Mining of epitopes on spike protein of SARS-CoV-2 from COVID-19 patients. Cell Res. 2020, 30, 702–704. [Google Scholar] [CrossRef] [PubMed]
- Starr, T.N.; Greaney, A.J.; Hilton, S.K.; Ellis, D.; Crawford, K.H.; Dingens, A.S.; Navarro, M.J.; Bowen, J.E.; Tortorici, M.A.; Walls, A.C.; et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell 2020, 182, 1295–1310.e20. [Google Scholar] [CrossRef] [PubMed]
- Mei, X.; Gu, P.; Shen, C.; Lin, X.; Li, J. Computer-Based Immunoinformatic Analysis to Predict Candidate T-Cell Epitopes for SARS-CoV-2 Vaccine Design. Front. Immunol. 2022, 13, 847617. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Wu, S.; Zhao, G.; He, Y.; Guo, X.; Zhang, Z.; Hou, J.; Ding, Y.; Cheng, A.; Wang, B. Identification of a promiscuous conserved CTL epitope within the SARS-CoV-2 spike protein. Emerg. Microbes Infect. 2022, 11, 730–740. [Google Scholar] [CrossRef] [PubMed]
- Zannella, C.; Chianese, A.; Greco, G.; Santella, B.; Squillaci, G.; Monti, A.; Doti, N.; Sanna, G.; Manzin, A.; Morana, A.; et al. Design of Three Residues Peptides against SARS-CoV-2 Infection. Viruses 2022, 14, 2103. [Google Scholar] [CrossRef] [PubMed]
- Nerli, S.; Sgourakis, N.G. Structure-Based Modeling of SARS-CoV-2 Peptide/HLA-A02 Antigens. Front. Med. Technol. 2020, 2, 553478. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.C.; Olsen, K.S.; Gentry, K.M.; Sambade, M.; Beck, W.; Garness, J.; Entwistle, S.; Willis, C.; Vensko, S.; Woods, A.; et al. Landscape and selection of vaccine epitopes in SARS-CoV-2. Genome Med. 2021, 13, 101. [Google Scholar] [CrossRef]
- Poran, A.; Harjanto, D.; Malloy, M.; Arieta, C.M.; Rothenberg, D.A.; Lenkala, D.; van Buuren, M.M.; Addona, T.A.; Rooney, M.S.; Srinivasan, L.; et al. Sequence-based prediction of SARS-CoV-2 vaccine targets using a mass spectrometry-based bioinformatics predictor identifies immunogenic T cell epitopes. Genome Med. 2020, 12, 70. [Google Scholar] [CrossRef]
- Galdiero, M.; Galdiero, M.; Folliero, V.; Zannella, C.; De Filippis, A.; Mali, A.; Rinaldi, L.; Franci, G. SARS-CoV-2 vaccine development: Where are we? Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 2752–2784. [Google Scholar] [CrossRef] [PubMed]
