Microfluidic System for Cell Mixing and Particle Focusing Using Dean Flow Fractionation
Abstract
:1. Introduction
2. Materials and Methods
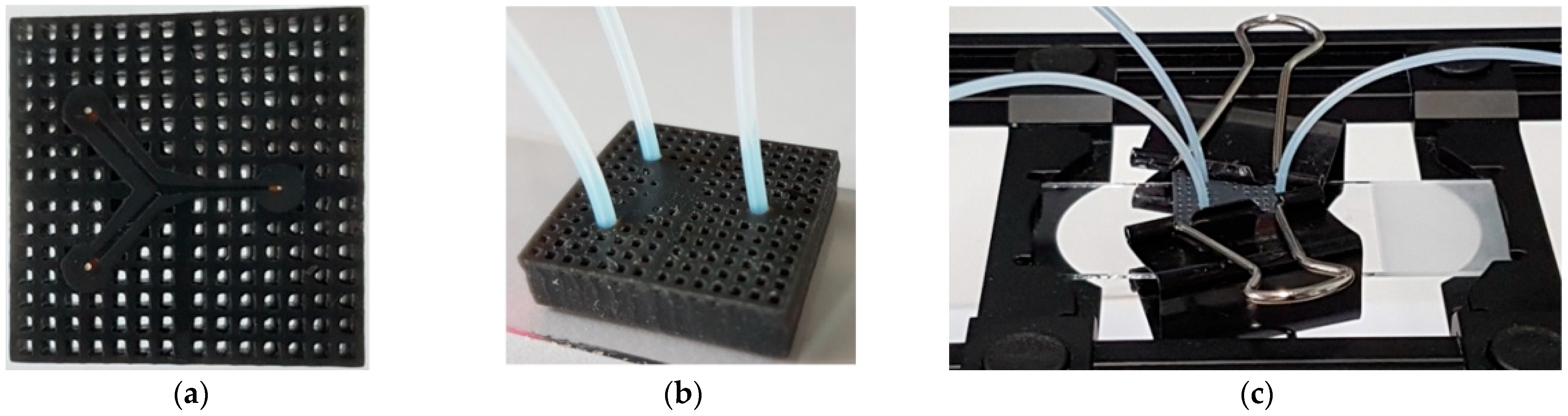
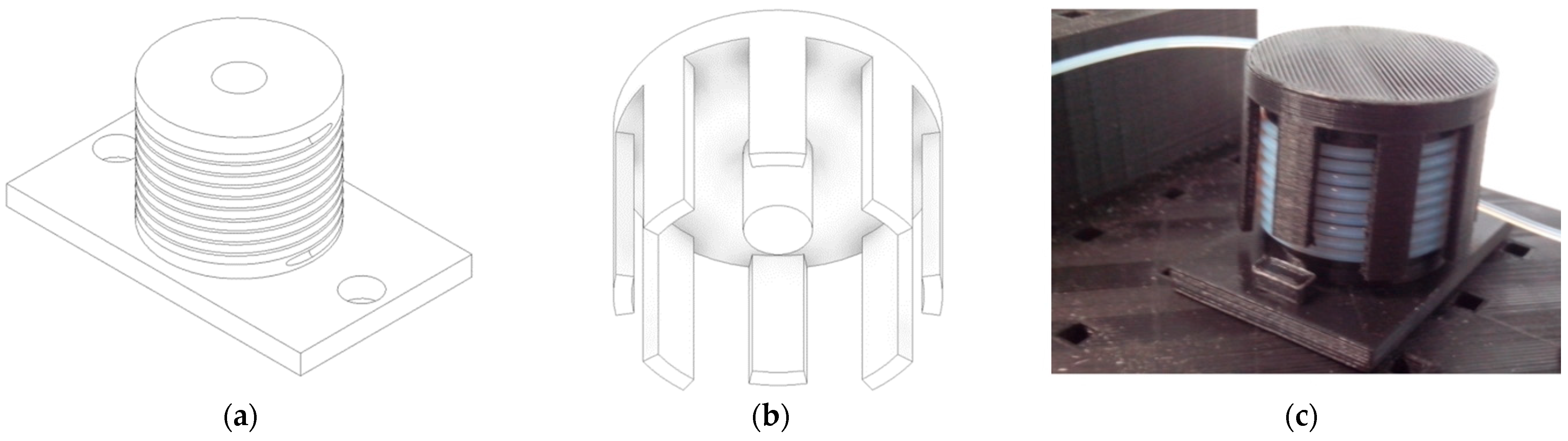
2.1. Microfluidic Component Printing and Assembly
2.2. Cell Culture, Fluorescence Labeling, and Live–Dead Staining
2.3. Bacterial Culture
2.4. Fluorescence Microscopy
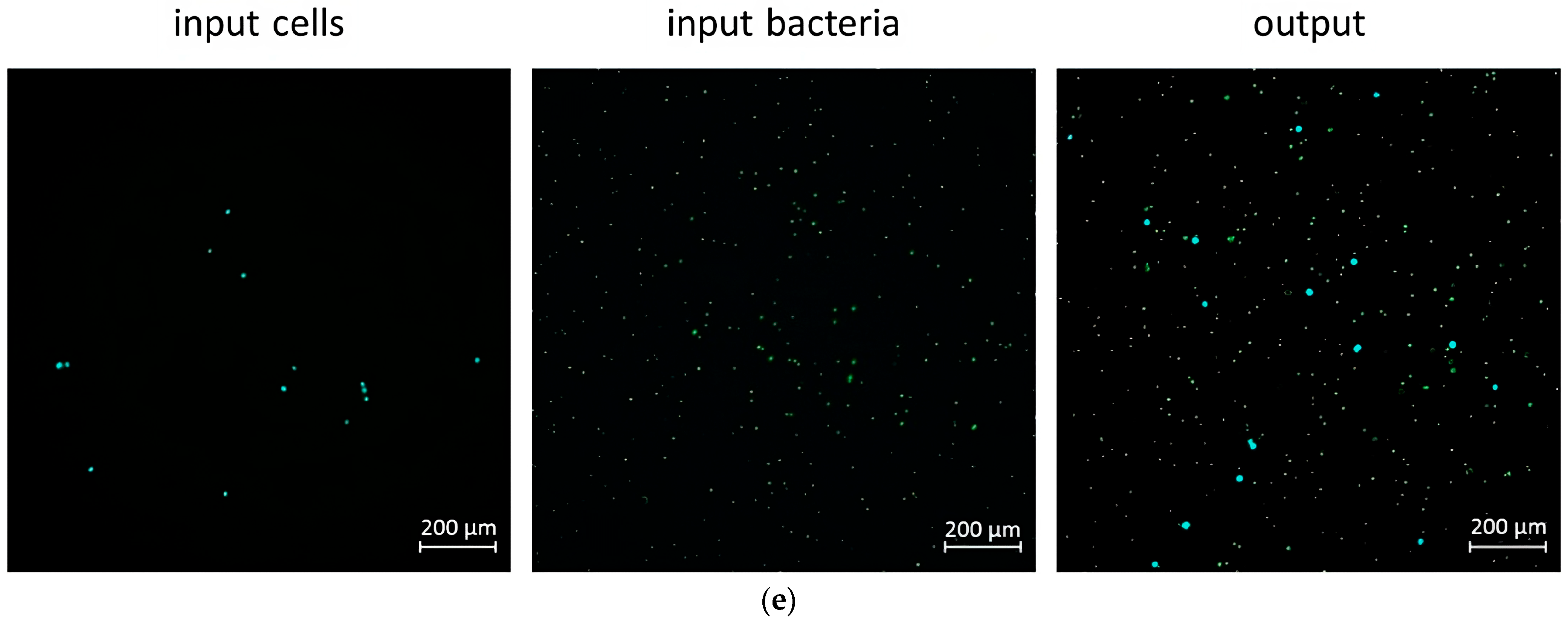
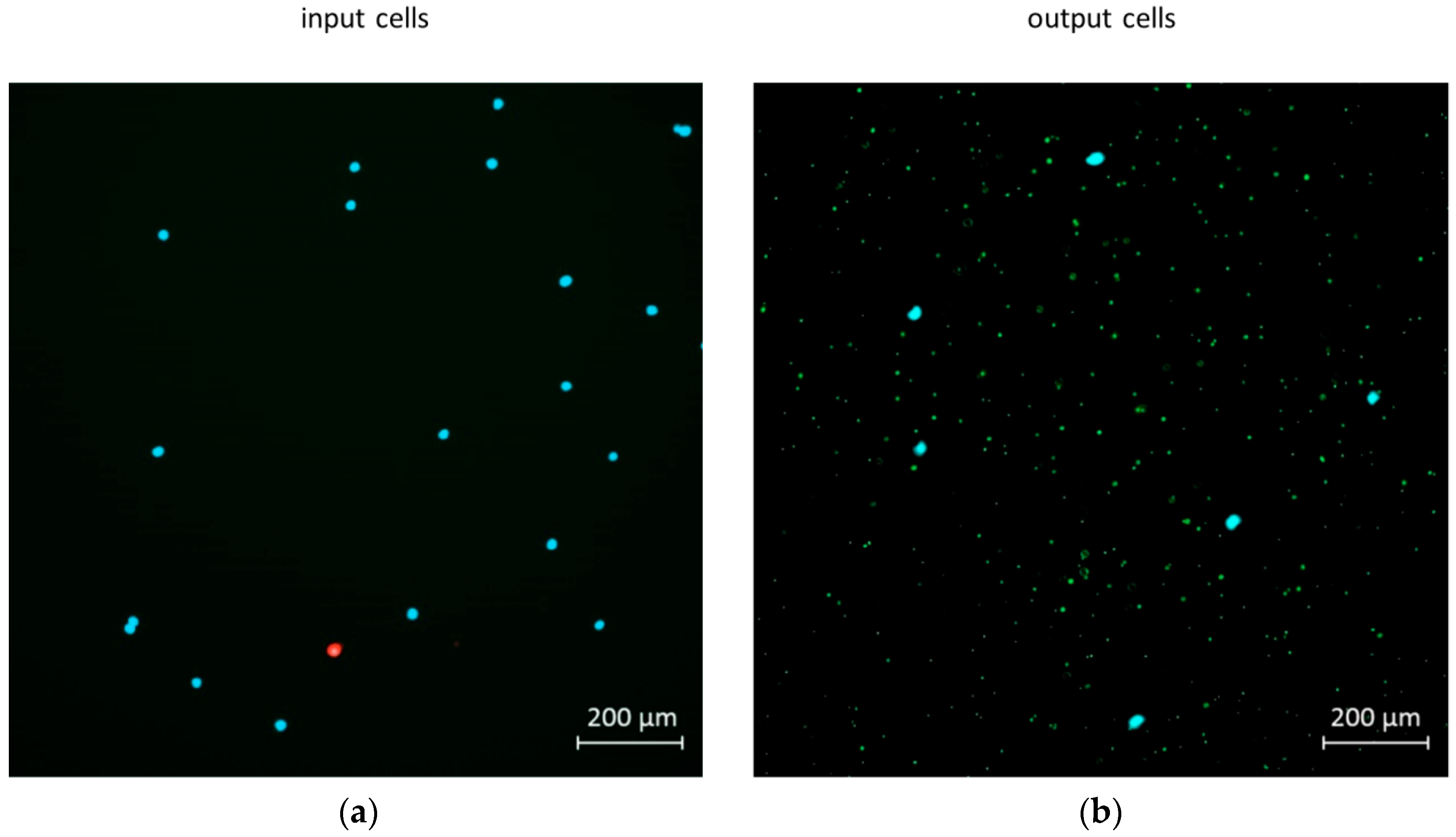
2.5. Mixing and Separation Particles of Cells with Bacteria
3. Results
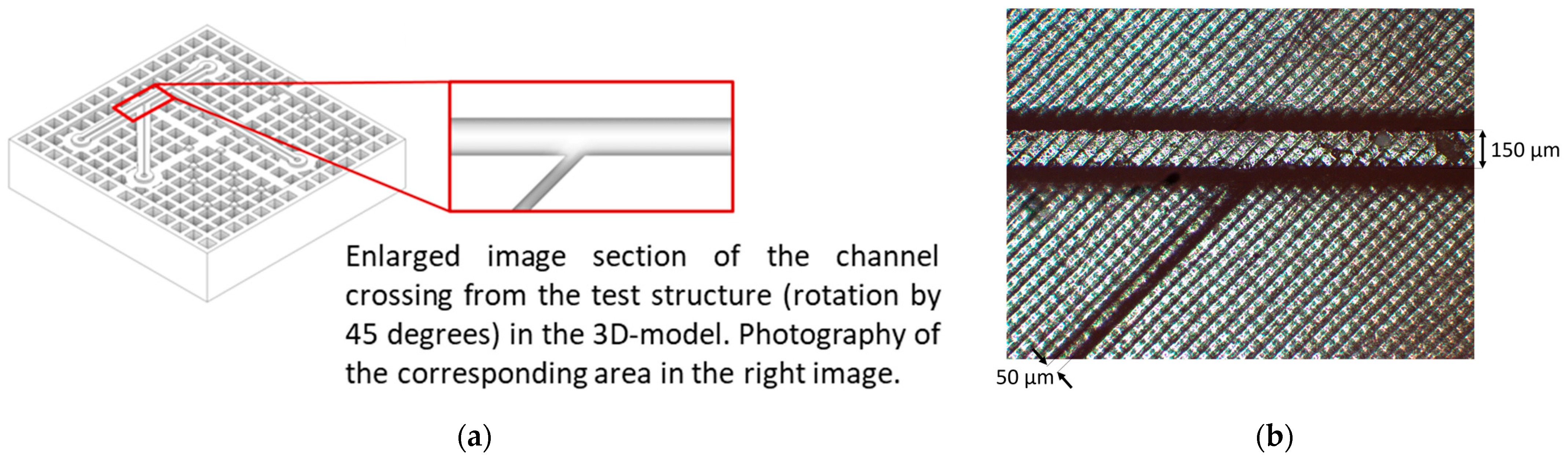
3.1. Microfluidic Channels
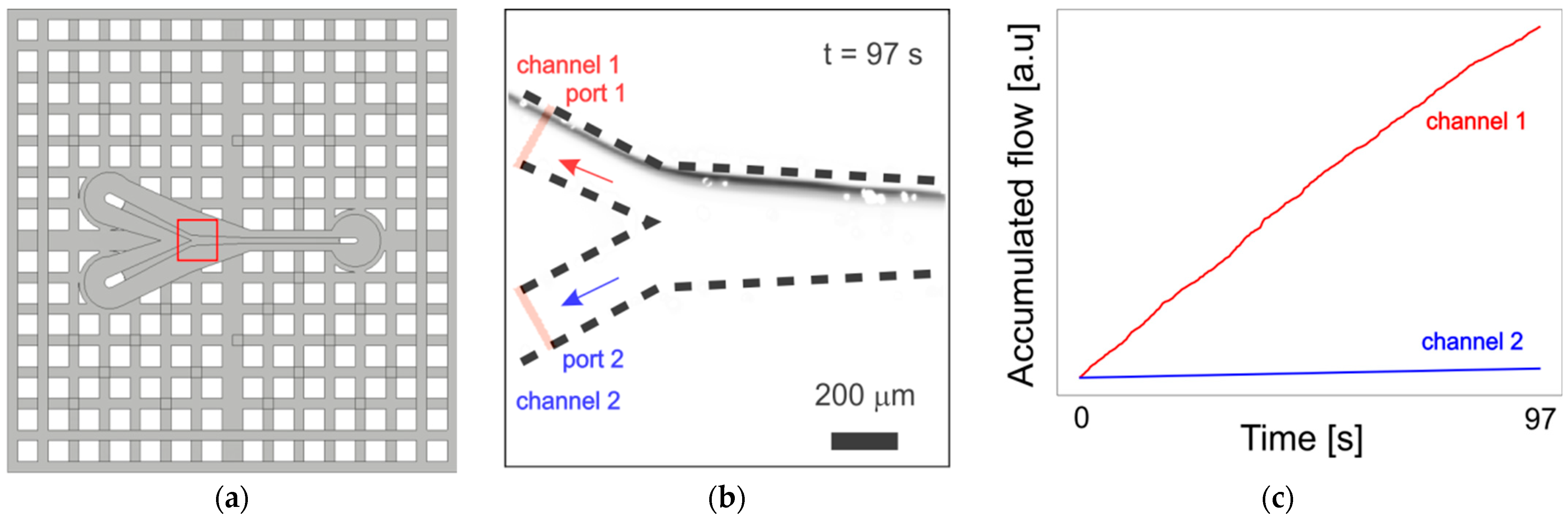
3.2. Mixer
3.3. Dean Flow Helix
3.3.1. Establishment of Microfluidic Components
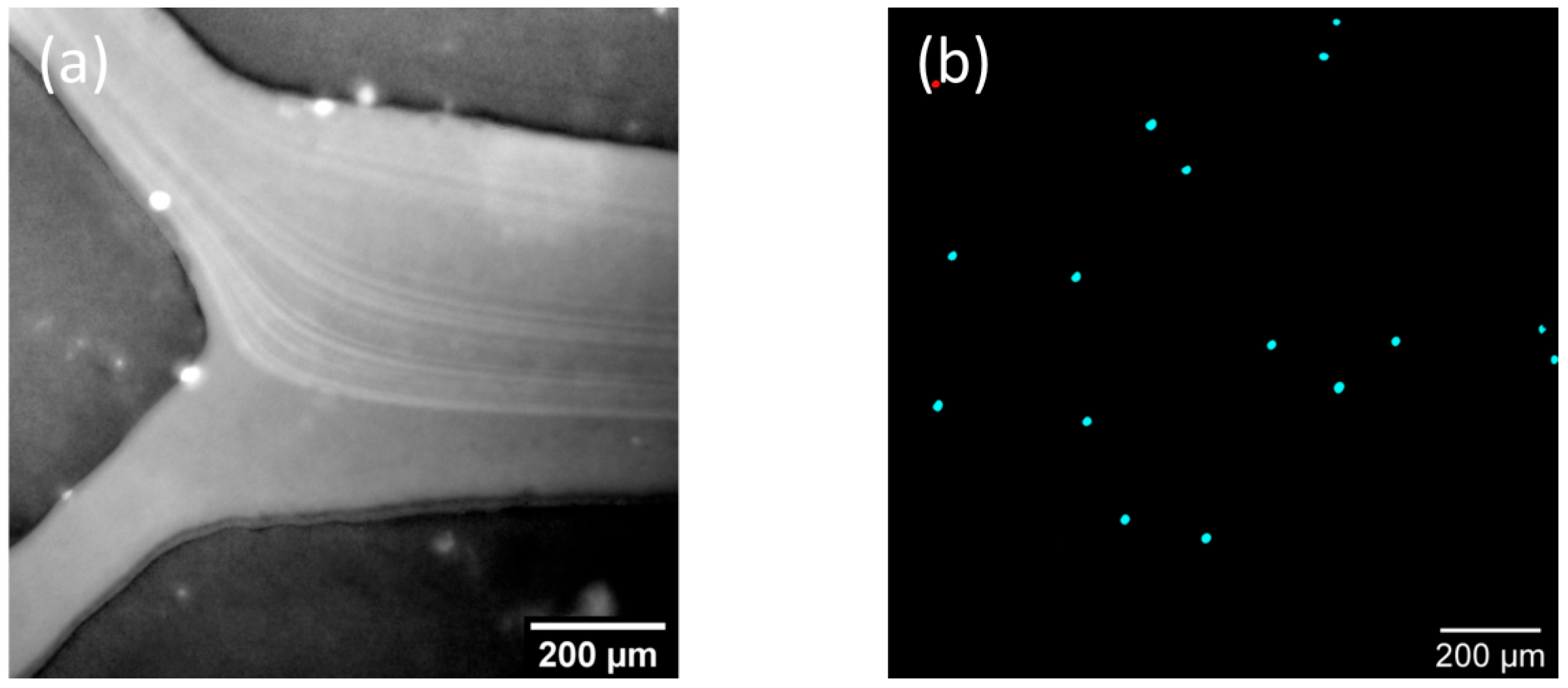
3.3.2. Application Examples: Separating Small and Large Particles in Dean’s Flow Helix
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lenshof, A.; Laurell, T. Continuous separation of cells and particles in microfluidic systems. Chem. Soc. Rev. 2010, 39, 1203–1217. [Google Scholar] [CrossRef]
- Bhagat, A.A.; Bow, H.; Hou, H.W.; Tan, S.J.; Han, J.; Lim, C.T. Microfluidics for cell separation. Med. Biol. Eng. Comput. 2010, 48, 999–1014. [Google Scholar] [CrossRef]
