Employing Active Learning in Medium Optimization for Selective Bacterial Growth
Abstract
:1. Introduction
2. Results
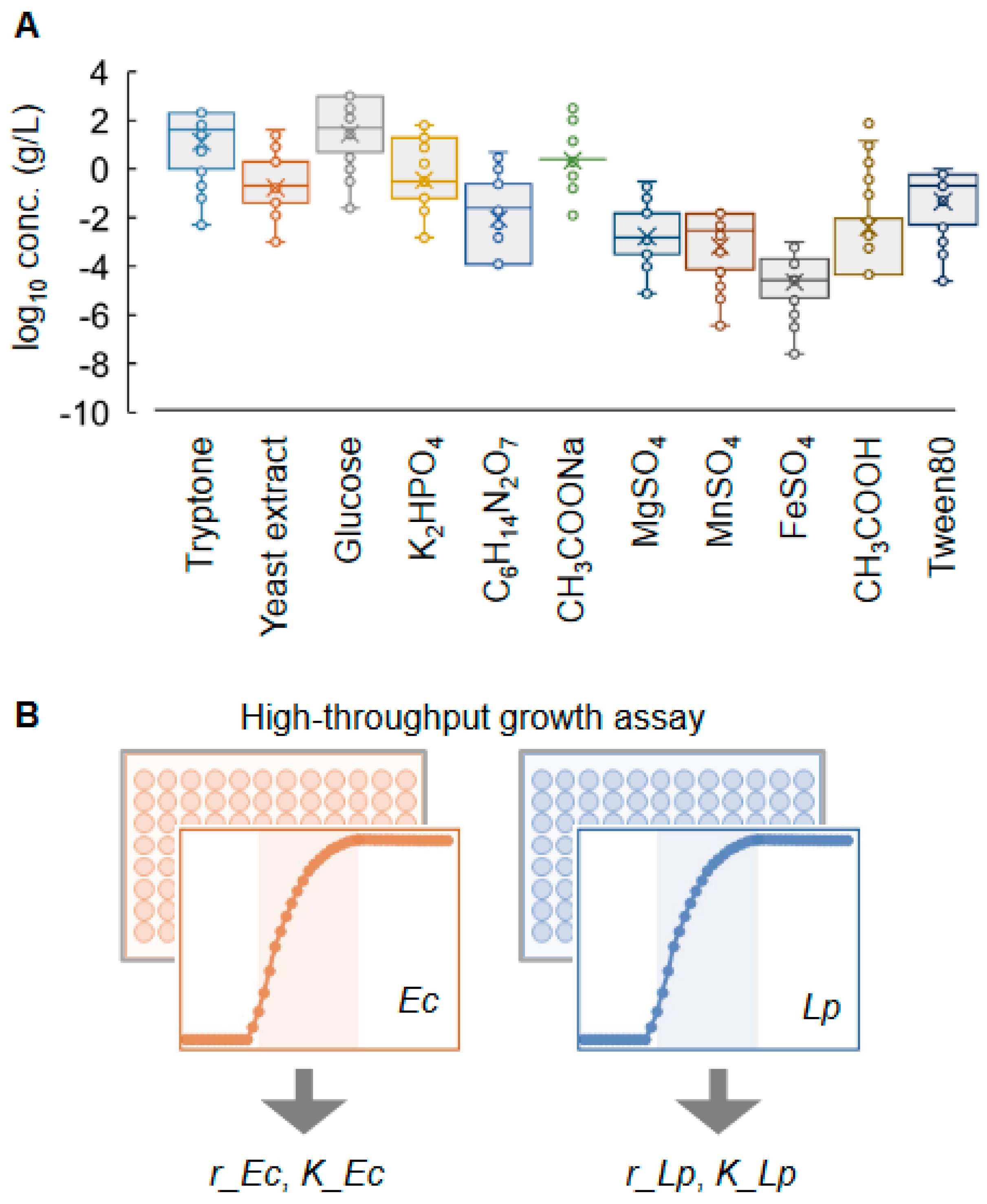
2.1. Experimental and Computational Design of Active Learning for Medium Optimization
2.2. Active Learning Successfully Fine-Tuned the Media for Selective Bacterial Growth
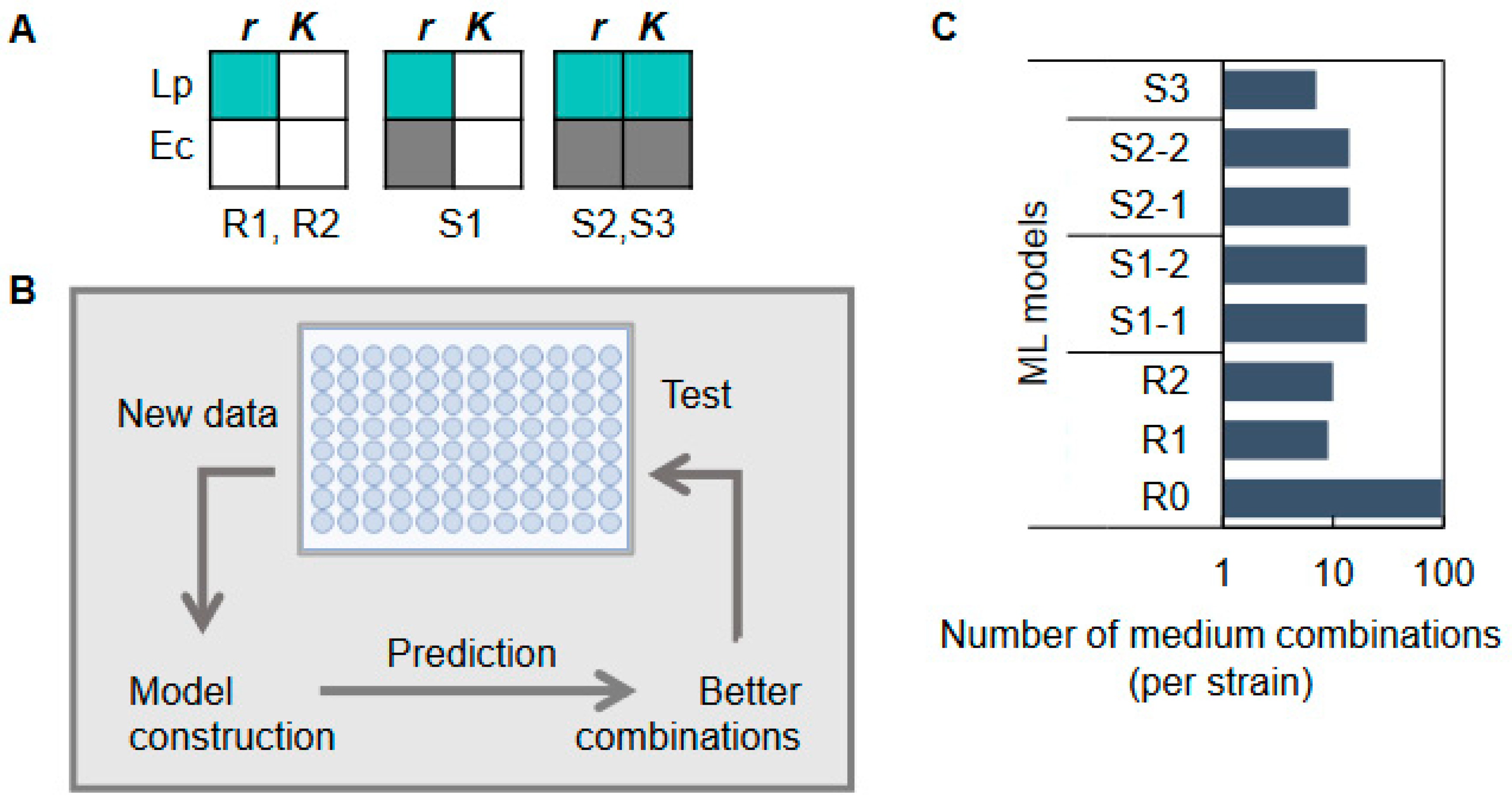
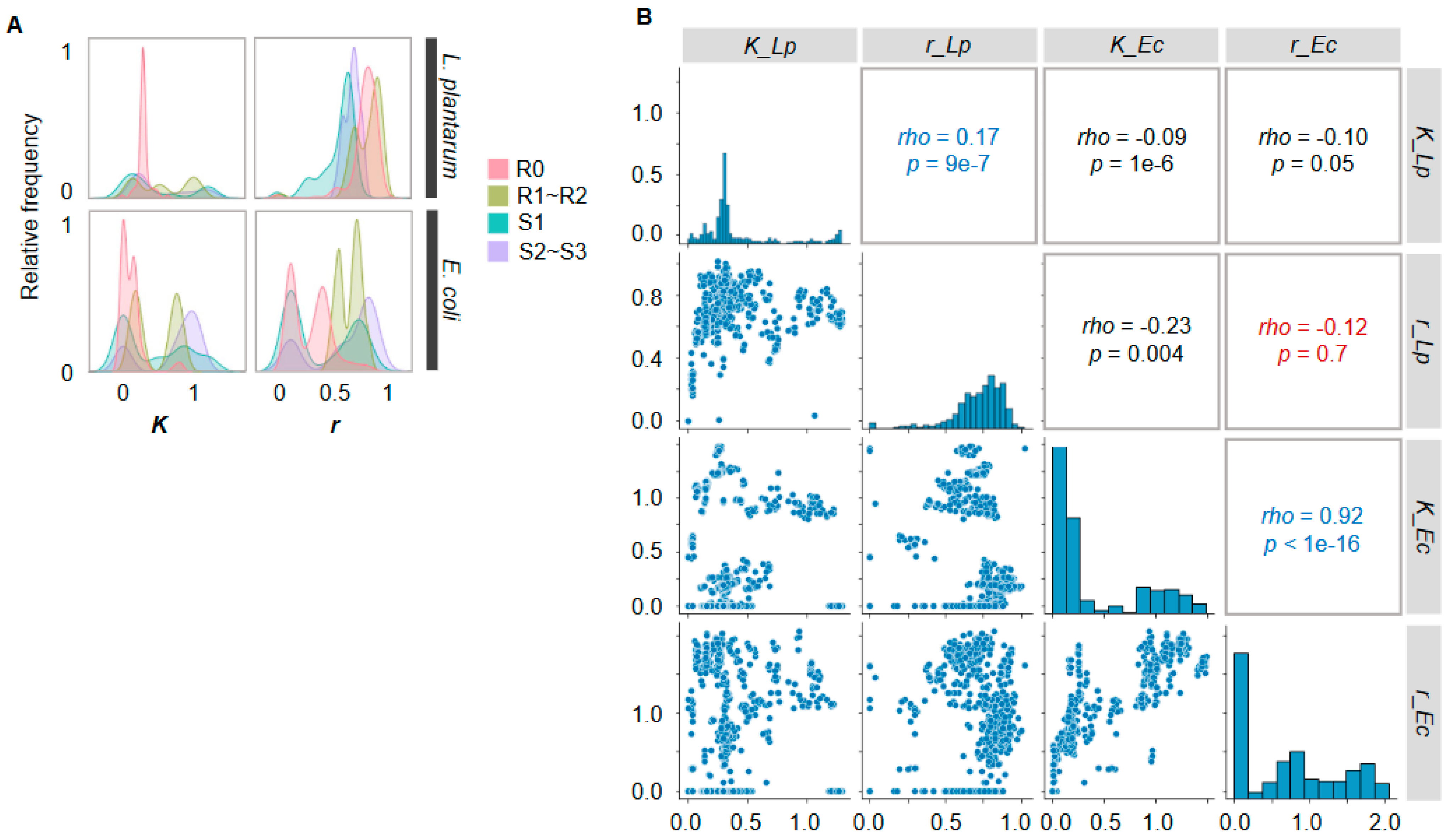
2.3. Changes in Growth Parameters Revealed the Effectiveness of Active Learning
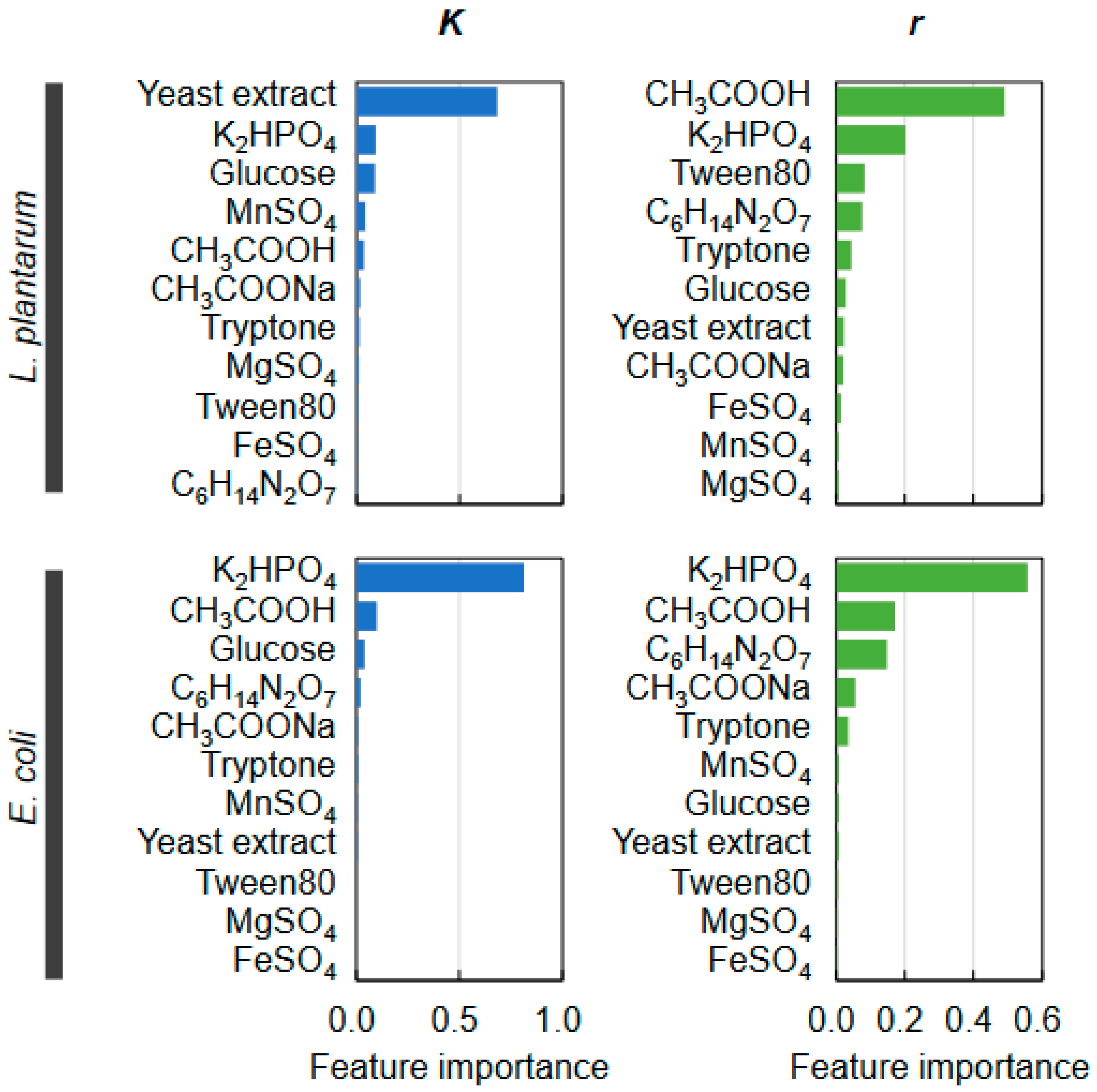
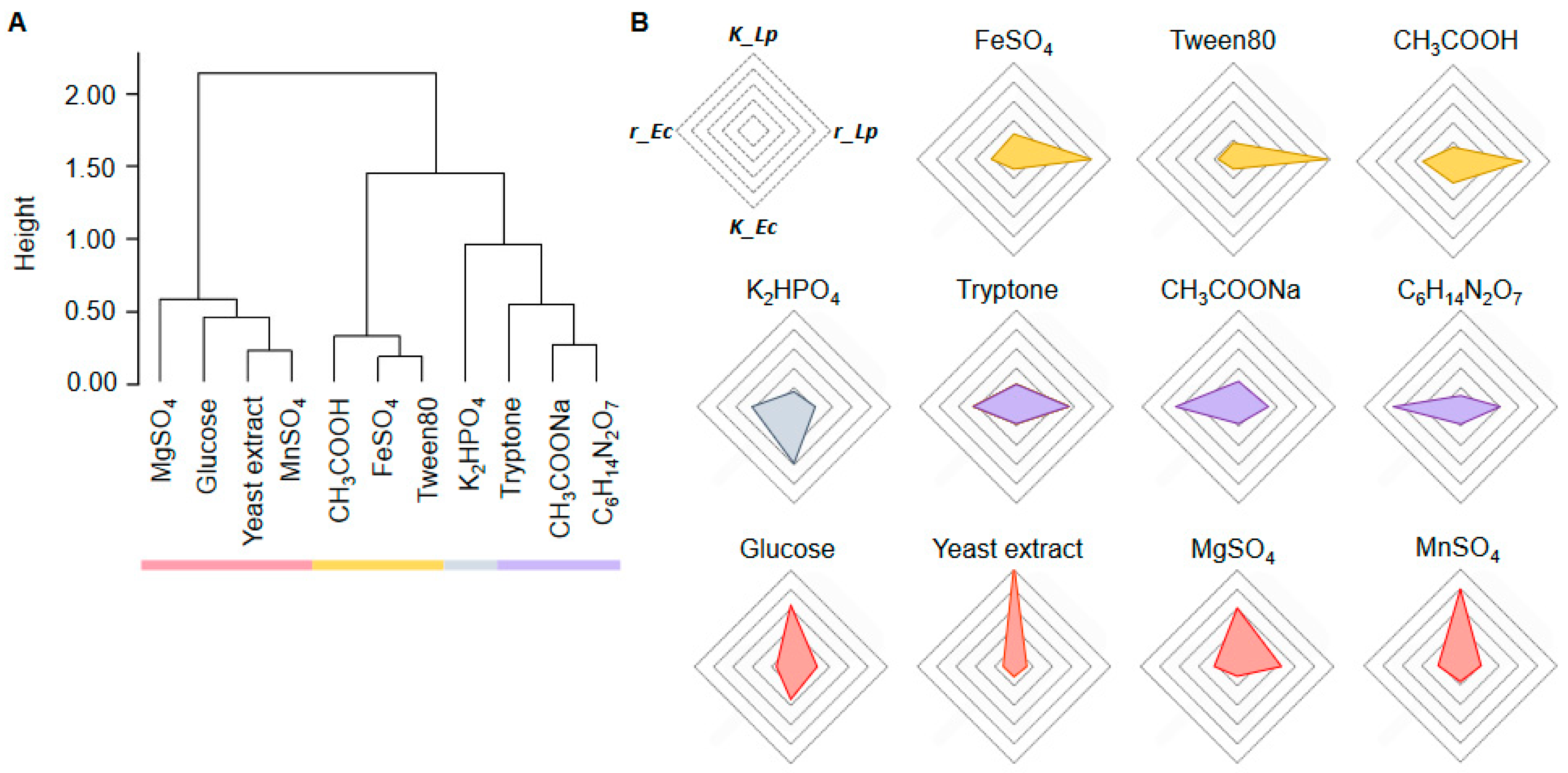
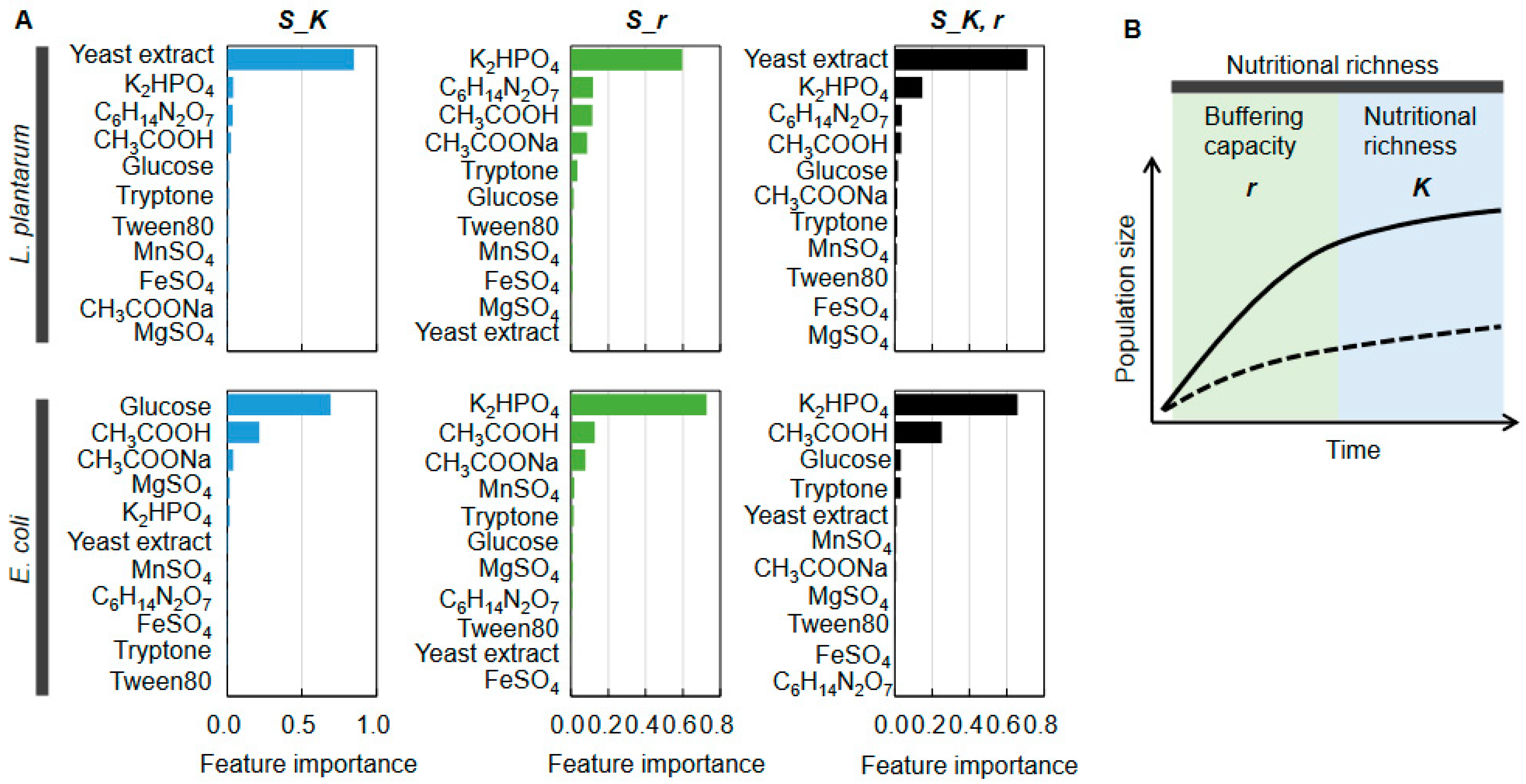
2.4. Decision-Making Medium Components for the Changes in Bacterial Growth
2.5. Medium Components Adjusted via Active Learning for Bacterial Growth Specificity
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains
4.2. Medium Combinations
4.3. Growth Assay and Calculation
4.4. Machine Learning and Computational Analyses
4.5. Model Construction and Active Learning
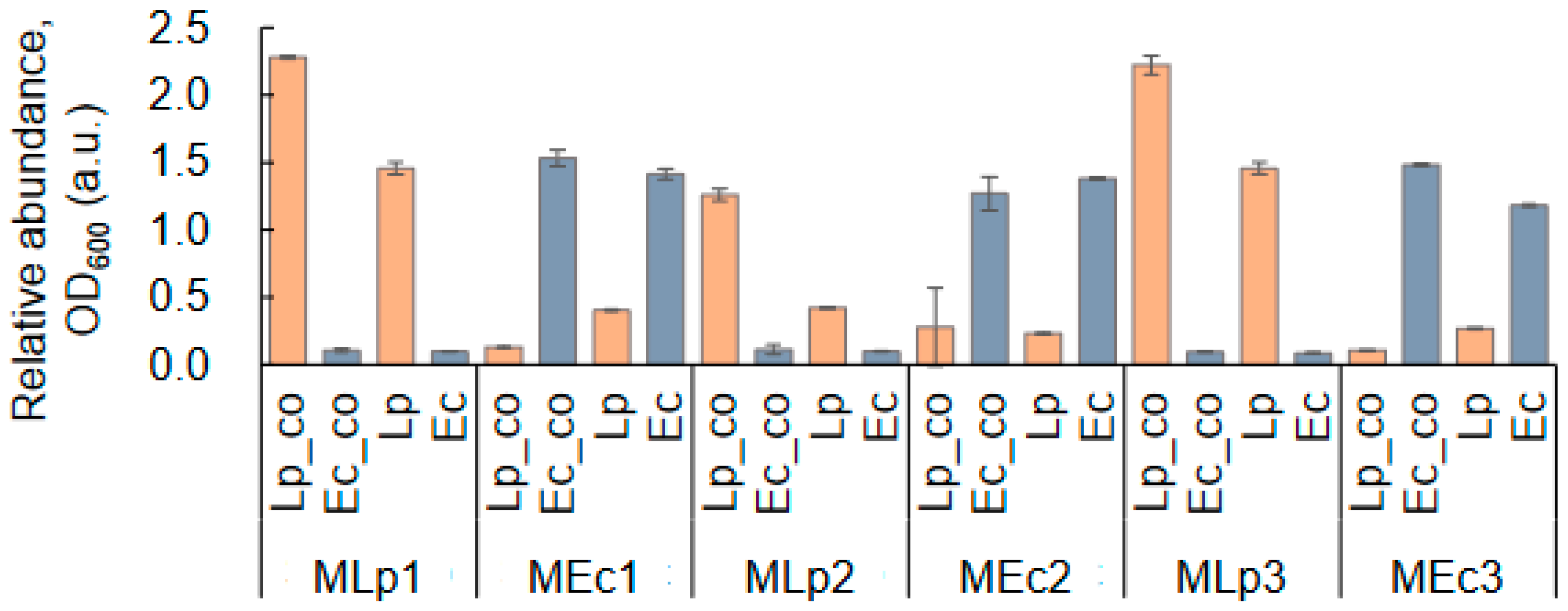
4.6. Co-Culture Verification
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Hugon, P.; Dufour, J.C.; Colson, P.; Fournier, P.E.; Sallah, K.; Raoult, D. A comprehensive repertoire of prokaryotic species identified in human beings. Lancet Infect. Dis. 2015, 15, 1211–1219. [Google Scholar] [CrossRef] [PubMed]
- Lagier, J.C.; Armougom, F.; Million, M.; Hugon, P.; Pagnier, I.; Robert, C.; Bittar, F.; Fournous, G.; Gimenez, G.; Maraninchi, M.; et al. Microbial culturomics: Paradigm shift in the human gut microbiome study. Clin. Microbiol. Infect. 2012, 18, 1185–1193. [Google Scholar] [CrossRef] [PubMed]
- Lagier, J.C.; Dubourg, G.; Million, M.; Cadoret, F.; Bilen, M.; Fenollar, F.; Levasseur, A.; Rolain, J.M.; Fournier, P.E.; Raoult, D. Culturing the human microbiota and culturomics. Nat. Rev. Microbiol. 2018, 16, 540–550. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, M.; Lagier, J.C.; Raoult, D.; Khelaifia, S. Bacterial culture through selective and non-selective conditions: The evolution of culture media in clinical microbiology. New Microbes New Infect. 2020, 34, 100622. [Google Scholar] [CrossRef] [PubMed]