- Lippi, G.; Plebani, M.; Henry, B.M. Thrombocytopenia is Associated With Severe Coronavirus Disease 2019 (COVID-19) Infections: A Meta-Analysis. Clin. Chim. Acta 2020, 506, 145–148. [Google Scholar] [CrossRef]
- Thachil, J. What do Monitoring Platelet Counts in COVID-19 Teach Us? J. Thromb. Haemost. 2020, 18, 2071–2072. [Google Scholar] [CrossRef]
- Klok, F.A.; Pai, M.; Huisman, M.V.; Makris, M. Vaccine induced immune thrombotic thrombocytopenia. Lancet Haematol. 2022, 9, e73–e80. [Google Scholar] [CrossRef]


| Peptide | Allele | Population Coverage | Cumulative Population Coverage | VaxiJen Score | Docking Score Kcal/mol |
|---|---|---|---|---|---|
| P1: FTISVTTEI | HLA-A*68:02, HLA-B*58:01, HLA-A*02:06, HLA-A*26:01, HLA-B*51:01, HLA-A*02:01, HLA-A*02:03 | 53.24% | 95.26% | 0.8535 | −162.402 |
| P2: FAMQMAYRF | HLA-B*35:01, HLA-B*53:01, HLA-A*23:01, HLA-B*58:01, HLA-A*24:02, HLA-B*08:01 | 47.12% | 1.0278 | −173.416 | |
| P3: YQPYRVVVLSF | HLA-A*23:01, HLA-A*24:02, HLA-B*07:02, HLA-B*51:01, HLA-B*15:01 | 46.30% | 0.8648 | −178.359 | |
| P4: SLIDLQELGK | HLA-A*11:01, HLA-A*03:01, HLA-A*01:01 | 45.19% | 1.0275 | −131.419 | |
| P5: KLNDLCFTNV | HLA-A*02:03, HLA-A*32:01, HLA-A*02:01 | 43.40% | 2.6927 | −166.504 | |
| P6: FVFLVLLPLV | HLA-A*02:06, HLA-A*02:01, HLA-A*02:03, HLA-A*02:06, HLA-A*02:01, HLA-A*68:02 | 43.26% | 0.8044 | −170.516 | |
| P7: KIADYNYKL | HLA-A*32:01, HLA-A*02:01 | 42.66% | 1.6639 | −164.515 | |
| P8: YIWLGFIAGL | HLA-A*02:01, HLA-A*02:06 | 40.60% | 0.5798 | −182.395 | |
| P9: GLIAIVMVTI | HLA-A*02:03, HLA-A*02:01 | 39.84% | 1.0813 | −159.635 | |
| P10: FELLHAPATV | HLA-A*02:01 | 39.08% | 0.5982 | −167.465 |
| Peptide | Allele | Population Coverage | Cumulative Population Coverage | VaxiJen Score | Docking Score Kcal/mol |
|---|---|---|---|---|---|
| P1: IRASANLAA | HLA-DPA1*02:01,HLA-DPB1*14:01,HLA-DQA1*01:02,HLA-DQB1*06:02,HLA-DRB1*09:01,HLA-DRB1*04:05,HLA-DPA1*01:03,HLA-DPB1*04:01,HLA-DRB1*11:01,HLA-DRB1*04:01,HLA-DRB1*01:01,HLA-DRB1*08:02,HLA-DPA1*03:01,HLA-DPB1*04:02,HLA-DQA1*01:01,HLA-DQB1*05:01,HLA-DRB1*12:01,HLA-DRB1*13:02,HLA-DRB1*07:01,HLA-DQA1*05:01,HLA-DQB1*03:01,HLA-DRB1*03:01,HLA-DQA1*04:01,HLA-DQB1*04:02,HLA-DQA1*03:01,HLA-DQB1*03:02 | 99.94% | 99.99% | 0.4455 | −160.809 |