- Gossett, D.R.; Weaver, W.M.; Mach, A.J.; Hur, S.C.; Tse, H.T.K.; Lee, W.; Amini, H.; Di Carlo, D. Label-free cell separation and sorting in microfluidic systems. Anal. Bioanal. Chem. 2010, 397, 3249–3267. [Google Scholar] [CrossRef]
- Whitesides, G.M. The origins and the future of microfluidics. Nature 2006, 442, 368–373. [Google Scholar] [CrossRef]
- Zhou, J.; Ellis, A.V.; Voelcker, N.H. Recent developments in PDMS surface modification for microfluidic devices. Electrophoresis 2010, 31, 2–16. [Google Scholar] [CrossRef]
- Eddings, M.A.; Johnson, M.A.; Gale, B.K. Determining the optimal PDMS-PDMS bonding technique for microfluidic devices. J. Micromechanics Microeng. 2008, 18, 67001. [Google Scholar] [CrossRef]
- Au, A.K.; Bhattacharjee, N.; Horowitz, L.F.; Chang, T.C.; Folch, A. 3D-printed microfluidic automation. Lab Chip 2015, 15, 1934–1941. [Google Scholar] [CrossRef]
- Bhattacharjee, N.; Urrios, A.; Kang, S.; Folch, A. The upcoming 3D-printing revolution in microfluidics. Lab Chip 2016, 16, 1720–1742. [Google Scholar] [CrossRef]
- Nielsen, A.V.; Beauchamp, M.J.; Nordin, G.P.; Woolley, A.T. 3D Printed Microfluidics. Annu. Rev. Anal. Chem. 2020, 13, 45–65. [Google Scholar] [CrossRef]
- Au, A.K.; Huynh, W.; Horowitz, L.F.; Folch, A. 3D-Printed Microfluidics. Angew. Chem. Int. Ed. 2016, 55, 3862–3881. [Google Scholar] [CrossRef]
- Waheed, S.; Cabot, J.M.; Macdonald, N.P.; Lewis, T.; Guijt, R.M.; Paull, B.; Breadmore, M.C. 3D printed microfluidic devices: Enablers and barriers. Lab Chip 2016, 16, 1993–2013. [Google Scholar] [CrossRef] [PubMed]