- Rappé, M.S.; Connon, S.A.; Vergin, K.L.; Giovannoni, S.J. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 2002, 418, 630–633. [Google Scholar] [CrossRef] [PubMed]
- Prasad, M.; Shetty, S.K.; Nair, B.G.; Pal, S.; Madhavan, A. A novel and improved selective media for the isolation and enumeration of Klebsiella species. Appl. Microbiol. Biotechnol. 2022, 106, 8273–8284. [Google Scholar] [CrossRef]
- Inoue, Y. Three semi-selective media for Pseudomonas syringae pv. maculicola and P. cannabina pv. alisalensis. Appl. Microbiol. Biotechnol. 2022, 106, 5741–5755. [Google Scholar] [CrossRef]
- Cook, B.S.; Beddow, J.G.; Manso-Silván, L.; Maglennon, G.A.; Rycroft, A.N. Selective medium for culture of Mycoplasma hyopneumoniae. Vet. Microbiol. 2016, 195, 158–164. [Google Scholar] [CrossRef]
- Chon, J.W.; Hyeon, J.Y.; Park, J.H.; Song, K.Y.; Kim, J.H.; Seo, K.H. Improvement of mannitol-yolk-polymyxin B agar by supplementing with trimethoprim for quantitative detection of Bacillus cereus in foods. J. Food Prot. 2012, 75, 1342–1345. [Google Scholar] [CrossRef]
- Kim, D.H.; Kim, H.; Chon, J.W.; Moon, J.S.; Song, K.Y.; Seo, K.H. Development of blood-yolk-polymyxin B-trimethoprim agar for the enumeration of Bacillus cereus in various foods. Int. J. Food Microbiol. 2013, 165, 144–147. [Google Scholar] [CrossRef]
- Azubuike, C.C.; Edwards, M.G.; Gatehouse, A.M.R.; Howard, T.P. Applying Statistical Design of Experiments to Understanding the Effect of Growth Medium Components on Cupriavidus necator H16 Growth. Appl. Environ. Microbiol. 2020, 86, e00705-20. [Google Scholar] [CrossRef]
- Schwarz, H.; Lee, K.; Castan, A.; Chotteau, V. Optimization of medium with perfusion microbioreactors for high density CHO cell cultures at very low renewal rate aided by design of experiments. Biotechnol. Bioeng. 2023, 120, 2523–2541. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Haque, S.; Niwas, R.; Srivastava, A.; Pasupuleti, M.; Tripathi, C.K. Strategies for Fermentation Medium Optimization: An In-Depth Review. Front. Microbiol. 2016, 7, 2087. [Google Scholar] [CrossRef]
- Hemalatha, M.; Subathra Devi, C. A statistical optimization by response surface methodology for the enhanced production of riboflavin from Lactobacillus plantarum-HDS27: A strain isolated from bovine milk. Front. Microbiol. 2022, 13, 982260. [Google Scholar] [CrossRef]