| P2: FLHVTYVPA | HLA-DPA1*01:03,HLA-DPA1*02:01,HLA-DPA1*03:01,HLA-DPB1*01:01,HLA-DPB1*02:01,HLA-DPB1*04:01,HLA-DPB1*04:02,HLA-DPB1*05:01,HLA-DPB1*14:01,HLA-DQA1*01:01,HLA-DQA1*01:02,HLA-DQA1*03:01,HLA-DQA1*04:01,HLA-DQA1*05:01,HLA-DQB1*02:01,HLA-DQB1*03:02,HLA-DQB1*04:02,HLA-DQB1*05:01,HLA-DQB1*06:02,HLA-DRB1*04:01 | 99.90% | 1.3346 | −204.550 | |
| P3: FNATRFASV | HLA-DRB1*01:01, HLA-DPA1*01:03, HLA-DPB1*04:01, HLA-DPA1*02:01, HLA-DPB1*14:01, HLA-DRB1*15:01, HLA-DPB1*05:01, HLA-DRB1*04:01, HLA-DPA1*03:01, HLA-DPB1*04:02, HLA-DQA1*05:01, HLA-DQB1*03:01, HLA-DQA1*03:01, HLA-DQB1*03:02, HLA-DQA1*01:02, HLA-DQB1*06:02, HLA-DRB1*03:01, HLA-DQB1*02:01 | 99.85% | 0.5609 | −199.001 | |
| P4: FTISVTTEI | HLA-DPA1*01:03,HLA-DPA1*02:01,HLA-DPA1*03:01,HLA-DPB1*01:01,HLA-DPB1*04:01,HLA-DPB1*04:02,HLA-DQA1*01:02,HLA-DQA1*03:01,HLA-DQA1*05:01,HLA-DQB1*03:01,HLA-DQB1*03:02,HLA-DQB1*06:02,HLA-DRB1*01:01,HLA-DRB1*03:01,HLA-DRB1*04:01,HLA-DRB1*04:05,HLA-DRB1*07:01,HLA-DRB1*08:02,HLA-DRB1*11:01,HLA-DRB3*02:02 | 99.76% | 0.8535 | −140.288 | |
| P5: FLPFFSNVT | HLA-DPA1*01:03, HLA-DPA1*02:01, HLA-DPA1*03:01, HLA-DPB1*01:01, HLA-DPB1*02:01, HLA-DPB1*04:01, HLA-DPB1*04:02, HLA-DPB1*05:01, HLA-DPB1*14:01, HLA-DQA1*03:01, HLA-DQA1*05:01, HLA-DQB1*02:01, HLA-DQB1*03:01, HLA-DQB1*03:02, HLA-DRB1*01:01 | 99.68% | 0.4400 | −183.885 | |
| P6: FSNVTWFHA | HLA-DPA1*01:03, HLA-DPA1*02:01, HLA-DPA1*03:01, HLA-DPB1*01:01, HLA-DPB1*04:01, HLA-DPB1*04:02, HLA-DPB1*05:01, HLA-DPB1*14:01, HLA-DQA1*01:01, HLA-DQA1*04:01, HLA-DQA1*05:01, HLA-DQB1*03:01, HLA-DQB1*04:02, HLA-DQB1*05:01, HLA-DRB1*01:01, HLA-DRB1*03:01, HLA-DRB1*04:05 | 99.49% | 0.8156 | −238.837 | |
| P7: FGAISSVLN | HLA-DPA1*01:03, HLA-DPA1*02:01, HLA-DPA1*03:01, HLA-DPB1*02:01, HLA-DPB1*04:01, HLA-DPB1*04:02, HLA-DPB1*05:01, HLA-DPB1*14:01, HLA-DQA1*01:01, HLA-DQA1*01:02, HLA-DQA1*03:01, HLA-DQB1*03:02, HLA-DQB1*05:01, HLA-DQB1*06:02, HLA-DRB1*01:01 | 99.47% | 0.5435 | −164.450 | |
| P8: FGAGAALQI | HLA-DPA1*01:03, HLA-DPA1*02:01, HLA-DPA1*03:01, HLA-DPB1*01:01, HLA-DPB1*02:01, HLA-DPB1*04:01, HLA-DPB1*04:02, HLA-DPB1*05:01, HLA-DPB1*14:01, HLA-DQA1*01:02, HLA-DQA1*05:01, HLA-DQB1*03:01, HLA-DQB1*06:02, HLA-DRB1*09:01, HLA-DRB3*01:01 | 99.45% | 0.6377 | −163.317 | |
| P9: FKIYSKHTP | HLA-DPA1*01:03, HLA-DPA1*02:01, HLA-DPA1*03:01, HLA-DPB1*01:01, HLA-DPB1*02:01, HLA-DPB1*04:02, HLA-DPB1*14:01, HLA-DQA1*01:01, HLA-DQA1*03:01, HLA-DQA1*05:01,HLA-DQB1*03:01,HLA-DQB1*03:02,HLA-DQB1*05:01,HLA-DRB1*01:01,HLA-DRB1*03:01 | 99.44% | 0.9886 | −171.594 | |