- Su, R.; Wang, F.; McAlpine, M.C. 3D printed microfluidics: Advances in strategies, integration, and applications. Lab Chip 2023, 23, 1279–1299. [Google Scholar] [CrossRef] [PubMed]
- Monia Kabandana, G.K.; Zhang, T.; Chen, C. Emerging 3D printing technologies and methodologies for microfluidic development. Anal. Methods 2022, 14, 2885–2906. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, H.K.; Badar, F.; Doeven, E.H.; Novak, J.I.; Merenda, A.; Dumée, L.F.; Loy, J.; Guijt, R.M. 3D Printing: An Alternative Microfabrication Approach with Unprecedented Opportunities in Design. Anal. Chem. 2021, 93, 350–366. [Google Scholar] [CrossRef]
- Ho, C.M.B.; Ng, S.H.; Li, K.H.H.; Yoon, Y.J. 3D printed microfluidics for biological applications. Lab Chip 2015, 15, 3627–3637. [Google Scholar] [CrossRef]
- Macdonald, N.P.; Cabot, J.M.; Smejkal, P.; Guijt, R.M.; Paull, B.; Breadmore, M.C. Comparing Microfluidic Performance of Three-Dimensional (3D) Printing Platforms. Anal. Chem. 2017, 89, 3858–3866. [Google Scholar] [CrossRef]
- Gonzalez, G.; Roppolo, I.; Pirri, C.F.; Chiappone, A. Current and emerging trends in polymeric 3D printed microfluidic devices. Addit. Manuf. 2022, 55, 102867. [Google Scholar] [CrossRef]