- Nettoor Veettil, V.; A, V.C. Optimization of bacteriocin production by Lactobacillus plantarum using Response Surface Methodology. Cell. Mol. Biol. 2022, 68, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Khan, M.; Khan, S.; Tripathi, C.K. Optimization of actinomycin V production by Streptomyces triostinicus using artificial neural network and genetic algorithm. Appl. Microbiol. Biotechnol. 2009, 82, 379–385. [Google Scholar] [CrossRef]
- Camacho, D.M.; Collins, K.M.; Powers, R.K.; Costello, J.C.; Collins, J.J. Next-Generation Machine Learning for Biological Networks. Cell 2018, 173, 1581–1592. [Google Scholar] [CrossRef]
- Bao, Z.; Bufton, J.; Hickman, R.J.; Aspuru-Guzik, A.; Bannigan, P.; Allen, C. Revolutionizing drug formulation development: The increasing impact of machine learning. Adv. Drug Deliv. Rev. 2023, 202, 115108. [Google Scholar] [CrossRef]
- Vamathevan, J.; Clark, D.; Czodrowski, P.; Dunham, I.; Ferran, E.; Lee, G.; Li, B.; Madabhushi, A.; Shah, P.; Spitzer, M.; et al. Applications of machine learning in drug discovery and development. Nat. Rev. Drug Discov. 2019, 18, 463–477. [Google Scholar] [CrossRef] [PubMed]
- AlQuraishi, M. Machine learning in protein structure prediction. Curr. Opin. Chem. Biol. 2021, 65, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.; Jude, K.M.; Lai, B.; Minot, M.; Kocyla, A.M.; Glassman, C.R.; Nishimiya, D.; Kim, Y.S.; Reddy, S.T.; Khan, A.A.; et al. Deploying synthetic coevolution and machine learning to engineer protein-protein interactions. Science 2023, 381, eadh1720. [Google Scholar] [CrossRef] [PubMed]
- Stefanis, C.; Giorgi, E.; Kalentzis, K.; Tselemponis, A.; Nena, E.; Tsigalou, C.; Kontogiorgis, C.; Kourkoutas, Y.; Chatzak, E.; Dokas, I.; et al. Sentiment analysis of epidemiological surveillance reports on COVID-19 in Greece using machine learning models. Front. Public Health 2023, 11, 1191730. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Cai, G.Y.; Fang, W.; Li, H.Y.; Wang, S.Y.; Chen, L.; Yu, Y.; Liu, D.; Xu, S.; Cui, P.F.; et al. Machine learning based early warning system enables accurate mortality risk prediction for COVID-19. Nat. Commun. 2020, 11, 5033. [Google Scholar] [CrossRef] [PubMed]