| P10: FRVQPTESI | HLA-DPA1*02:01,HLA-DPA1*03:01,HLA-DPB1*01:01,HLA-DPB1*04:02,HLA-DPB1*05:01,HLA-DPB1*14:01,HLA-DQA1*01:01,HLA-DQA1*01:02,HLA-DQA1*03:01,HLA-DQA1*04:01,HLA-DQA1*05:01,HLA-DQB1*03:01,HLA-DQB1*03:02,HLA-DQB1*04:02,HLA-DQB1*05:01,HLA-DQB1*06:02,HLA-DRB1*01:01,HLA-DRB1*03:01,HLA-DRB1*04:01,HLA-DRB1*04:05 | 99.19% | 0.9396 | −155.782 |
| Peptide | Start-End | Allele | Population Coverage | Cumulative Population Coverage | VaxiJen Score |
|---|---|---|---|---|---|
| P1: YQPYRVVVLSF | 505–515 | HLA-A*23:01, HLA-A*24:02, HLA-B*07:02, HLA-B*51:01, HLA-B*15:01 | 46.30% | 94.54% | 0.8648 |
| P2: KLNDLCFTNV | 386–395 | HLA-A*32:01, HLA-A*02:03, HLA-A*02:01 | 43.40% | 2.6927 | |
| P3: KIADYNYKL | 417–425 | HLA-A*32:01, HLA-A*02:01 | 42.66% | 1.6639 | |
| P4: FELLHAPATV | 515–524 | HLA-A*02:01 | 39.08% | 0.5982 | |
| P5: FASVYAWNRK | 347–356 | HLA-A*68:01, HLA-A*33:01, HLA-A*11:01, HLA-A*31:01 | 27.17% | 0.5868 | |
| P6: CYFPLQSYGFQ | 488–498 | HLA-A*24:02, HLA-A*23:01 | 26.18% | 0.7378 | |
| P7: VYAWNRKRI | 350–358 | HLA-A*24:02, HLA-A*23:01 | 26.18% | 0.5003 | |
| P8: RQIAPGQTGK | 408–417 | HLA-B*15:01, HLA-A*03:01 | 23.84% | 1.7893 | |
| P9: QTGKIADYNY | 414–423 | HLA-A*30:02, HLA-A*01:01 | 19.55% | 1.5116 | |
| P10: NLDSKVGGNY | 440–449 | HLA-A*01:01 | 17.34% | 0.7882 |
| Peptide | Start-End | Allele | Population Coverage | Cumulative Population Coverage | VaxiJen Score |
|---|---|---|---|---|---|
| P1: FNATRFASV | 342–350 | HLA-DRB1*01:01, HLA-DPA1*01:03, HLA-DPB1*04:01, HLA-DPA1*02:01, HLA-DPB1*14:01, HLA-DRB1*15:01, HLA-DPB1*05:01, HLA-DRB1*04:01, HLA-DPA1*03:01, HLA-DPB1*04:02, HLA-DQA1*05:01, HLA-DQB1*03:01, HLA-DQA1*03:01, HLA-DQB1*03:02, HLA-DQA1*01:02, HLA-DQB1*06:02, HLA-DRB1*03:01, HLA-DQB1*02:01 | 99.85% | 99.99% | 0.5609 |
| P2: VVVLSFELLH | 510–519 | HLA-DPA1*01:03, HLA-DPA1*02:01, HLA-DPA1*03:01, HLA-DPB1*01:01, HLA-DPB1*02:01, HLA-DPB1*04:02, HLA-DPB1*05:01, HLA-DQA1*01:01, HLA-DQA1*05:01, HLA-DQB1*02:01, HLA-DQB1*05:01, HLA-DRB1*03:01, HLA-DRB1*04:05, HLA-DRB1*09:01, HLA-DRB4*01:01 | 98.89% | 1.2274 | |
| P3: RVVVLSFEL | 509–517 | HLA-DPA1*01:03, HLA-DPA1*02:01, HLA-DPA1*03:01, HLA-DPB1*02:01, HLA-DPB1*04:01, HLA-DPB1*04:02, HLA-DPB1*05:01, HLA-DPB1*14:01, HLA-DRB1*07:01, HLA-DRB1*15:01 | 98.24% | 1.1918 | |