- Do, J.; Zhang, J.Y.; Klapperich, C.M. Maskless writing of microfluidics: Rapid prototyping of 3D microfluidics using scratch on a polymer substrate. Robot. Comput. Integr. Manuf. 2011, 27, 245–248. [Google Scholar] [CrossRef]
- Del Rosario, M.; Heil, H.S.; Mendes, A.; Saggiomo, V.; Henriques, R. The Field Guide to 3D Printing in Optical Microscopy for Life Sciences. Adv. Biol. 2022, 6, 2100994. [Google Scholar] [CrossRef]
- Dalili, A.; Samiei, E.; Hoorfar, M. A review of sorting, separation and isolation of cells and microbeads for biomedical applications: Microfluidic approaches. Analyst 2019, 144, 87–113. [Google Scholar] [CrossRef]
- Bayareh, M. An updated review on particle separation in passive microfluidic devices. Chem. Eng. Process. Process Intensif. 2020, 153, 107984. [Google Scholar] [CrossRef]
- Lee, W.; Kwon, D.; Choi, W.; Jung, G.Y.; Au, A.K.; Folch, A.; Jeon, S. 3D-Printed Microfluidic Device for the Detection of Pathogenic Bacteria Using Size-based Separation in Helical Channel with Trapezoid Cross-Section. Sci. Rep. 2015, 5, 7717. [Google Scholar] [CrossRef]
- Hahn, Y.K.; Hong, D.; Kang, J.H.; Choi, S. A Reconfigurable Microfluidics Platform for Microparticle Separation and Fluid Mixing. Micromachines 2016, 7, 139. [Google Scholar] [CrossRef] [PubMed]