- Cosenza, Z.; Block, D.E.; Baar, K. Optimization of muscle cell culture media using nonlinear design of experiments. Biotechnol. J. 2021, 16, e2100228. [Google Scholar] [CrossRef]
- Hashizume, T.; Ying, B.W. Challenges in developing cell culture media using machine learning. Biotechnol. Adv. 2023, 70, 108293. [Google Scholar] [CrossRef] [PubMed]
- Hashizume, T.; Ozawa, Y.; Ying, B.W. Employing active learning in the optimization of culture medium for mammalian cells. NPJ Syst. Biol. Appl. 2023, 9, 20. [Google Scholar] [CrossRef]
- Jang, J.; Hur, H.G.; Sadowsky, M.J.; Byappanahalli, M.N.; Yan, T.; Ishii, S. Environmental Escherichia coli: Ecology and public health implications-a review. J. Appl. Microbiol. 2017, 123, 570–581. [Google Scholar] [CrossRef]
- Huleani, S.; Roberts, M.R.; Beales, L.; Papaioannou, E.H. Escherichia coli as an antibody expression host for the production of diagnostic proteins: Significance and expression. Crit. Rev. Biotechnol. 2022, 42, 756–773. [Google Scholar] [CrossRef]
- Colautti, A.; Orecchia, E.; Comi, G.; Iacumin, L. Lactobacilli, a Weapon to Counteract Pathogens through the Inhibition of Their Virulence Factors. J. Bacteriol. 2022, 204, e0027222. [Google Scholar] [CrossRef]
- van de Wijgert, J.; Verwijs, M.C. Lactobacilli-containing vaginal probiotics to cure or prevent bacterial or fungal vaginal dysbiosis: A systematic review and recommendations for future trial designs. Bjog 2020, 127, 287–299. [Google Scholar] [CrossRef]
- Aida, H.; Uchida, K.; Nagai, M.; Hashizume, T.; Masuo, S.; Takaya, N.; Ying, B.W. Machine learning-assisted medium optimization revealed the discriminated strategies for improved production of the foreign and native metabolites. Comput. Struct. Biotechnol. J. 2023, 21, 2654–2663. [Google Scholar] [CrossRef] [PubMed]
- Aida, H.; Hashizume, T.; Ashino, K.; Ying, B.W. Machine learning-assisted discovery of growth decision elements by relating bacterial population dynamics to environmental diversity. eLife 2022, 11, e76846. [Google Scholar] [CrossRef] [PubMed]
- Nestor, E.; Toledano, G.; Friedman, J. Interactions between Culturable Bacteria Are Predicted by Individual Species’ Growth. mSystems 2023, 8, e0083622. [Google Scholar] [CrossRef] [PubMed]