| P4: YRVVVLSFE | 508–516 | HLA-DPA1*01:03, HLA-DPA1*02:01, HLA-DPB1*04:01, HLA-DPB1*05:01, HLA-DPB1*14:01, HLA-DQA1*03:01, HLA-DQA1*05:01, HLA-DQB1*02:01, HLA-DQB1*03:02, HLA-DRB1*04:05, HLA-DRB4*01:01 | 98.06% | 1.2096 | |
| P5: YQPYRVVVL | 505–513 | HLA-DPA1*01:03, HLA-DPA1*02:01, HLA-DPA1*03:01, HLA-DPB1*01:01, HLA-DPB1*02:01, HLA-DPB1*04:01, HLA-DPB1*04:02, HLA-DRB1*01:01, HLA-DRB1*07:01, HLA-DRB1*15:01, HLA-DRB3*01:01 | 97.88% | 0.5964 | |
| P6: FRKSNLKPF | 456–464 | HLA-DPA1*01:03, HLA-DPA1*02:01, HLA-DPA1*03:01, HLA-DPB1*04:01, HLA-DPB1*04:02, HLA-DPB1*05:01, HLA-DPB1*14:01, HLA-DRB1*07:01, HLA-DRB1*08:02, HLA-DRB1*09:01, HLA-DRB1*11:01, HLA-DRB1*12:01, HLA-DRB1*13:02, HLA-DRB3*01:01, HLA-DRB3*02:02, HLA-DRB5*01:01 | 97.72% | 0.6280 | |
| P7: VLYNSASFS | 367–375 | HLA-DPA1*01:03, HLA-DPA1*02:01, HLA-DPB1*02:01, HLA-DPB1*14:01, HLA-DQA1*05:01, HLA-DQB1*03:01, HLA-DRB1*03:01, HLA-DRB1*08:02, HLA-DRB1*12:01, HLA-DRB1*13:02, HLA-DRB3*02:02 | 96.51% | 0.4029 | |
| P8: VLSFELLHA | 512–520 | HLA-DPA1*01:03, HLA-DPA1*02:01, HLA-DPB1*04:01, HLA-DPB1*14:01, HLA-DQA1*01:01, HLA-DQB1*05:01, HLA-DRB1*01:01, HLA-DRB1*04:01, HLA-DRB1*12:01, HLA-DRB1*15:01 | 96.07% | 1.0776 | |
| P9: LCFTNVYAD | 390–398 | HLA-DPA1*01:03, HLA-DPB1*04:01, HLA-DQA1*01:02, HLA-DQA1*03:01, HLA-DQA1*04:01, HLA-DQB1*03:02, HLA-DQB1*04:02, HLA-DQB1*06:02, HLA-DRB1*07:01 | 95.16% | 0.9994 | |
| P10: FNCYFPLQS | 486–494 | HLA-DPA1*01:03, HLA-DPB1*04:01, HLA-DQA1*01:01, HLA-DQB1*05:01, HLA-DRB1*01:01, HLA-DRB1*04:01, HLA-DRB1*04:05, HLA-DRB1*09:01, HLA-DRB1*15:01, HLA-DRB5*01:01 | 92.24% | 1.0649 |
| HLA Allele | No of Hits | Epitopes | HLA Allele | No of Hits | Epitopes |
|---|---|---|---|---|---|
| HLA-DPA1*01:03 | 10 | P1-P10 | HLA-DPB1*02:01 | 4 | P2, P3, P5, P7 |
| HLA-DPB1*04:01 | 8 | P1, P3-P6, P8-P10 | HLA-DQA1*01:01 | 3 | P2, P8, P10 |
| HLA-DPA1*02:01 | 8 | P1-P8 | HLA-DQB1*02:01 | 3 | P1, P2, P4 |
| HLA-DPB1*14:01 | 6 | P1, P3, P4, P6-P8 | HLA-DRB1*04:05 | 3 | P2, P4, P10 |
| HLA-DRB1*15:01 | 5 | P1, P3, P5, P8, P10 | HLA-DRB1*04:01 | 3 | P1, P8, P10 |
| HLA-DPB1*05:01 | 5 | P1-P4, P6 | HLA-DRB1*12:01 | 3 | P6-P8 |
| HLA-DPA1*03:01 | 5 | P1-P3, P5, P6 | HLA-DRB1*09:01 | 3 | P2, P6, P10 |