- Razavi Bazaz, S.; Rouhi, O.; Raoufi, M.A.; Ejeian, F.; Asadnia, M.; Jin, D.; Ebrahimi Warkiani, M. 3D Printing of Inertial Microfluidic Devices. Sci. Rep. 2020, 10, 5929. [Google Scholar] [CrossRef] [PubMed]
- Russom, A.; Gupta, A.K.; Nagrath, S.; Di Carlo, D.; Edd, J.F.; Toner, M. Differential inertial focusing of particles in curved low-aspect-ratio microchannels. New J. Phys. 2009, 11, 75025. [Google Scholar] [CrossRef] [PubMed]
- Kuntaegowdanahalli, S.S.; Bhagat, A.A.S.; Kumar, G.; Papautsky, I. Inertial microfluidics for continuous particle separation in spiral microchannels. Lab Chip 2009, 9, 2973–2980. [Google Scholar] [CrossRef] [PubMed]
- Johnston, I.D.; McDonnell, M.B.; Tan, C.K.L.; McCluskey, D.K.; Davies, M.J.; Tracey, M.C. Dean flow focusing and separation of small microspheres within a narrow size range. Microfluid. Nanofluid 2014, 17, 509–518. [Google Scholar] [CrossRef]
- Nivedita, N.; Papautsky, I. Continuous separation of blood cells in spiral microfluidic devices. Biomicrofluidics 2013, 7, 054101. [Google Scholar] [CrossRef]
- Zhang, J.; Yan, S.; Yuan, D.; Alici, G.; Nguyen, N.-T.; Ebrahimi Warkiani, M.; Li, W. Fundamentals and applications of inertial microfluidics: A review. Lab Chip 2016, 16, 10–34. [Google Scholar] [CrossRef]
- Di Carlo, D.; Irimia, D.; Tompkins, R.G.; Toner, M. Continuous inertial focusing, ordering, and separation of particles in microchannels. Proc. Natl. Acad. Sci. USA 2007, 104, 18892–18897. [Google Scholar] [CrossRef]