- Aditya, A.; Rahaman, S.O.; Biswas, D. Impact of Lactobacillus-originated metabolites on enterohemorrhagic E. coli in rumen fluid. FEMS Microbiol. Ecol. 2022, 98, fiac128. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Lu, Y.; Wu, X.; Yu, P.; Lan, P.; Wu, X.; Jiang, Y.; Li, Q.; Pi, X.; Liu, W.; et al. Anticolonization of Carbapenem-Resistant Klebsiella pneumoniae by Lactobacillus plantarum LP1812 through Accumulated Acetic Acid in Mice Intestinal. Front. Cell. Infect. Microbiol. 2021, 11, 804253. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.; Ealy, J.; Huang, Y.; Benites, N.C.; Polk, M.; Basan, M. Coexisting ecotypes in long-term evolution emerged from interacting trade-offs. Nat. Commun. 2023, 14, 3805. [Google Scholar] [CrossRef] [PubMed]
- Tao, Z.; Yuan, H.; Liu, M.; Liu, Q.; Zhang, S.; Liu, H.; Jiang, Y.; Huang, D.; Wang, T. Yeast Extract: Characteristics, Production, Applications and Future Perspectives. J. Microbiol. Biotechnol. 2023, 33, 151–166. [Google Scholar] [CrossRef]
- Jiang, M.; Chen, K.; Liu, Z.; Wei, P.; Ying, H.; Chang, H. Succinic acid production by Actinobacillus succinogenes using spent brewer’s yeast hydrolysate as a nitrogen source. Appl. Biochem. Biotechnol. 2010, 160, 244–254. [Google Scholar] [CrossRef]
- Fraise, A.P.; Wilkinson, M.A.; Bradley, C.R.; Oppenheim, B.; Moiemen, N. The antibacterial activity and stability of acetic acid. J. Hosp. Infect. 2013, 84, 329–331. [Google Scholar] [CrossRef]
- Ge, J.; Kang, J.; Ping, W. Effect of Acetic Acid on Bacteriocin Production by Gram-Positive Bacteria. J. Microbiol. Biotechnol. 2019, 29, 1341–1348. [Google Scholar] [CrossRef]
- Feng, L.; Xu, M.; Zeng, W.; Zhang, X.; Wang, S.; Yao, Z.; Zhou, T.; Shi, S.; Cao, J.; Chen, L. Evaluation of the antibacterial, antibiofilm, and anti-virulence effects of acetic acid and the related mechanisms on colistin-resistant Pseudomonas aeruginosa. BMC Microbiol. 2022, 22, 306. [Google Scholar] [CrossRef]
- Blomberg, A. Measuring growth rate in high-throughput growth phenotyping. Curr. Opin. Biotechnol. 2011, 22, 94–102. [Google Scholar] [CrossRef]