| HLA-DPB1*04:02 | 5 | P1-P3, P5, P6 | HLA-DQA1*03:01 | 3 | P1, P4, P9 |
| HLA-DQA1*05:01 | 4 | P1, P2, P4, P7 | HLA-DQB1*03:02 | 3 | P1, P4, P9 |
| HLA-DRB1*01:01 | 4 | P1, P5, P8, P10 | HLA-DQB1*05:01 | 3 | P2, P8, P10 |
| HLA-DRB1*07:01 | 4 | P3, P5, P6, P9 | HLA-DRB1*03:01 | 3 | P1, P2, P7 |
| HLA-DRB5*01:01 | 2 | P6, P10 | HLA-DPB1*01:01 | 2 | P2, P5 |
| HLA-DRB3*01:01 | 2 | P5, P6 | HLA-DQB1*06:02 | 2 | P1, P9 |
| HLA-DRB1*08:02 | 2 | P6, P7 | HLA-DRB3*02:02 | 2 | P6, P7 |
| HLA-DQB1*03:01 | 2 | P1, P7 | HLA-DQA1*04:01 | 1 | P9 |
| HLA-DRB4*01:01 | 2 | P2, P4 | HLA-DRB1*11:01 | 1 | P6 |
| HLA-DQA1*01:02 | 2 | P1, P9 | HLA-DQB1*04:02 | 1 | P9 |
| HLA-DRB1*13:02 | 2 | P6, P7 |
| Sl. No. | Start | End | Peptide | Peptide Length | VaxiJen Score (Cut Off = 0.4) |
|---|---|---|---|---|---|
| 1 | 405 | 413 | DEVRQIAPG | 9 | 0.7216 |
| 2 | 415 | 426 | TGKIADYNYKLP | 12 | 1.1956 |
| 3 | 440 | 468 | NLDSKVGGNYNYLYRLFRKSNLKPFERDI | 29 | 0.3934 |
| 4 | 470 | 481 | TEIYQAGSTPCN | 12 | 0.0966 |
| 5 | 491 | 505 | PLQSYGFQPTNGVGY | 15 | 0.3415 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarma, K.; Bali, N.K.; Sarmah, N.; Borkakoty, B. In Silico Screening of Prospective MHC Class I and II Restricted T-Cell Based Epitopes of the Spike Protein of SARS-CoV-2 for Designing of a Peptide Vaccine for COVID-19. COVID 2022, 2, 1731-1747. https://doi.org/10.3390/covid2120124
Sarma K, Bali NK, Sarmah N, Borkakoty B. In Silico Screening of Prospective MHC Class I and II Restricted T-Cell Based Epitopes of the Spike Protein of SARS-CoV-2 for Designing of a Peptide Vaccine for COVID-19. COVID. 2022; 2(12):1731-1747. https://doi.org/10.3390/covid2120124
Chicago/Turabian StyleSarma, Kishore, Nargis K. Bali, Neelanjana Sarmah, and Biswajyoti Borkakoty. 2022. "In Silico Screening of Prospective MHC Class I and II Restricted T-Cell Based Epitopes of the Spike Protein of SARS-CoV-2 for Designing of a Peptide Vaccine for COVID-19" COVID 2, no. 12: 1731-1747. https://doi.org/10.3390/covid2120124
APA StyleSarma, K., Bali, N. K., Sarmah, N., & Borkakoty, B. (2022). In Silico Screening of Prospective MHC Class I and II Restricted T-Cell Based Epitopes of the Spike Protein of SARS-CoV-2 for Designing of a Peptide Vaccine for COVID-19. COVID, 2(12), 1731-1747. https://doi.org/10.3390/covid2120124