- Dutz, S.; Hayden, M.E.; Schaap, A.; Stoeber, B.; Hafeli, U.O. A microfluidic spiral for size-dependent fractionation of magnetic microspheres. J. Magn. Magn. Mater. 2012, 324, 3791–3798. [Google Scholar] [CrossRef]
- Di Carlo, D. Inertial microfluidics. Lab Chip 2009, 9, 3038–3046. [Google Scholar] [CrossRef] [PubMed]
- Bhagat, A.A.S.; Kuntaegowdanahalli, S.S.; Papautsky, I. Inertial microfluidics for continuous particle filtration and extraction. Microfluid. Nanofluid. 2009, 7, 217–226. [Google Scholar] [CrossRef]
- Gao, H.; Zhou, J.; Naderi, M.M.; Peng, Z.; Papautsky, I. Evolution of focused streams for viscoelastic flow in spiral microchannels. Microsyst. Nanoeng. 2023, 9, 73. [Google Scholar] [CrossRef]
- Martel, J.M.; Toner, M. Particle Focusing in Curved Microfluidic Channels. Sci. Rep. 2013, 3, 3340. [Google Scholar] [CrossRef]
- Tuchscherr, L.; Heitmann, V.; Hussain, M.; Viemann, D.; Roth, J.; von Eiff, C.; Peters, G.; Becker, K.; Loffler, B. Staphylococcus aureus Small-Colony Variants Are Adapted Phenotypes for Intracellular Persistence. J. Infect. Dis. 2010, 202, 1031–1040. [Google Scholar] [CrossRef]
- Gong, H.; Beauchamp, M.; Perry, S.; Woolley, A.T.; Nordin, G.P. Optical approach to resin formulation for 3D printed microfluidics. RSC Adv. 2015, 5, 106621–106632. [Google Scholar] [CrossRef]