- Peleg, M.; Corradini, M.G. Microbial growth curves: What the models tell us and what they cannot. Crit. Rev. Food Sci. Nutr. 2011, 51, 917–945. [Google Scholar] [CrossRef]
- Jingjing, E.; Lili, M.; Zichao, C.; Rongze, M.; Qiaoling, Z.; Ruiyin, S.; Zongbai, H.; Junguo, W. Effects of buffer salts on the freeze-drying survival rate of Lactobacillus plantarum LIP-1 based on transcriptome and proteome analyses. Food Chem. 2020, 326, 126849. [Google Scholar] [CrossRef] [PubMed]
- Jingjing, E.; Rongze, M.; Zichao, C.; Caiqing, Y.; Ruixue, W.; Qiaoling, Z.; Zongbai, H.; Ruiyin, S.; Junguo, W. Improving the freeze-drying survival rate of Lactobacillus plantarum LIP-1 by increasing biofilm formation based on adjusting the composition of buffer salts in medium. Food Chem. 2021, 338, 128134. [Google Scholar] [CrossRef] [PubMed]
- De Bruyn, I.N.; Holzapfel, W.H.; Visser, L.; Louw, A.I. Glucose metabolism by Lactobacillus divergens. Microbiology 1988, 134, 2103–2109. [Google Scholar] [CrossRef]
- Mogodiniyai Kasmaei, K.; Schlosser, D.; Sträuber, H.; Kleinsteuber, S. Does glucose affect the de-esterification of methyl ferulate by Lactobacillus buchneri? Microbiologyopen 2020, 9, e971. [Google Scholar] [CrossRef] [PubMed]
- Aida, H.; Ying, B.W. Efforts to Minimise the Bacterial Genome as a Free-Living Growing System. Biology 2023, 12, 1170. [Google Scholar] [CrossRef] [PubMed]
- Radivojevic, T.; Costello, Z.; Workman, K.; Garcia Martin, H. A machine learning Automated Recommendation Tool for synthetic biology. Nat. Commun. 2020, 11, 4879. [Google Scholar] [CrossRef]
- Settles, B. Active Learning Literature Survey; Computer Sciences Technical Report 1648; University of Wisconsin-Madison: Madison, WI, USA, 2009. [Google Scholar]
- Vargas-Garcia, C.A.; Ghusinga, K.R.; Singh, A. Cell size control and gene expression homeostasis in single-cells. Curr. Opin. Syst. Biol. 2018, 8, 109–116. [Google Scholar] [CrossRef]
- Orland, C.; Emilson, E.J.S.; Basiliko, N.; Mykytczuk, N.C.S.; Gunn, J.M.; Tanentzap, A.J. Microbiome functioning depends on individual and interactive effects of the environment and community structure. ISME J. 2018, 13, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, I.; Kurokawa, M.; Liu, L.; Ying, B.W. Coordinated Changes in Mutation and Growth Rates Induced by Genome Reduction. mBio 2017, 8, e00676-17. [Google Scholar] [CrossRef] [PubMed]
- Rivett, D.W.; Scheuerl, T.; Culbert, C.T.; Mombrikotb, S.B.; Johnstone, E.; Barraclough, T.G.; Bell, T. Resource-dependent attenuation of species interactions during bacterial succession. ISME J. 2016, 10, 2259–2268. [Google Scholar] [CrossRef] [PubMed]
- Engen, S.; Saether, B.E. r- and K-selection in fluctuating populations is determined by the evolutionary trade-off between two fitness measures: Growth rate and lifetime reproductive success. Evolution 2017, 71, 167–173. [Google Scholar] [CrossRef]
- Luckinbill, L.S. r and K Selection in Experimental Populations of Escherichia coli. Science 1978, 202, 1201–1203. [Google Scholar] [CrossRef]
- Baichman-Kass, A.; Song, T.; Friedman, J. Competitive interactions between culturable bacteria are highly non-additive. eLife 2023, 12, e83398. [Google Scholar] [CrossRef]
- Gould, A.L.; Zhang, V.; Lamberti, L.; Jones, E.W.; Obadia, B.; Korasidis, N.; Gavryushkin, A.; Carlson, J.M.; Beerenwinkel, N.; Ludington, W.B. Microbiome interactions shape host fitness. Proc. Natl. Acad. Sci. USA 2018, 115, E11951–E11960. [Google Scholar] [CrossRef]
- Piccardi, P.; Vessman, B.; Mitri, S. Toxicity drives facilitation between 4 bacterial species. Proc. Natl. Acad. Sci. USA 2019, 116, 15979–15984. [Google Scholar] [CrossRef]
- Palmer, J.D.; Foster, K.R. Bacterial species rarely work together. Science 2022, 376, 581–582. [Google Scholar] [CrossRef]
- Ma, Y.; Ramoneda, J.; Johnson, D.R. Timing of antibiotic administration determines the spread of plasmid-encoded antibiotic resistance during microbial range expansion. Nat. Commun. 2023, 14, 3530. [Google Scholar] [CrossRef]
- Nordholt, N.; Kanaris, O.; Schmidt, S.B.I.; Schreiber, F. Persistence against benzalkonium chloride promotes rapid evolution of tolerance during periodic disinfection. Nat. Commun. 2021, 12, 6792. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.; Warsi, O.M.; Andersson, D.I. Pervasive Selection for Clinically Relevant Resistance and Media Adaptive Mutations at Very Low Antibiotic Concentrations. Mol. Biol. Evol. 2023, 40, msad010. [Google Scholar] [CrossRef]