- Takenaga, S.; Schneider, B.; Erbay, E.; Biselli, M.; Schnitzler, T.; Schöning, M.J.; Wagner, T. Fabrication of biocompatible lab-on-chip devices for biomedical applications by means of a 3D-printing process. Phys. Status Solidi (A) 2015, 212, 1347–1352. [Google Scholar] [CrossRef]
- Chia, H.N.; Wu, B.M. Recent advances in 3D printing of biomaterials. J. Biol. Eng. 2015, 9, 4. [Google Scholar] [CrossRef]
- Cai, G.Z.; Xue, L.; Zhang, H.L.; Lin, J.H. A Review on Micromixers. Micromachines 2017, 8, 27. [Google Scholar] [CrossRef]
- Bayareh, M.; Ashani, M.N.; Usefian, A. Active and passive micromixers: A comprehensive review. Chem. Eng. Process. Process Intensif. 2020, 147, 107771. [Google Scholar] [CrossRef]
- Shallan, A.I.; Smejkal, P.; Corban, M.; Guijt, R.M.; Breadmore, M.C. Cost-Effective Three-Dimensional Printing of Visibly Transparent Microchips within Minutes. Anal. Chem. 2014, 86, 3124–3130. [Google Scholar] [CrossRef] [PubMed]
- Bhagat, A.A.S.; Hou, H.W.; Li, L.D.; Lim, C.T.; Han, J. Pinched flow coupled shear-modulated inertial microfluidics for high-throughput rare blood cell separation. Lab Chip 2011, 11, 1870–1878. [Google Scholar] [CrossRef] [PubMed]
- Ebnesajjad, S. Chapter 12—Adhesion Promoters. In Surface Treatment of Materials for Adhesive Bonding, 2nd ed.; Ebnesajjad, S., Ed.; William Andrew Publishing: Oxford, UK, 2014; pp. 301–329. [Google Scholar]