- Zheng, E.J.; Andrews, I.W.; Grote, A.T.; Manson, A.L.; Alcantar, M.A.; Earl, A.M.; Collins, J.J. Modulating the evolutionary trajectory of tolerance using antibiotics with different metabolic dependencies. Nat. Commun. 2022, 13, 2525. [Google Scholar] [CrossRef] [PubMed]
- Ashino, K.; Sugano, K.; Amagasa, T.; Ying, B.W. Predicting the decision making chemicals used for bacterial growth. Sci. Rep. 2019, 9, 7251. [Google Scholar] [CrossRef] [PubMed]
- Kurokawa, M.; Ying, B.W. Precise, High-throughput Analysis of Bacterial Growth. J. Vis. Exp. 2017, 127, 56197. [Google Scholar] [CrossRef]

| (g/L) | MLp1 | MEc1 | MLp2 | MEc2 | MLp3 | MEc3 | MRS |
|---|---|---|---|---|---|---|---|
| Tryptone | 10 | 10 | 10 | 10 | 10 | 10 | 10 |
| Yeast extract | 5 | 1.35 | 1.35 | 0.05 | 5 | 1.35 | 5 |
| Glucose | 16.2 | 0.6 | 0.6 | 0.2 | 16.2 | 0.2 | 20 |
| K2HPO4 | 0.18 | 4.86 | 6 | 6 | 0.18 | 6 | 6 |
| C6H14N2O7 | 1.62 | 0.54 | 0.54 | 0.18 | 1.62 | 0.18 | 2 |
| CH3COONa | 4.05 | 0.15 | 0.15 | 0.15 | 4.05 | 0.15 | 15 |
| MgSO4 | 0.6 | 0.6 | 0.486 | 0.006 | 0.6 | 0.018 | 0.6 |
| MnSO4 | 0.16 | 0.1296 | 0.1296 | 0.0016 | 0.16 | 0.0144 | 0.16 |
| FeSO4 | 0.0005 | 0.0045 | 0.0045 | 0.0135 | 0.0005 | 0.0135 | 0.05 |
| CH3COOH | 0.013 | 0.013 | 3.159 | 0.013 | 0.013 | 0.039 | 1.3 |
| Tween80 | 1 | 0.27 | 0.27 | 0.81 | 1 | 0.81 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, S.; Aida, H.; Ying, B.-W. Employing Active Learning in Medium Optimization for Selective Bacterial Growth. Appl. Microbiol. 2023, 3, 1355-1369. https://doi.org/10.3390/applmicrobiol3040091
Zhang S, Aida H, Ying B-W. Employing Active Learning in Medium Optimization for Selective Bacterial Growth. Applied Microbiology. 2023; 3(4):1355-1369. https://doi.org/10.3390/applmicrobiol3040091
Chicago/Turabian StyleZhang, Shuyang, Honoka Aida, and Bei-Wen Ying. 2023. "Employing Active Learning in Medium Optimization for Selective Bacterial Growth" Applied Microbiology 3, no. 4: 1355-1369. https://doi.org/10.3390/applmicrobiol3040091
APA StyleZhang, S., Aida, H., & Ying, B.-W. (2023). Employing Active Learning in Medium Optimization for Selective Bacterial Growth. Applied Microbiology, 3(4), 1355-1369. https://doi.org/10.3390/applmicrobiol3040091