| Flow rate in µL/min | 50 | 100 | 200 |
| Flow velocity in m/s | 0.017 | 0.034 | 0.068 |
| Reynolds number | 4.2 | 8.5 | 16.9 |
| Deans number (Equation (1)) | 0.39 | 0.77 | 1.54 |
| Particle Reynolds number (Ø 1.95 µm) | 0.0003 | 0.0005 | 0.0010 |
| Particle Reynolds number (Ø 11.95 µm) | 0.0096 | 0.0193 | 0.0386 |
| Focusing of large particles achieved | No | No | Yes |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wiede, A.; Stranik, O.; Tannert, A.; Neugebauer, U. Microfluidic System for Cell Mixing and Particle Focusing Using Dean Flow Fractionation. Micro 2023, 3, 671-685. https://doi.org/10.3390/micro3030047
Wiede A, Stranik O, Tannert A, Neugebauer U. Microfluidic System for Cell Mixing and Particle Focusing Using Dean Flow Fractionation. Micro. 2023; 3(3):671-685. https://doi.org/10.3390/micro3030047
Chicago/Turabian StyleWiede, Alexander, Ondrej Stranik, Astrid Tannert, and Ute Neugebauer. 2023. "Microfluidic System for Cell Mixing and Particle Focusing Using Dean Flow Fractionation" Micro 3, no. 3: 671-685. https://doi.org/10.3390/micro3030047
APA StyleWiede, A., Stranik, O., Tannert, A., & Neugebauer, U. (2023). Microfluidic System for Cell Mixing and Particle Focusing Using Dean Flow Fractionation. Micro, 3(3), 671-685. https://doi.org/10.3390/micro3030047







